Bacterial flagella are dynamic, not only because they rotate and reverse, but also because some of their components exchange on a short time scale. In PNAS, Delalez et al. (1) show that signal-dependent turnover of FliM subunits in the motor–switch complex may be part of the chemotaxis response in Escherichia coli.
E. coli swims in a 3D random walk in which “runs” of several seconds are punctuated by brief, reorienting “tumbles.” Chemotaxis occurs when cells extend their runs up attractant gradients or down repellent gradients.
The flagellar motor was the first biological rotary device discovered (2). Flagella spin at several hundred to >1,000 revolutions per second in different bacteria. In E. coli, counterclockwise (CCW) flagellar rotation causes runs, and clockwise (CW) flagellar rotation causes tumbles. The default direction is CCW. CW rotation ensues when phosphorylated CheY (CheY-P) binds to the motor. CheY is phosphorylated at the chemoreceptor patch, and its dephosphorylation is accelerated by the CheZ phosphatase. When attractants bind to receptors, they inhibit the activity of the CheY kinase (CheA), thereby lowering CheY-P levels.
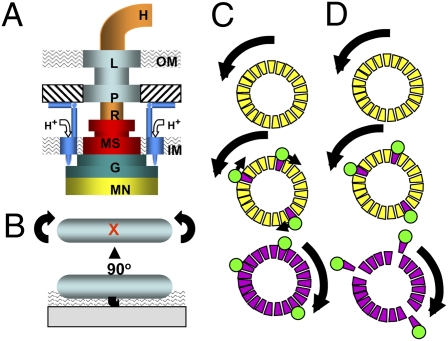
The motor contains two structural elements: stator and rotor (Fig. 1A). The L and P rings serve as a bushing for the rotating rod in the cell envelope. The stator includes up to 11 (3) MotA4MotB2 complexes (4) that anchor to the cell wall (5); they undergo proton-driven conformational changes to drive flagellar rotation (6).
Fig. 1.
(A) Schematic view of the E. coli flagellar motor. Rotor elements are as follows: MN (FliMN), G (FliG), MS (MS ring), R (rod), and H (hook). Stationary elements are P (P ring), L (L ring), and M (MotA4MotB2) complex. Mot complexes attach to the cell wall (hatched), conduct H+ through the membrane, and interact with FliG. IM and OM denote the inner and outer membranes. (B) Cartoon of a tethered cell. A sheared flagellar filament is affixed by using antifilament antibody. The cell body counter rotates to the direction of motor rotation. The red X marks the point at which the tethering flagellum is located. The wavy pattern indicates the evanescent wave generated by TIRF. (C) The domino model for cooperative switching within the motor. The motor at the top is spinning CCW. CheY-P binding to FliM (trapezoids) initiates a wave of conformational change, shown as CW-facing arrows, to place the C ring in the CW conformation. (D) FliM destabilization model for cooperative switching. CheY-P (green circle) binding to FliM destabilizes the FliM ring, which allows the motor to “relax” into the CW conformation. Both models are speculative, and the scenario shown in D exists only in the imagination of the author.
The rotor includes a cytoplasmic structure known as the C ring, which contains ≈26 FliG proteins, ≈34 FliM proteins (7), and ≥100 FliN proteins (8). It is connected to the rod via the MS ring. The distal end of the rod is attached to the helical flagellar filament via a flexible hook. Filament growth decreases with length, and a broken filament can regenerate. Unfolded flagellin subunits diffuse through the hollow center of the filament and assemble at its distal tip (9). Filaments extend several cell lengths and are quite fragile; their dynamic nature is necessary.
Each flagellar motor functions for the lifetime of its cell. Mot complexes can assemble around preexisting rotors (10), and introduction of each Mot complex increases the rotational velocity of a tethered cell (Fig. 1B). The high torque required to turn a flagellum under heavy load (11) requires that Mot complexes attach firmly to the cell wall.
Despite its anchoring, the stator is surprisingly dynamic (12). Fluorescence recovery after photobleaching (FRAP) showed that individual Mot complexes have a half-life of ≈30 s in an E. coli motor. Detached Mot complexes diffuse freely in the cell membrane, and their H+-conducting channels are plugged by a short, amphipathic helix (13). Pom complexes, the Mot equivalents in Na+-driven motors, assemble in a Na+-dependent fashion (14). In Shewanella oneidensis, a single rotor can be driven by proton-conducting Mot complexes or Na+-conducting Pom complexes (15). This ability to exchange stator elements probably arose by transfer of mot genes when a marine bacterium with a Na+-driven motor became isolated in a freshwater habitat.
Recent FRAP studies (16) have shown that FliM and FliN in the rotor turn over much more rapidly than FliG, in keeping with their peripheral location on the C ring. Delalez et al. (1) used total internal reflection fluorescence (TIRF), coupled with FRAP and fluorescence loss in photobleaching (FLIP), to quantify FliM turnover in tethered and immobilized E. coli cells.
Cells expressing YFP-labeled FliM (FliM-YPet) are chemotactic. Fluorescence–decay curves allow a calculation of 30 ± 6 FliM-YPet molecules at the base of each tethered flagellum, a value that agrees with prior estimates of the number of copies of FliM per C ring (7).
The distribution of FliM within immobilized whole cells was also assessed. Extrapolation from the fluorescent spots in TIRF images indicates that there are ≈24 FliM-YPet spots per cell. Of these spots, 40% have ≈32 FliM-YPet molecules, the same number as in tethered motors. The remaining 60% form a unimodal population with ≈18 FliM-YPet molecules.
E. coli cells typically have four to eight flagella, some of which are presumably still being assembled. The function of the spots with ≈18 copies of FliM-YPet is unknown. The sum of FliM-YPet molecules in all spots is ≈600. An equal number of FliM-YPet proteins diffuse freely within the cells, providing a pool of replacement parts.
Reciprocal FRAP and FLIP measurements show that FliM turnover is complete within 10 min and occurs only in intact C rings. Approximately two thirds of the FliM-YPet molecules in C rings turn over, with a half-life of ≈40 s. This ratio may reflect two different populations of FliM that have been described (17): ≈26 FliM exchangeable molecules in a peripheral location and a core of ≈8 FliM molecules. It seems likely that FliN associated with FliM (18) turns over as well.
FRAP and FLIP data show no FliM–YPet exchange in cells that do not contain CheY. Exchange occurs in cells that produce constitutively active CheY (the D13K, Y106W mutant) but decreases in cells containing unphosphorylated CheY (the D57A mutant). These results suggest that CheY binding induces turnover of FliM.
One of the most notable properties of the E. coli flagellar motor is its high degree of cooperativity (19). One model to explain this cooperativity envisions FliM subunits acting like a dynamic circle of “dominos” (20). If a few tip in the CW direction after binding CheY-P, the whole ring goes CW (Fig. 1C).
The work of Delalez et al. (1) suggests an alternative model. An intact C ring, with all FliM subunits in place, may rotate CCW. The nonexchanging population of FliM might stabilize the C ring; fliM knockout mutants fail to assemble flagella (21). CheY-P binding could destabilize the peripheral array of FliM subunits, which are in ≈1:1 stoichiometry with FliG. FliG would then assume the conformation needed to generate CW rotation (Fig. 1D).
Parts exchange in the stator and rotor may just be routine maintenance, and the aggregates of 18 FliM molecules could be storage devices rather than assembly intermediates. The authors are suitably cautious about speculating whether FliM turnover is involved in the switch function of the C ring, emphasizing that the exchange of FliM subunits could be either a cause or effect of motor reversal.
FliM turnover could be affected by fliG, fliM, and fliN switch mutations (21), by the absence of Mot complexes, and by addition of attractants and repellents. Fluorescence resonance energy transfer (FRET) between FliM–YPet and motor proteins with suitable fluorescent tags may be able to track movements of rotor components. Colocalization studies with fluorescently tagged proteins could identify FliM's neighbors in the 18-subunit spots. Further studies of this type will undoubtedly lead to exciting new revelations about the inner workings of the elegant molecular machinery of the flagellar motor.
Footnotes
The author declares no conflict of interest.
See companion article on page 11347.
References
- 1.Delalez NJ, et al. Signal-dependent turnover of the bacterial flagellar switch protein FliM. Proc Natl Acd Sci USA. 2010;107:11347–11351. doi: 10.1073/pnas.1000284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg HC, Anderson RA. Bacteria swim by rotating their flagellar filaments. Nature. 1973;245:380–382. doi: 10.1038/245380a0. [DOI] [PubMed] [Google Scholar]
- 3.Reid SW, et al. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci USA. 2006;103:8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kojima S, Blair DF. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry. 2004;43:26–34. doi: 10.1021/bi035405l. [DOI] [PubMed] [Google Scholar]
- 5.Kojima S, et al. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol Microbiol. 2009;73:710–718. doi: 10.1111/j.1365-2958.2009.06802.x. [DOI] [PubMed] [Google Scholar]
- 6.Kojima S, Blair DF. Conformational change in the stator of the bacterial flagellar motor. Biochemistry. 2001;40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- 7.Zhao R, Amsler CD, Matsumura P, Khan S. FliG and FliM distribution in the Salmonella typhimurium cell and flagellar basal bodies. J Bacteriol. 1996;178:258–265. doi: 10.1128/jb.178.1.258-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao R, Pathak N, Jaffe H, Reese TS, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- 9.Yonekura K, et al. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science. 2000;290:2148–2152. doi: 10.1126/science.290.5499.2148. [DOI] [PubMed] [Google Scholar]
- 10.Blair DF, Berg HC. Restoration of torque in defective flagellar motors. Science. 1988;242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Berg HC. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophys J. 2000;78:1036–1041. doi: 10.1016/S0006-3495(00)76662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leake MC, et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 13.Hosking ER, Vogt C, Bakker EP, Manson MD. The Escherichia coli MotAB proton channel unplugged. J Mol Biol. 2006;364:921–937. doi: 10.1016/j.jmb.2006.09.035. [DOI] [PubMed] [Google Scholar]
- 14.Fukuoka H, Wada T, Kojima S, Ishijima A, Homma M. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol Microbiol. 2009;71:825–835. doi: 10.1111/j.1365-2958.2008.06569.x. [DOI] [PubMed] [Google Scholar]
- 15.Paulick A, et al. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol Microbiol. 2009;71:836–850. doi: 10.1111/j.1365-2958.2008.06570.x. [DOI] [PubMed] [Google Scholar]
- 16.Fukuoka H, Inoue Y, Terasawa S, Takahashi H, Ishijima A. Exchange of rotor components in functioning bacterial flagellar motor. Biochem Biophys Res Commun. 2010;394:130–135. doi: 10.1016/j.bbrc.2010.02.129. [DOI] [PubMed] [Google Scholar]
- 17.Brown PN, Terrazas M, Paul K, Blair DF. Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J Bacteriol. 2007;189:305–312. doi: 10.1128/JB.01281-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar MK, Paul K, Blair DF. Subunit organization and reversal-associated movements in the flagellar switch of Escherichia coli. J Biol Chem. 2010;285:675–684. doi: 10.1074/jbc.M109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 20.Duke TA, Le Novère N, Bray D. Conformational spread in a ring of proteins: a stochastic approach to allostery. J Mol Biol. 2001;308:541–553. doi: 10.1006/jmbi.2001.4610. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi S, Fujita H, Ishihara A, Aizawa S, Macnab RM. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986;166:187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]