Abstract
The human ATM and ATR proteins participate in the DNA damage and DNA replication checkpoint pathways and are critical to maintaining genome stability. The Saccharomyces cerevisiae homologs of ATM and ATR are Tel1p and Mec1p, respectively. Haploid tel1 mec1 strains have very short telomeres and very high rates of chromosomal aberrations. Here, we examine genetic stability in tel1 mec1 diploid cells. In the absence of induced DNA damage, these yeast strains had very high frequencies of aneuploidy (both trisomy and monosomy) in addition to elevated rates of chromosome rearrangements. Although we found the aneuploidy in the tel1 mec1 diploids mimicked that observed in bub1 diploids, the tel1 mec1 diploids had a functional spindle assembly checkpoint. Restoration of wild-type telomere lengths in the tel1 mec1 strain substantially reduced the rate of chromosome rearrangements but had no effect on the frequency of aneuploidy.
Keywords: chromosome nondisjunction, spindle assembly checkpoint, telomeres
Two types of genetic instability are associated with cells derived from solid tumors (1): elevated rates of single-base substitutions/microsatellite alterations and high frequencies of chromosome rearrangements and aneuploidy. The first type of instability is associated with tumors deficient in nucleotide excision repair and DNA mismatch repair. Tumors with elevated rates of chromosome aberrations (deletions, duplications, and translocations) and/or aneuploidy have been termed “chromosome instability” (CIN) tumors (2). Although some CIN tumors have mutations in genes affecting the spindle assembly checkpoint (SAC) (3) or chromatid cohesion (4), the relevant genome-destabilizing mutations have not been identified in most CIN tumors.
Many yeast mutants have been identified that have elevated rates of chromosome alterations and/or aneuploidy. One assay designed to detect gross chromosomal rearrangements (GCRs) involves selecting for the deletion of two closely linked markers near the end of chromosome V (5). Mutations that elevate GCRs are found in genes controlling S-phase DNA damage checkpoints (6), DNA replication (7), telomere length regulation (8, 9), and chromatin assembly (10) as well as other pathways. Importantly, strains with mutations in the SAC (which result in elevated levels of chromosome nondisjunction) do not necessarily have elevated rates of GCRs (11), indicating that these two phenotypes are not intrinsically linked.
In Saccharomyces cerevisiae, the related kinases Tel1p (orthologous to the mammalian ATM gene) and Mec1p (orthologous to the mammalian ATR gene) have overlapping roles in the DNA damage/S-phase checkpoint and in telomere length regulation (12). In response to DNA damage such as a double-strand break (DSB), Tel1p and Mec1p are recruited to the site of the break, where they initiate a signal transduction cascade that results in phosphorylation and activation of effector proteins, transcription of DNA repair genes, and cell cycle arrest. The tel1 mec1 strains are more sensitive to DNA damaging agents than either single-mutant strain (13). Single-mutant tel1 and mec1 strains have short but stable telomeres, whereas double-mutant strains have a phenotype similar to a telomerase-negative strain (14). Mec1p (but not Tel1p) is an essential protein, but the lethality of the mec1 mutation can be suppressed by a mutation in SML1 (15). In addition, mec1 strains have elevated levels of DSBs in hard-to-replicate regions of the genome (16). These and other results suggest that the essential role of Mec1p may be to stabilize stalled replication forks (17).
Haploid yeast strains lacking both Tel1p and Mec1p have very high rates of chromosome rearrangements (6, 9). In haploid strains, the rate of deletions of CAN1 is elevated about 10,000-fold in the tel1 mec1 strain relative to WT. In some can1 strains, the deletion derivative of chromosome V (the location of CAN1) was fused to the telomere of a nonhomologous chromosome by a nonhomologous end-joining (NHEJ) event (9). In addition, the tel1 mec1 strains had very high rates of telomere–telomere fusions (TTFs) relative to tel1 or mec1 strains (18). Expression of a Cdc13-Est2 fusion protein allows extension of telomeres to WT length in a tel1 mec1 strain (19). This extension reduces the rate of CAN1 deletions about 20-fold, arguing that about 95% of the deletions/chromosome rearrangements are likely to reflect breakage of dicentric chromosomes formed by TTFs (18). It should be noted, however, that tel1 mec1 strains with the Cdc13-Est2 fusion protein have a deletion rate that is several hundred times higher than the very low rate (<10−9) observed in WT cells (18), possibly reflecting DSBs formed at stalled replication forks (16). We also observed a high frequency of unselected chromosome rearrangements in haploid tel1 mec1 strains (20), reflecting homologous recombination between nonallelic Ty elements.
As described above, CIN tumors have elevated rates of aneuploidy in addition to high rates of chromosome rearrangements. Yeast strains with high rates of aneuploidy often have a defect in the SAC. This checkpoint ensures that chromosomes establish bipolar orientation on the spindle before anaphase. There are two types of kinetochore defects that activate the checkpoint: lack of kinetochore occupancy by microtubules and lack of tension among sister kinetochores (21, 22). Although most SAC proteins (Mad1p–Mad3p, Bub1p, and Bub3p) are required to respond to both types of defects, separation-of-function alleles of bub1 have been obtained; strains that lack the kinase domain of Bub1p (bub1-ΔK) are defective only in the tension-sensing function of the SAC (23).
Although mutants affecting the DNA damage response usually have elevated levels of GCRs and mutants affecting the SAC have elevated levels of aneuploidy, connections between these two pathways exist. The elimination of the delay of the cell cycle in response to DNA damaging agents in yeast requires mutations in both the DNA damage checkpoint pathway and the SAC (24–27). Also, the DNA damage checkpoint and the SAC are both activated by defective telomeres (28) and nocodazole (29).
The connections between the DNA damage checkpoint and the SAC previously observed are in the presence of DNA damage. Below, we show that tel1 mec1 diploid strains have a very high rate of aneuploidy and chromosome rearrangements in the absence of induced DNA damage but that the SAC remains functional in these strains. We also show that these two phenotypes reflect two different cellular defects because the frequency of chromosome rearrangements (but not the frequency of aneuploidy) is reduced by correcting the telomere defect of the tel1 mec1 mutant strain.
Results
Rationale.
Previous studies of tel1 mec1 haploid yeast strains found high rates of chromosome aberrations. Because some genetic alterations are haploid-lethal, in this study, we examined genomic alterations in tel1 mec1 sml1 diploids. The sml1 mutation in necessary to suppress the lethality of mec1 null alleles (15); in our subsequent discussion, the tel1 mec1 sml1 genotype will be abbreviated to tel1 mec1. The tel1 mec1 diploids initially had a plasmid-borne MEC1 gene (details are provided in Tables S1–S3). The strains JLMy80–JLMy83 are isogenic tel1 mec1 diploids. We then selected derivatives that lost the MEC1 plasmid, subcultured these derivatives for 100 cell divisions, and retransformed them with the MEC1-containing plasmid (details provided in Materials and Methods). We examined the frequency of aneuploidy and other changes in gene dosage in these strains by comparative genome hybridization (CGH) microarrays and alterations in the sizes of chromosomal DNA molecules by contour-clamped homogeneous electrical field (CHEF) gel analysis. No alterations were present in the mutant diploids before subculturing.
Aneuploidy in tel1 mec1 Diploids.
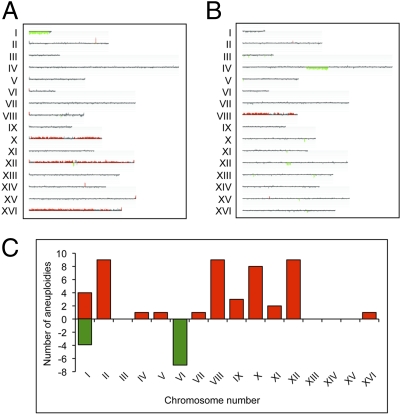
From our CGH analysis, it was clear that the tel1 mec1 subcultured diploids had very high frequencies of aneuploidy and other changes in gene dosage that reflected chromosome rearrangements. For example, strain JLMy80-1s (Fig. 1A) was monosomic for chromosome I and trisomic for chromosomes X, XII, and XVI, whereas strain JLMy81-1L (Fig. 1B) was trisomic for chromosome VIII and had an interstitial deletion on chromosome IV. Of the 20 subcultured diploids analyzed, 18 were trisomic for one or more chromosomes and half were monosomic for chromosomes I and/or VI (Table S4).
Fig. 1.
Microarray analysis of aneuploidy and chromosome rearrangements in tel1 mec1 diploids. Representative microarrays of two subcultured strains are shown (CGH Miner depiction). Each chromosome is shown as a single line with losses and gains of chromosomes or chromosome segments indicated in green and red, respectively. (A) The tel1 mec1 diploid JLMy80-1s is monosomic for chromosome I and trisomic for chromosomes X, XII, and XVI. (B) Strain JLMy81-1L is trisomic for chromosome VIII and contains an interstitial deletion on chromosome IV. (C) Distribution of trisomies (red) and monosomies (green) in 20 independently subcultured tel1 mec1 strains. Chromosomes II, VIII, X, and XII became trisomic with the highest frequencies.
Chromosomes II, VIII, X, and XII accounted for three-quarters of the trisomies, and chromosomes I and VI accounted for all the monosomies (Fig. 1C). By χ2 goodness-of-fit tests (with Yates’ correction), the distributions of trisomies and monosomies were very significantly (P < 0.001) different from random. Because chromosomes I and VI are the smallest chromosomes, it is likely that strains monosomic for other chromosomes have a selective growth disadvantage or that the small chromosomes have a lower rate of reduplication. Haploid strains that are disomic for chromosome VI are difficult to generate because of the imbalance created in the level of β-tubulin (encoded by a gene on chromosome VI) and α-tubulin (encoded by a gene on chromosome XIII) (30). Our results suggest that diploids may more readily tolerate this imbalance, a conclusion also supported by observations of Reid et al. (31).
We also examined the frequencies of aneuploid chromosomes in subcultured WT, tel1, and mec1 diploids and in tel1 mec1 haploids (Table 1 and Table S4). The tel1 mec1 diploid strain had significantly more aneuploidy than any of these other strains. Thus, as for a number of other phenotypes, the tel1 and mec1 mutations have a synergistic or additive interaction for the aneuploidy phenotype. It is likely that the higher frequency of trisomy in the diploid tel1 mec1 strains compared with the frequency of disomy in the haploid tel1 mec1 strain reflects an ability of the cell to tolerate a 1.5-fold imbalance in gene expression of chromosomes more readily than a 2-fold imbalance.
Table 1.
Number of trisomic and monosomic (in parentheses) chromosomes in strains with mutations in the DNA damage checkpoint after five cycles of subculturing*
| Chromosome no. | tel1 mec1 diploids | WT diploids | tel1 diploids | mec1 diploids | tel1 mec1 haploids | tel1 mec1 + pVL1107-URA3 diploids |
| I | 4 (4) | 0 | 1 | 0 | 0 | 4 (1) |
| II | 9 | 0 | 0 | 1 | 3 | 4 |
| III | 0 | 0 | 0 | 0 | 1 | 0 (1) |
| IV | 1 | 0 | 0 | 0 | 0 | 1 |
| V | 1 | 0 | 0 | 0 | 0 | 2 |
| VI | 0 (7) | 0 | 0 (1) | 0 | 0 | 0 |
| VII | 1 | 0 | 0 | 1 | 0 | 5 |
| VIII | 9 | 0 | 1 | 1 | 1 | 8 |
| IX | 3 | 0 | 0 | 1 | 0 | 4 |
| X | 8 | 0 | 0 | 2 | 0 | 2 |
| XI | 2 | 0 | 0 | 0 | 0 | 2 |
| XII | 9 | 0 | 0 | 0 | 2 | 5 |
| XIII | 0 | 0 | 0 | 0 | 0 | 3 |
| XIV | 0 | 0 | 0 | 1 | 0 | 0 |
| XV | 0 | 0 | 0 | 0 | 0 | 0 |
| XVI | 1 | 0 | 0 | 1 | 0 | 2 |
| Total no. aneuploid chromosomes | 48 (11) | 0 (0) | 2 (1) | 8 (0) | 7 (0) | 42 (2) |
| Total no. chromosomes analyzed† | 320 | 64 | 64 | 160 | 144 | 192 |
| Aneuploid/euploid chromosomes | 59/261 | 0/64‡ | 3/61§ | 8/152¶ | 7/137⊥ | 44/148 |
*After five cycles of subculturing, we examined aneuploidy using CGH microarrays. The numbers outside parentheses represent the number of strains in which trisomy was observed; the numbers inside parentheses represent the number of times we observed monosomy. The strain names for each genotype are provided in Table S4.
†Total number of chromosomes was calculated by multiplying the number of independent strains examined by 16, the number of homologs in S. cerevisiae.
‡Significant (P < 0.001) reduction in aneuploidy compared with the tel1 mec1 diploid (Fisher exact test).
§Significant (P < 0.05) reduction in aneuploidy compared with the tel1 mec1 diploid.
¶Significant (P < 0.0001) reduction in aneuploidy compared with the tel1 mec1 diploid.
⊥Significant (P < 0.005) reduction in disomy compared with trisomy in the tel1 mec1 diploid.
Chromosome Rearrangements in tel1 mec1 Diploids.
We also diagnosed chromosome rearrangements using microarrays (SI Text). Not including aneuploidy or changes involving the ribosomal RNA, CUP1, or Y′ genes, we found 20 large (>10 kb) genomic deletions or duplications among the 20 subcultured strains (Tables S4 and S5).
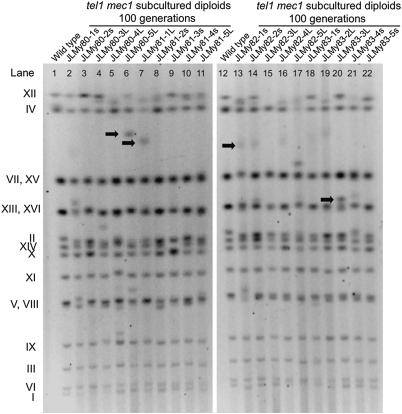
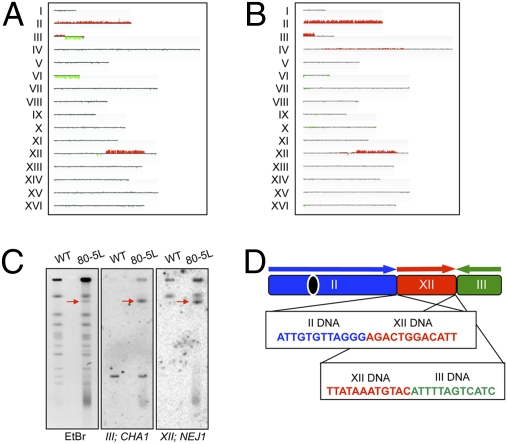
Some of the chromosome rearrangements were characterized in detail (SI Text), and a few of these are indicated by arrows in Fig. 2. The strain JMY80-5L had a unique chromosome of about 1,350 kb. By both band microarray analysis (excision of the chromosome from a CHEF gel, followed by hybridization to a microarray; Fig. 3B) and Southern blot analysis of the CHEF gel (Fig. 3C), we showed that this chromosome had all the DNA of chromosome II, a portion of the left arm of chromosome III, and an interstitial segment of chromosome XII. We used standard Southern blot analysis to determine the orientation and connectivity of the chromosome fragments and PCR analysis to amplify the fusion junctions (SI Text). Sequence analysis of fusion junctions (Fig. 3D) showed that the breakpoint on chromosome II is at the telomere. The sequence junctions share no homology, indicating that the tripartite chromosome was a consequence of two NHEJ events. Although NHEJ events are suppressed in MATa/MATα diploids (32), JLMy80-5L is hemizygous at the mating type locus (Fig. 3A).
Fig. 2.
CHEF gel analysis of DNA derived from subcultured tel1 mec1 diploids. Separated yeast chromosomes were visualized by staining of the gel with ethidium bromide and exposure to UV light. Relative to the WT strains (lanes 1 and 12), tel1 mec1 diploids often had chromosomes of altered size. Chromosomes indicated with arrows are discussed in the main text or in SI Text.
Fig. 3.
Mapping of a tripartite chromosome rearrangement. (A) Genomic microarray analysis showed that the strain JLMy80-5L had an interstitial duplication on chromosome XII, a tandem deletion/duplication on chromosome III, trisomy for chromosome II, and monosomy for chromosome VI. (B) The 1,350-kb altered chromosome was excised from the CHEF gel (indicated by an arrow in lane 6, Fig. 2) and analyzed by a microarray, delimiting the regions derived from chromosomes II, III, and XII. (C) The unique chromosome hybridized to probes derived from chromosomes III (CHA1) and XII (NEJ1). (D) PCR amplification and sequencing of the breakpoint junctions revealed that the tripartite chromosome was formed through two NHEJ events.
With the exception of JLMy80-5L, most chromosome rearrangements reflected changes in the lengths of tandemly repeated genes (e.g., the CUP1 gene array; Fig. S1) or intrachromosomal or interchromosomal exchanges in dispersed Ty retrotransposons (Fig. S2). These rearrangements are characterized in detail in SI Text. Deletions and duplications of chromosomal sequences were observed more frequently in the tel1 mec1 diploids than in the WT, tel1, or mec1 diploids (Tables S4 and S5).
Restoring WT-Length Telomeres to tel1 mec1 Strains Reduces the Frequency of Chromosome Rearrangements but Does Not Reduce the Frequency of Aneuploidy.
To examine the relationship between telomere length and the increased levels of aneuploidy and chromosome rearrangements in tel1 mec1 diploid cells, we introduced a plasmid (pVL1107-URA3) encoding a Cdc13-Est2 fusion protein (33) that allows telomere elongation independent of Tel1p and Mec1p (19). This plasmid restored WT-length telomeres to the tel1 mec1 diploid (Fig. S3A) and substantially reduced the level of TTFs (Fig. S3B). The tel1 mec1 strains with WT-length telomeres had the same frequency of aneuploidy as the tel1 mec1 strains with the short telomeres (Table 1). In contrast, only 1 of 12 tel1 mec1 diploids with WT-length telomeres had a chromosome alteration compared with 10 of 20 tel1 mec1 strains with short telomeres (Tables S4 and S5). In addition, there were a total of 20 chromosomes with deletions/duplications in the tel1 mec1 strains with short telomeres vs. a total of 2 chromosomes in the tel1 mec1 strains with long telomeres.
These results demonstrate two important points. First, the high frequencies of aneuploidy and chromosome rearrangements are caused by two different types of mechanisms. Second, in tel1 mec1 diploids, as in haploids (18), it is likely that many chromosome rearrangements are initiated by telomere fusions. This conclusion is somewhat surprising, because TTFs are a type of NHEJ, a recombination pathway that is suppressed in MATa/MATα diploids. Another observation that TTF formation and fusions between nontelomeric broken ends are regulated differently is that the Nej1 protein, which promotes NHEJ events among nontelomeric broken ends, prevents telomere fusions in diploid yeast strains (34).
Investigation of the Relationship Between the Elevated Levels of Aneuploidy in tel1 mec1 Diploids and the SAC.
One large class of genes involved in chromosome disjunction in both yeast and mammalian cells is that involved in the SAC. In addition, in response to DNA damage, Tel1p and Mec1p activate the SAC. To examine the relationship between Tel1p, Mec1p, and the SAC, we first examined the aneuploidy associated with mutations in the known SAC genes BUB1 and MAD2 using microarrays. In diploids with the bub1-Δ mutation, eight of eight independent unselected bub1-Δ diploids examined by microarrays were trisomic and/or monosomic for ≥1 chromosome (Table S4). Interestingly, 16 of the 25 trisomic chromosomes observed were chromosomes II, VIII, and X (Table 2); these 3 chromosomes plus chromosome XII were the most commonly observed trisomes in the tel1 mec1 diploid. Strains monosomic for chromosomes III, IX, XI, and XIV were observed, whereas only monosomy for chromosomes I and VI was seen in the tel1 mec1 diploids.
Table 2.
Number of trisomic/tetrasomic and monosomic (in parentheses) chromosomes in strains with mutations in the SAC genes and/or DNA damage checkpoint genes*
| Chromosome no. | bub1-Δ diploid (SC 0) | bub1-ΔK diploid (SC 5)‡ | tel1 mec1 bub1-ΔK diploid (SC 5)‡ | mad2 diploid (SC 5) |
| I | 1 | 1 | 8 | 1 (1) |
| II | 3 | 5 | 9 | 0 |
| III | 2 (1) | 4 | 2 | 0 |
| IV | 0 | 0 | 1 | 0 |
| V | 3 | 1 | 4 | 0 |
| VI | 0 | 2 | 0 | 0 |
| VII | 0 | 0 | 0 | 0 |
| VIII | 8 | 8 | 11 | 2 |
| IX | 0 (1) | 0 | 1 (1) | 0 |
| X | 5 | 5 | 11 | 0 |
| XI | 0 (2) | 0 (2) | 1 | 1 |
| XII | 0 | 0 | 4 | 0 |
| XIII | 0 | 1 | 1 | 0 |
| XIV | 1 (1) | 2 | 1 | 0 |
| XV | 0 | 0 | 0 | 0 |
| XVI | 2 | 6 | 7 | 0 |
| Total no. trisomic (monosomic) chromosomes | 25 (5) | 35 (2) | 61 (1) | 4 (1) |
| Total no. chromosomes analyzed† | 128 | 192 | 192 | 128 |
| Aneuploid/euploid chromosomes | 30/98 | 37/155 | 62/130 | 5/123 |
*We examined aneuploidy using microarrays. Strains with the null mutation of BUB1 (bub1-Δ) had very high frequencies of aneuploidy without subculturing (indicated as SC 0). Strains of the other three genotypes were subcultured five times (SC 5). The numbers outside parentheses represent the number of strains in which trisomy (or, rarely, tetrasomy) was observed for a given chromosome; the numbers inside parentheses represent the number of times we observed monosomy.
†Total number of chromosomes was calculated by multiplying the number of independent strains examined by 16, the number of homologs in S. cerevisiae.
‡Genotypes of bub1-ΔK and tel1 mec1 bub1-ΔK were not free of aneuploidy at SC 0; the data show all genetic alterations at SC 5 regardless of whether they existed at SC 0 or not. The complete spectra of aneuploidy for these strains at SC 0 and SC 5 are shown in Table S4.
We extended our analysis to examine diploids with the bub1-ΔK allele. This deletion of the kinase domain of Bub1p results in a strain that has a functional kinetochore-occupancy SAC but a defective tension-sensing SAC (23). After ≈100 cell divisions, 12 of 12 independent isolates were trisomic and/or monosomic for multiple chromosomes (Table S4). Both the frequencies of aneuploidy and the types of chromosomes that become trisomic are similar to the observations with the tel1 mec1 diploids, except that the bub1-ΔK strains have higher frequencies of trisomy for chromosome XVI and lower levels of trisomy for chromosome XII (Tables 1 and 2).
In contrast, subcultured mad2 diploids had much lower frequencies of aneuploidy (Table 2). Because mad2 diploids lack all SAC function, these observations argue that aneuploidy in tel1 mec1 and bub1 diploids reflects a defect in chromosome segregation that is, at least in part, independent of the SAC. We also examined aneuploidy in triple-mutant tel1 mec1 bub1-ΔK diploids. The frequency of aneuploidy exceeded the frequencies observed in tel1 mec1 and bub1-ΔK strains.
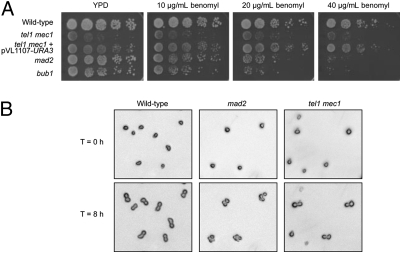
Because yeast strains with a defective SAC are sensitive to the microtubule-destabilizing drug benomyl (35, 36), we investigated the benomyl sensitivity of the tel1 mec1 strain. As shown in Fig. 4A, the tel1 mec1 strain grew poorly on all plates, including the plate lacking benomyl. The same strain with the pVL1107-URA3 plasmid, which restores WT-length telomeres to the tel1 mec1 strain, had better growth on all plates and did not exhibit obvious sensitivity to benomyl.
Fig. 4.
The tel1 mec1 cells have a functional SAC in response to the microtubule-destabilizing drug benomyl. (A) Ten-fold serial dilutions of cells from diploid WT (JLMy101), tel1 mec1 (JLMy80-Trp−), tel1 mec1 + pVL1107-URA3 (JLMy1262-Trp−), mad2 (JLMy366), and bub1 (JLMy156) strains were spotted onto solid medium containing various amounts of benomyl. (B) Haploid WT (JLMy62-7c), tel1 mec1 (JLMy62-18c-Trp−), and mad2 (JLMy309) strains were arrested in α-factor, sonicated, and released onto plates containing 70 μg/mL benomyl. The cell cycle progression of individual cells was followed for 8 h. Representative pictures show that whereas mad2 cells fail to arrest at G2/M following exposure to benomyl, WT and tel1 mec1 strains demonstrate robust cell cycle arrest. By microscopic examination, we determined that the small protuberances seen in the WT and tel1 mec1 cells represent “shmoos” (cell shapes produced by long incubation of cells in α-pheromone) rather than buds.
We also tested whether tel1 mec1 strains arrest the cell cycle in response to benomyl by examining individual cells in the presence of the drug. We treated haploid MATa WT, tel1 mec1, and mad2 strains with α-factor to arrest the cells in G1 phase. We then removed α-factor and plated the cells on rich solid growth medium (YPD) containing benomyl or lacking benomyl. We photographed cells that were unbudded (>80% after α-factor treatment) and reexamined the same cells after 8 h at room temperature. In the absence of benomyl, about 80% of the WT and mad2 cells budded more than once. In the presence of benomyl, 97% of the WT cells arrested as doublets, as expected for cells with a functional SAC (Fig. 4B and Table S6). In contrast, most (78%) of the mad2 cells continued to divide. Less than half of the tel1 mec1 cells produced a bud in the presence or absence of benomyl, as expected, because tel1 mec1 cultures contain many cells incapable of division (14). Of those cells producing a bud, however, most (97%) arrested as doublets in the presence of benomyl. Thus, tel1 mec1 strains have a functional SAC.
Discussion
As discussed in the introductory section, CIN tumors have very elevated rates of chromosome rearrangements and aneuploidy. Our observations of diploid tel1 mec1 strains show that both of these phenotypes can be produced by a single genotype. The main conclusions from our study are as follows: (i) the elevated frequencies of chromosome rearrangements and aneuploidy in tel1 mec1 strains reflect two different mechanisms, (ii) the patterns of aneuploidy in the tel1 mec1 and bub1 strains are similar, and (iii) the aneuploidy in tel1 mec1 strains is not a consequence of a defective SAC.
Chromosome Rearrangements.
As in our previous study of haploids (20), most of the chromosome rearrangements in the diploid tel1 mec1 strain are a consequence of homologous recombination among Ty elements. The frequency of chromosome rearrangements was substantially reduced in tel1 mec1 strains that expressed the Cdc13-Est2 protein. Because these strains have fewer TTFs than strains without the Cdc13-Est2 fusion protein (Fig. S3B), one simple interpretation of this result is that most chromosome rearrangements in tel1 mec1 strains are initiated as a consequence of breakage of dicentric chromosomes resulting from telomere fusions. DSB formation in a Ty element could be repaired by recombination with an ectopic Ty element, leading to chromosome rearrangements (20). An alternative model is that chromosomes in the tel1 mec1 background are degraded from the ends until a Ty element is rendered single-stranded and recombinogenic (37).
Frequencies of Aneuploidy in tel1 mec1 and in SAC-Deficient Strains.
Strains with both tel1 and mec1 mutations had significantly elevated frequencies of aneuploidy compared with either single-mutant strain, although the mec1 strain also had a high frequency of aneuploidy, as expected (9, 38). We interpret this result as indicating that Tel1p and Mec1p have functionally redundant roles in chromosome disjunction similar to their functionally redundant roles in DNA damage checkpoints and telomere length regulation. Because there is considerable heterogeneity in the growth rates of various aneuploid strains and many tel1 mec1 cells fail to give rise to viable colonies, it is difficult to calculate an accurate rate of chromosome nondisjunction from the frequency data. As a crude estimate, we calculate that the probability of detecting aneuploidy in 100 cell divisions, if the rate of nondisjunction per cell division is 10−3, is 1 − (0.999)100 or about 0.1; if the rate of nondisjunction is 10−2, the probability of detecting aneuploidy in 100 divisions is 1 − (0.99)100 or about 0.7. Thus, the rate of nondisjunction per chromosome per cell division is about 10−2 to 10−3 for the most frequently lost chromosomes.
The frequency of nondisjunction was also very high in the bub1-Δ and bub1-ΔK strains but not in the mad2 strain. Warren et al. (39) previously observed that bub1 mutants had about 10-fold higher chromosome loss rates than mad2 strains and suggested that Bub1p (but not Mad2p) had a role in chromosome segregation in the absence of induced spindle damage. A variety of experiments have indicated that Bub1p is important in establishing the correct inner and outer kinetochore structures (40), and cells with low levels or no Bub1p have defective associations between the kinetochore and microtubules, decreased levels of chromosomes that are bioriented at the spindle, and decreased sister chromatid cohesion at the centromere (41). Thus, loss of Bub1p (unlike the loss of Mad2p) leads to defects in the mechanics of chromosome segregation in addition to defects in the SAC, resulting in very high levels of aneuploidy.
Nonrandom Recovery of Trisomic Chromosomes in tel1 mec1 and in SAC-Deficient Strains.
There is a preference for the recovery of chromosomes II, VIII, X, and XII as trisomes in the tel1 mec1 strain (Table 1). If we perform a χ2 goodness-of-fit test for these same four chromosomes in the bub1-Δ and bub1-ΔK strains, there is a very significant (P < 0.001) nonrandomness in their recovery (Table 2). There are three plausible explanations for the preferential recovery of certain chromosomes as trisomes. First, certain chromosomes, when present in three copies in a diploid, may greatly reduce cell growth rates. This explanation is unlikely for two reasons: in nine strains with trisomic chromosomes resulting from γ-irradiation of diploids, a broad spectrum of trisomies was observed that did not include chromosome II or X (42) and Torres et al. (30) observed that all yeast chromosomes, except chromosome VI, were recoverable as disomes in haploids. Second, the presence of certain trisomic chromosomes may alleviate some of the negative effects of tel1 mec1 and/or bub1 mutations. For example, we previously showed that chromosome VIII disomy in tel1 mec1-21 haploids was suppressed by DNA2, a gene located on chromosome VIII (20). A third possibility is that the defect in chromosome segregation observed in the tel1 mec1 and bub1 strains affects some chromosomes more than others. We suggest that the nonrandom recovery of trisomes is likely to reflect the two latter explanations, possibly arguing a similarity in the mechanistic chromosome disjunction defects in the tel1 mec1 and bub1 strains.
Cellular Roles of Tel1p and Mec1p in Regulating Aneuploidy.
Our analysis demonstrates that Tel1p and Mec1p have an important role in chromosome segregation in addition to their previously demonstrated roles in DNA repair and telomere length regulation. Elevated rates of chromosome loss have been observed in mutants affecting kinetochores, microtubules, the spindle pole body, sister chromatid cohesion, DNA replication/repair, and the SAC. Our results rule out an involvement of Tel1p and Mec1p in the SAC, although we cannot define what other role these proteins have in chromosome segregation.
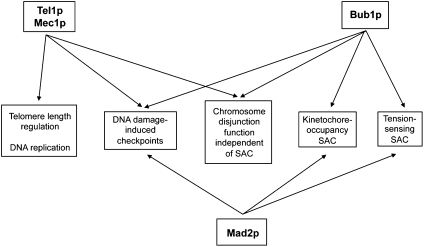
One possibility is that Tel1p and Mec1p affect chromosome segregation in the same pathways affected by Bub1p related to kinetochore function (Fig. 5). An argument in favor of this possibility is the similar patterns of trisomies observed in tel1 mec1 and bub1 strains. An argument against this possibility is that the chromosome segregation defect of the triple-mutant tel1 mec1 bub1-ΔK strain is more extreme than the defects of the tel1 mec1 or bub1-ΔK strain. Although this result could be interpreted as indicating that Tel1p/Mec1p and Bub1p affect different chromosome disjunction mechanisms, it is also possible that the Tel1p/Mec1p effects are partially redundant with those of Bub1p, acting in the same pathway. Finally, we point out that direct investigations of kinetochore function are very difficult in tel1 mec1 strains because this genotype results in a high rate of nonviable cells and heterogeneity in growth rates among the viable cells (14).
Fig. 5.
Pathways involving Tel1p, Mec1p, and SAC proteins. The indicated associations of proteins and pathways are based on information provided in the main text.
Materials and Methods
Strain Construction and Subculturing of Yeast Strains.
All strains were constructed in the MS71 background (MATα ade5-1 leu2-3 trp1-289 ura3-52 his7-2 LEU2) (43) using standard techniques (44). Because the lethality of a mec1 null allele can be rescued by a concurrent deletion of SML1 (15), all mec1::NAT strains in our study had a sml1::HYG deletion as well. Details of the constructions are provided in SI Materials and Methods and in Tables S1–S3. The plasmid pVL1107 contains LEU2 and a gene encoding a Cdc13-Est2 fusion protein (33). In the related plasmid (pVL1107-URA3), the LEU2 gene was replaced by URA3 (SI Materials and Methods).
Because tel1 mec1 strains are genomically unstable, the diploid strains were constructed with a WT copy of MEC1 on a TRP1-marked plasmid (pSAD3-3b/MEC1) (45). Four isogenic diploids (JLMy80–83) were constructed by mating spores derived from JLMy62 (details are provided in Table S2). To examine genomic instability, we identified derivatives of each diploid that had lost the plasmid (subculture 0) and subcultured these derivatives by streaking the cells on YPD (46), allowing the cells to grow for 2 to 3 days at 30 °C (subculture 1). Five individual colonies were picked from subculture 1 of these Trp− derivatives of JLMy80–83 and separately subcultured four more times on plates containing YPD, picking cells from the heavy growth region of the plate. Each subculture equals ≈20 cell divisions. The 20 subcultured derivatives were retransformed with pSAD3-3b/MEC1 before analysis.
Physical Analysis of Chromosome Rearrangements and Aneuploidy.
The sizes of rearranged chromosomes were determined by CHEF gel analysis and Southern blotting techniques, as previously described (47). We also used CGH microarrays to diagnose the breakpoints of chromosome rearrangements and to examine changes in chromosome number. The details of the gel and microarray analyses are given in SI Text.
Statistical Analysis of the Frequency of Aneuploidy in tel1 mec1 and Related Strains.
For independently subcultured strains of each genotype, we calculated the number of aneuploid and euploid chromosomes. These numbers were compared in strains of different genotypes using the Fisher exact probability test or χ2 analysis tool on the VassarStats Web site (http://faculty.vassar.edu/lowry/VassarStats.html).
Assay of Benomyl Sensitivity and Assay of the SAC in Response to Benomyl.
We examined the ability of WT, tel1, mec1, and tel1 mec1 cells to grow on YPD plates containing concentrations of benomyl (Sigma) of 10, 20, or 40 μg/mL (details provided in SI Text). We also determined the fraction of cells (unbudded at the start of treatment with 70 μg/mL benomyl) that arrested as doublets after 8 h of treatment.
Supplementary Material
Acknowledgments
We thank members of the Petes laboratory for useful discussions; S. Elledge (Harvard Medical School, Boston, MA) and V. Lundblad (Salk Institute for Biological Studies, La Jolla, CA) for supplying plasmids; and J.L. Argueso, P. Mieczkowski, and J. Lieb for help in preparing DNA microarrays. We thank D. Burke, D. Lew, and S. Haase for discussions concerning the SAC. This research was supported by National Institutes of Health Grant GM52319 (to T.D.P.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE17903).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006281107/-/DCSupplemental.
References
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 3.Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer. 2003;3:695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 4.Barber TD, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci USA. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460:984–989. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 7.Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 8.Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 9.Craven RJ, Greenwell PW, Dominska M, Petes TD. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics. 2002;161:493–507. doi: 10.1093/genetics/161.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myung K, Pennaneach V, Kats ES, Kolodner RD. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci USA. 2003;100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myung K, Smith S, Kolodner RD. Mitotic checkpoint function in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2004;101:15980–15985. doi: 10.1073/pnas.0407010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison JC, Haber JE. Surviving the breakup: The DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 13.Morrow DM, Tagle DA, Shiloh Y, Collins FS, Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 14.Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 16.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 17.Cimprich KA, Cortez D. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mieczkowski PA, Mieczkowska JO, Dominska M, Petes TD. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100:10854–10859. doi: 10.1073/pnas.1934561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukamoto Y, Taggart AK, Zakian VA. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol. 2001;11:1328–1335. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- 20.Vernon M, Lobachev K, Petes TD. High rates of “unselected” aneuploidy and chromosome rearrangements in tel1 mec1 haploid yeast strains. Genetics. 2008;179:237–247. doi: 10.1534/genetics.107.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annu Rev Genet. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- 22.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 23.Fernius J, Hardwick KG. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 2007;3:e213. doi: 10.1371/journal.pgen.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collura A, Blaisonneau J, Baldacci G, Francesconi S. The fission yeast Crb2/Chk1 pathway coordinates the DNA damage and spindle checkpoint in response to replication stress induced by topoisomerase I inhibitor. Mol Cell Biol. 2005;25:7889–7899. doi: 10.1128/MCB.25.17.7889-7899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garber PM, Rine J. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics. 2002;161:521–534. doi: 10.1093/genetics/161.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim EM, Burke DJ. DNA damage activates the SAC in an ATM/ATR-dependent manner, independently of the kinetochore. PLoS Genet. 2008;4:e1000015. doi: 10.1371/journal.pgen.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugimoto I, Murakami H, Tonami Y, Moriyama A, Nakanishi M. DNA replication checkpoint control mediated by the spindle checkpoint protein Mad2p in fission yeast. J Biol Chem. 2004;279:47372–47378. doi: 10.1074/jbc.M403231200. [DOI] [PubMed] [Google Scholar]
- 28.Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clémenson C, Marsolier-Kergoat MC. The spindle assembly checkpoint regulates the phosphorylation state of a subset of DNA checkpoint proteins in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:9149–9161. doi: 10.1128/MCB.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 31.Reid RJ, et al. Chromosome-scale genetic mapping using a set of 16 conditionally stable Saccharomyces cerevisiae chromosomes. Genetics. 2008;180:1799–1808. doi: 10.1534/genetics.108.087999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kegel A, Sjöstrand JO, Aström SU. Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr Biol. 2001;11:1611–1617. doi: 10.1016/s0960-9822(01)00488-2. [DOI] [PubMed] [Google Scholar]
- 33.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 34.Liti G, Louis EJ. NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol Cell. 2003;11:1373–1378. doi: 10.1016/s1097-2765(03)00177-1. [DOI] [PubMed] [Google Scholar]
- 35.Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 36.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 37.Hackett JA, Greider CW. End resection initiates genomic instability in the absence of telomerase. Mol Cell Biol. 2003;23:8450–8461. doi: 10.1128/MCB.23.23.8450-8461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein HL. Spontaneous chromosome loss in Saccharomyces cerevisiae is suppressed by DNA damage checkpoint functions. Genetics. 2001;159:1501–1509. doi: 10.1093/genetics/159.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warren CD, et al. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol Biol Cell. 2002;13:3029–3041. doi: 10.1091/mbc.E02-04-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams GL, Roberts TM, Gjoerup OV. Bub1: Escapades in a cellular world. Cell Cycle. 2007;6:1699–1704. doi: 10.4161/cc.6.14.4493. [DOI] [PubMed] [Google Scholar]
- 41.Logarinho E, Bousbaa H. Kinetochore-microtubule interactions “in check” by Bub1, Bub3 and BubR1: The dual task of attaching and signalling. Cell Cycle. 2008;7:1763–1768. doi: 10.4161/cc.7.12.6180. [DOI] [PubMed] [Google Scholar]
- 42.Argueso JL, et al. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci USA. 2008;105:11845–11850. doi: 10.1073/pnas.0804529105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strand M, Earley MC, Crouse GF, Petes TD. Mutations in the MSH3 gene preferentially lead to deletions within tracts of simple repetitive DNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:10418–10421. doi: 10.1073/pnas.92.22.10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 45.Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 47.Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell. 2006;125:1283–1296. doi: 10.1016/j.cell.2006.04.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.