Abstract
Many small molecular anticancer agents are often ineffective at detecting or treating cancer due to their poor pharmacokinetics. Using nanoparticles as carriers can improve this because their large size reduces clearance and improves retention within tumors, but it also slows their rate of transfer from circulation into the tumor interstitium. Here, we demonstrate an alternative strategy whereby a molecular contrast agent and engineered nanoparticle undergo in vivo molecular assembly within tumors, combining the rapid influx of the smaller and high retention of the larger component. This strategy provided rapid tumor accumulation of a fluorescent contrast agent, 16- and 8-fold faster than fluorescently labeled macromolecule or nanoparticle controls achieved. Diagnostic sensitivity was 3.0 times that of a passively targeting nanoparticle, and this improvement was achieved 3 h after injection. The advantage of the in vivo assembly approach for targeting is rapid accumulation of small molecular agents in tumors, shorter circulation time requirements, possible systemic clearance while maintaining imaging sensitivity in the tumor, and nanoparticle anchors in tumors can be utilized to alter the pharmacokinetics of contrast agents, therapeutics, and other nanoparticles. This study demonstrates molecular assembly of nanoparticles within tumors, and provides a new basis for the future design of nanomaterials for medical applications.
Determining the correct prognosis and therapeutic options for cancer requires accurate staging and surveillance of tumors. Current detection strategies typically combine sensitive imaging modalities with contrast agents (1, 2). Yet these approaches fail to detect lesions in many cases, typically because poor imaging contrast is achieved (2). This can be improved through tumor targeting strategies that link contrast agents to a polymer or nanoparticle. Nanoparticles are well suited to act as tumor targeting vehicles because their in vivo behavior is determined by their design, and they are able to leak into and accumulate in tumors via the enhanced permeability and retention effect (3–7). Despite these advantages, several obstacles limit effective tumor detection with nanoparticle-based targeting strategies. Passive targeting requires particles with large diameters, but this simultaneously restricts transport into tumors and accumulation occurs only after many hours in circulation (8–10). Actively targeting nanoparticle designs can achieve faster accumulation (11–13), but may not be appropriate for detecting lesions whose antigens are uncharacterized or are heterogeneous and therefore unreliable. Finally, nanoparticles have long circulation and persistence times in the body, raising potential concerns of diagnostic or therapeutic agent toxicity. It would therefore be advantageous to develop targeting strategies that can rapidly accumulate contrast agent into tumors without relying on antigen characterization and without causing long-term persistence in the body.
The movement of nanoparticles through tumor extracellular matrix is primarily dependent on diffusion (8). A recent in vivo study from our lab demonstrated that diffusive transport is limited with larger particle diameters and becomes negligible by 100 nm. Those with diameters of 80 nm were found to penetrate slowly into the interstitium and became localized within several cell lengths of leaky vasculature 24 h after injection (4). We hypothesized that this design-dependent localization within tumors would make the particles highly accessible to molecular contrast agents in circulation. Moreover, antigens present on the particle surface would then provide a universal anchor for active targeting. This strategy might be highly effective because transport of a lower molecular weight contrast agent into the tumor would be rapid (i.e., high Kin), at which point its retention would be determined by the large nanoparticle diameter (i.e., low Kout).
Here, we demonstrate a targeting strategy whereby a molecular contrast agent and engineered nanoparticle undergo in vivo assembly within tumor interstitium. Prior studies have demonstrated that multicomponent nanoparticle systems can provide either improved or additional functionalities for tumor targeting applications (14–16), but the possibility of delivering synthetic nanoparticle antigens to tumors for rapid assembly with contrast agents has not been examined. Using an optical detection platform and fluorescent contrast agent, we compared the rate and extent of accumulation from assembly to that achieved by passive macromolecule and nanoparticle accumulation. We also demonstrate that the assembly process can be competitively inhibited, attenuating its effect on tumor accumulation. Intratumor assembly was found to provide the fastest accumulation kinetics and highest signal-to-noise. These results describe a strategy for nanoparticle-based tumor targeting, and provide a conceptual demonstration of how in vivo assembly can be applied to overcome in vivo barriers to targeting.
Results and Discussion
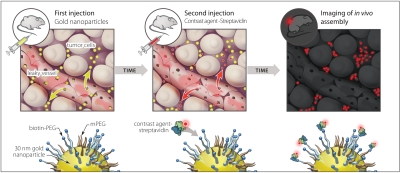
From a rationale of improving tumor accumulation by reducing Kout without impacting Kin, we designed a hybrid targeting system of passive and active targeting components that are injected in sequence and assembled in vivo. In our approach, a nonfluorescent metal nanoparticle “anchor” is administered and accumulates passively in tumor tissue over a 24-h period. This provides a synthetic molecular ligand for active targeting by a fluorescent contrast agent injected at an appropriate time (see schematic, Fig. 1).
Fig. 1.
Schematic of nanoparticles assembling with contrast agent in vivo. Gold nanoparticles stabilized with biotinylated PEG (denoted as biotin-PEG anchor) are injected as a first step. These enter tumors through leaky vasculature and passively accumulate in the extracellular matrix over 24 h. Fluorescently labeled streptavidin is injected, leaks into tumors, and interacts with biotin on the gold nanoparticles in the interstitium. This favorably alters the contrast agent’s tumor accumulation kinetics.
Previous research provided guidelines for designing the nanoparticle anchor component (4, 7, 17). We used gold nanoparticles (GNP) stabilized with a thiolated-poly-(ethylene glycol) surface brush layer, consisting of both biotin- and methoxy-terminating (mPEG) molecules (Fig. S1 shows nanoparticle characterization). Biotin-PEG provides ligands for assembling streptavidin-fluorophore conjugates onto the biotin-gold nanoparticle (biotin-GNP) surface. The biotin-GNP anchors were optimized to bind a large number of streptavidin conjugates (approximately 500 streptavidin constructs per particle; Fig. S2 A and B), and we verified that the metal nanoparticle did not quench the dye’s fluorescence upon binding (Fig. S2 C and D) (18, 19).
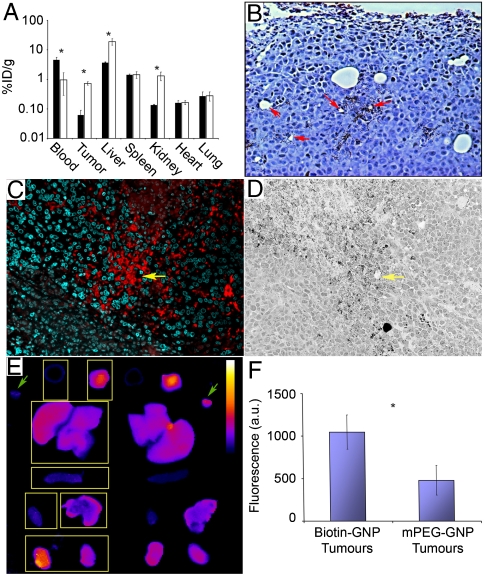
We engineered the nanoparticle geometry and surface chemistry toward specific in vivo behavioral criteria (i.e., pharmacokinetic and intratumor distribution), validated by profiling its biodistribution (Fig. 2A). By 24 h postinjection (HPI), the biotin-GNP had a significant accumulation in tumor tissue (0.72 ± 0.09 vs. 0.06 ± 0.03% ID/g at 6 HPI), and was nearly cleared from circulation (0.97 ± 0.68 vs. 4.67 ± 0.86% ID/g at 6 HPI). Its 70–80-nm hydrodynamic diameter permitted extravasation into the tumor interstitium but restricted permeation through the tumor extracellular matrix, causing the biotin-GNP to localize in perivascular spaces in close proximity to leaky blood vessels (Fig. 2B) (4, 20). The biotin-streptavidin system was chosen because it undergoes near-irreversible binding, but also because prior antibody-based targeting studies had clearly demonstrated the binding functionality of this system within the tumor interstitium (21–26).
Fig. 2.
Biodistribution, tumor localization, and assembly of the two components. (A) Biodistribution of the biotin-GNP anchor at 6 (▪) and 24 (□) HPI showing a decrease in blood concentration and an increase in tumor, liver, and kidney tissue. (B) Silver enhancement staining of tumor tissue 24 HPI of biotin-GNP showing perivascular localization of the anchor component. Red arrows mark examples of leaky blood vessels. (C) Fluorescence microscopy showing an overlapping distribution of streptavidin-Alexa568 (red) with the biotin-GNP anchors (shown in D) at 6 HPI of contrast agent. Nuclei are stained with DAPI (green), and yellow arrows mark a blood vessel. (E) FRET images of tissues harvested from mice 2 HPI of strept-A568 + 750 (excitation wavelength of 570, emission wavelength of 790). Control mPEG-GNP organs on the left are marked with boxes (Left to Right and Top to Bottom: blood, tumor, liver, spleen, heart, lung, kidneys). Green arrows mark an aliquot of the streptavidin conjugates mixed with mPEG-GNP (Left) and biotin-GNP (Right), clearly showing a higher signal from FRET in the latter. Calibration bar = 500–7,500 arbitrary unit (a.u.). (F) Quantification of fluorescence found a higher signal in tumors of biotin-GNP vs. mPEG-GNP injected mice at 2 HPI. Data points are the mean ± SEM from n = 3 animals. Asterisk, p < 0.05 Student’s t test between 6 and 24 h (biodistribution data), p = 0.050 1-tailed Student’s t test vs. mPEG control (FRET data).
Assuming the molecular component chases the biotin-GNP anchor into the tumor interstitium, assembly should occur upon colocalization. The spatial distribution of both components was visualized by injecting a single 2.5 uM bolus of streptavidin-Alexa fluor 568 (strept-A568) into tumor-bearing mice 24 HPI of GNP-biotin and collecting tissue for histology after a further 6 h. There was a similar pattern of contrast agent (Fig. 2C) and biotin-GNP (Fig. 2D) distribution away from blood vessels within the tumor interstitium. To further validate assembly of the two components in vivo, we tested whether two streptavidin-Alexa fluor dye conjugates (strept-A568 and -A750) could simultaneously bind biotin-GNP, causing Förster resonance energy transfer (FRET) between the two dyes by bringing them within close proximity. After achieving FRET on biotin-GNP in vitro (Fig. S3), we injected tumor-bearing mice with a single strept-A568/750 bolus 24 HPI of biotin-GNP. Organs were harvested and imaged 2 HPI of contrast agents for FRET-specific fluorescence (Fig. 2E). Autofluorescence-subtracted signal in tumors from biotin-GNP mice was more than twice as high as in control mPEG-GNP mice (Fig. 2F). This and the colocalization data strongly support that assembly of the two components can occur within tumors.
We were then motivated to test whether in vivo assembly could alter a contrast agent’s tumor accumulation kinetics in a favorable manner. Streptavidin-Alexa fluor 750 was injected 24 HPI of biotin-GNP, and fluorescence data were collected for nearly 2 h. We opted for single-fluorophore detection over FRET because longer wavelength excitation light provides better tissue penetration (1, 27). We compared the kinetics of our assembly strategy against a nonassembling control (mPEG-GNP injection followed 24 h later by strept-A750) and against a passive targeting fluorescent nanoparticle. This was constructed by assembling biotin-GNP and strept-A750 prior to injection. As a final control, we competitively inhibited our system by injecting nonfluorescent streptavidin 30 min before injection of strept-A750.
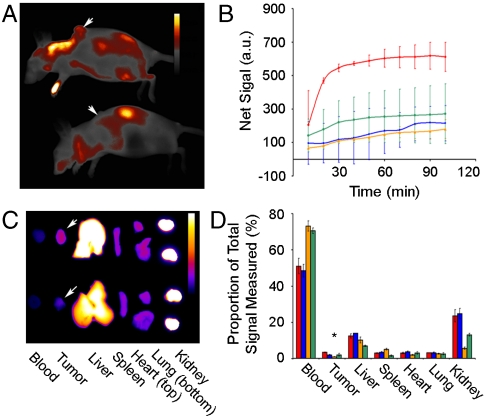
Region-of-interest (ROI) analysis (described in Fig. S4) revealed intriguing differences in contrast agent behavior between in vivo assembly and control mice. To examine the nontarget (background) signal within the mice, we applied an ROI that encompassed the whole animal side profile excluding the tumor ROI. The mean signal intensity (I) change over time (I/I0) in the nontarget ROI was identical in biotin-GNP and mPEG-GNP injected mice (Fig. S5). Both reached a maximum signal approximately 1 HPI followed by a reduction in background that may coincide with contrast agent clearance from circulation. Competitive inhibition of assembly appeared to reduce the clearance of Strept-A750, as background fluorescence continued to increase over the entire imaging session. Mice injected with passive targeting GNP-A750 displayed a linear rate of background signal increase lower than other experimental groups. This may reflect differences in the rate of nanoparticle and macromolecule diffusion into peripheral blood vessels most visible by fluorescence imaging.
We then examined the net tumor accumulation of the contrast agent by subtracting mean nontarget ROI signal from tumor signal (Fig. 3 A and B). The most remarkable difference revealed by this was the increased rate of accumulation from in vivo assembly versus controls. Tumors of biotin-GNP injected animals displayed a 265.8% net increase within 30 min, a rate 16.0, 8.1, and 4.6 times that of the mPEG-GNP, GNP-streptA750, and competitive inhibition controls, respectively. This suggests that perivascular localization of the nanoparticle anchor component allowed rapid colocalization and assembly. By 90 min, net tumor signal in biotin-GNP animals was 2.83, 3.67, and 2.32 times that in mPEG-GNP, GNP-A750, and competitive inhibition controls.
Fig. 3.
Optical detection of fluorescence and analysis of tumor accumulation over 2 HPI of assembling contrast agent. (A) Representative fluorescence image at 2 HPI of biotin-GNP (Top) and mPEG-GNP (Bottom) injected mice. Arrows mark tumor location, and calibration bar = 500–6,000 a.u. (B) Tumor accumulation (tumor-body) of contrast agent over time in biotin-GNP (red line) injected mice, as well as control mPEG-GNP (blue line), GNP-A750 (orange line), and competitive inhibition (green line) injected mice. (C) Representative fluorescence image of organs harvested from biotin-GNP (Top) and mPEG-GNP (Bottom) injected mice 2 HPI of strept-A750. Arrows mark the tumors, and calibration bar = 100–4,500 a.u. (D) Quantification of tissue fluorescence from biotin-GNP (red squares), mPEG-GNP (blue squares), GNP-A750 (orange squares), and competitive inhibition (green squares) mice. Data points are the mean ± SEM from n = 3 animals. Asterisk, p < 0.05 for biotin-GNP vs. mPEG-GNP and competitive inhibition controls, and p < 0.01 for biotin-GNP vs. GNP-A750 using ANOVA and Dunnet’s post hoc test.
The kinetic data were then cross-validated against biodistribution of the contrast agent in harvested organs (Fig. 3 C and D). With the exception of tumors, biodistribution of strept-A750 was identical in biotin-GNP and mPEG-GNP treated animals. Mice that were used for competitive inhibition of assembly showed a nonsignificant reduction in liver, spleen, and kidney accumulation. Passively targeted fluorescent GNP-A750 showed higher spleen (p < 0.05) and lower kidney (p < 0.01) uptake versus the assembly strategy.
The tumors of biotin-GNP-treated mice contained 3.4% of the total fluorescence measured across all tissues. This is higher than the 2.1%, 0.9%, and 2.1% in mPEG-GNP, passive targeting GNP-A750 and competitive inhibition controls, respectively. The passively targeting nanoparticle’s low tumor accumulation by 2 h may result from its relatively restricted rate of transport into the tumors (Kin). Accumulation in mPEG-GNP and competitive inhibition controls was identical, suggesting that assembly was successfully blocked in the latter. Accumulation in biotin-GNP tumors was 1.3% higher than in corresponding mPEG-GNP and competitive inhibition controls. We can estimate whether this increase can be attributed to assembly by calculating the capture potential of the biotin-GNP localized in tumor tissue. Based on a capture potential of 500 streptavidin per particle and accumulation of 0.72% of the total biotin-GNP injected dose, we would expect an increase in signal up to 1.93%. The measured increase is therefore within the expected range.
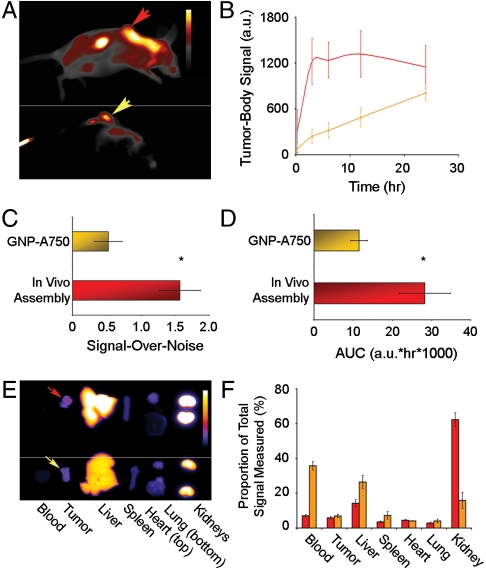
Finally, we compared the accumulation kinetics of in vivo assembly to that of passively targeting nanoparticles (GNP-A750) over a 24 h period (Fig. 4A). There was a striking difference in contrast agent retention in circulation and the background signal that resulted from this (Fig. S6). Background signal from streptA750 conjugate injected for in vivo assembly increased to 35% above that at T0 before dropping back. Background signal in GNP-A750 mice increased continuously and did not reach a plateau by 24 HPI. This suggests that assembly of strept-A750 onto the pegylated nanoparticle greatly reduced the rate of contrast agent clearance and provided an opportunity for tumor accumulation.
Fig. 4.
Optical detection of fluorescence and analysis of tumor accumulation over 24 HPI of streptavidin-Alexa fluor 750. (A) Representative fluorescence image at 24 HPI of biotin-GNP (Top) and GNP-A750 (Bottom) injected mice. Arrows mark tumor location, and calibration bar = 500–6,000 a.u. (B) Tumor accumulation of contrast agent over time in biotin-GNP (red line) and GNP-A750 (orange line) injected mice. (C) Signal-over-noise (S/N) at 3 HPI, and (D) total area-under-the-curve (AUC) achieved with in vivo assembly (red squares) vs. GNP-A750 (orange squares) of components. (E) Representative fluorescence image of organs harvested from mice injected with the in vivo assembly system (Top) and GNP-A750 construct (Bottom) at 24 HPI. Arrows mark tumors, and calibration bar = 50–3,000 a.u. (F) Quantification of tumor fluorescence from mice injected with the in vivo assembly system (red squares) vs. GNP-A750 (orange squares). Data points are the mean ± SEM from n = 3 animals. Asterisks, 2-tailed Student’s t test for in vivo assembly vs. passive targeting GNP-A750 of S/N (p = 0.048) and AUC (p = 0.036). Note that animal and organ panels (A and E) are products of separate images that underwent identical processing.
In vivo assembly provided a nearly 200-fold higher rate of tumor accumulation than passive targeting GNP-A750 within the first 3 HPI, and a 5.1-times higher tumor signal (p < 0.05, Fig. 4B). The net signal intensity then remained constant between 3 and 24 h. Net intensity of passive targeting GNP-A750 increased linearly until reaching a statistically similar level to that of in vivo assembly at 24 HPI (p = 0.31). In vivo assembly therefore achieved comparable signal to a passively targeting nanoparticle, but eight times faster.
As a measure of diagnostic sensitivity, the signal-over-noise achieved by in vivo assembly was triple that of passive targeting nanoparticles by 3 HPI (p < 0.05, Fig. 4C and Fig. S7). As well, the total area-under-the-curve in the tumor achieved by in vivo assembly was calculated to be 2.46 times that of passive targeting, offering a greater window of opportunity for detecting tumors (Fig. 4D).
We again cross-validated the kinetic data by profiling contrast agent biodistribution in organs harvested 24 HPI (Fig. 4 E and F). Mice injected with preassembled GNP-A750 had higher signal remaining in the blood (p < 0.001) and greater uptake in the liver (p < 0.05) and lower uptake in kidneys (p < 0.01) at 24 HPI. Tumor signal was comparable by 24 HPI (p < 0.5), supporting what we observed in vivo. Attachment of fluorescent streptavidin onto pegylated GNP can therefore maintain it within the circulation for long enough to achieve high levels of tumor accumulation via the enhanced permeability and retention effect 24 h after administration. However, a comparable level of signal was achieved by in vivo assembly at 3 HPI.
Conclusion
There are currently a number of major challenges in using nanoparticles to deliver contrast agents into tumors. Ideally, delivery systems would specifically and rapidly deliver a high fraction of the injected dose to diseased sites while minimizing retention and background signal in the rest of the body. In an early study, Gao et al. conclusively showed that a conventional active targeting approach can achieve rapid accumulation of fluorescent quantum dots in tumors (28). More recently, Choi et al. demonstrated that actively targeting quantum dots can be designed to allow renal clearance within 4 h of injection and that decreased retention in the body resulted in improved specific signal over background (29). However, conventional active targeting for tumor surveillance may be limited in many cases by the need to profile molecular targets within each individual patient’s tumor (30).
Our strategy therefore has two clear advantages. First, it avoids relying on molecular targets on cells and tissues and enables one to artificially design a molecular target that accumulates in the tumor by the enhanced permeability and retention effect. Second, it rapidly achieves a high specific signal in tumors and does not require the contrast agent to remain in circulation for many hours. This approach could allow the retention properties of the contrast agent to be optimized to further reduce background signal. Our data support the argument put forward by Choi et al. (29) that rapid tumor accumulation and low body retention should achieve the highest sensitivity. Using in vivo assembly to mediate targeting would be an alternative method to achieve good sensitivity and allow clearance but without the need to identify targeting receptors on tumors.
The specific nanoparticle design that we used for our targeting strategy was based on fundamental knowledge provided in earlier research (4, 7). We used the biotin-streptavidin binding system because of its strong binding affinity. Future studies on alternative molecular binding systems and optimization of nanoparticle size, shape, and chemistry as anchors could further improve the overall targeting efficiency and the signal-over-noise ratio. This unique assembly strategy could also be used for targeting alternative contrast agents (i.e., gadolinium, radioisotopes) or therapeutics in vivo. The results presented here clearly demonstrate the concept and advantage of assembling targeting components in vivo to improving tumor delivery of diagnostic agents.
Materials and Methods
Synthesis of Gold Nanoparticles.
The GNP were synthesized according to the method developed by Frens (31). Briefly, 300 μL of chloroauric acid (Sigma-Aldrich) was added to 300 mL ultrapurified H2O. The solution was brought to a boil, and 3 mL of 1% sodium citrate tribasic dihydrate (Sigma-Aldrich) was added while stirring rapidly. The solution was removed from heat after 5 min, cooled to room temperature, and filtered through a 0.22-μm, 500-mL filter (Nalgene).
Characterization and Pegylation of GNP.
Synthesized particles were characterized by ultraviolet and visible range absorption spectroscopy using a UV-1601PC spectrophotometer (Shimadzu). transmission electron microscope (TEM) images were obtained by adding 10 μL of a 1∶5 (stock: H2O) dilution of the particle solution onto carbon-coated copper grids, then imaged on a Hitachi HD2000 STEM (Hitachi Corp.). ImageJ (version 1.42q, National Institutes of Health) was used to measure the particle sizes by first setting the scale according to the TEM image scale bar, then Adjust-Threshold to create a binary image, and finally Analyze-Analyze Particles to determine Feret’s diameter. Several images were measured and the first 200 results were used to determine particle diameter.
The GNP concentration was determined by inductively-coupled plasma mass spectrometry (ICP-MS) (see below), allowing us to calculate the available surface area per milliliter of GNP stock. Then, we examined the PEG-blend dependence on binding of streptavidin-Alexa fluor 750 (Invitrogen) to the particles. Based on our prior work, addition of approximately 0.3× molecules of mPEG-SH (Mr = 5,000 g/mol, NOF Corp.) per square nanometer of surface area is enough to stabilize GNP of 15–50 nm (4). Keeping this constant, we added a serial dilution of biotin-PEG-SH (Mr = 10,000 g/mol, Nanocs) at a ratio of 10, 5, 2.5, 1.25, 0.6, 0.3, 0.15, and 0× per square nanometer, to 1 mL aliquots of the GNP stock. After immediate mixing by vortexing, the particles were incubated at room temperature (RT) for 30 min on an inversion mixer. 10× mPEG 5000 was then added to fully saturate the particle surface, incubating a further 30 min at RT and mixing by inversion. The eight treatments were then centrifuged at 15,000 × g for 5 min, the supernatant was removed, and 1 mL of H2O was added to wash. This was repeated five times total, and finally the particles were resuspended in 100 μL of PBS. Twenty microliters of the pegylated particle concentrates was then mixed with an excess of streptavidin-Alexa fluor 750. The products were separated on a 0.7% agarose gel in 0.5× Tris/Borate/EDTA buffer at 100 V for 20 min (Fig. S2A). Particle-bound streptavidin was determined by measuring mean fluorescence in the background-corrected gel image using ImageJ. A ratio of 0.6× biotin-PEG-SH was chosen based on a compromise between high streptavidin-Alexa fluor binding while leaving most of the particle surface covered with mPEG to stealth the particles from immune recognition. This should provide approximately 2,000 biotins per particle, or one biotin per square nanometer of surface area. Based on the tetramer structure of streptavidin and its hydrodynamic diameter (7 nm), we assume that one biotin-GNP can bind 500 streptavidin molecules.
The remaining particle stock was pegylated by scaling up the PEG added. Control mPEG particles were synthesized identically, replacing the biotin-PEG-SH (Mr = 10,000 g/mol) with methoxy-PEG-SH (Mr = 10,000 g/mol). Hydrodynamic diameter of the particles was measured by dynamic light scattering using a Nano-SZ Zetasizer (Malvern). The final pegylated stock was centrifuged at 12,000 × g for 30 min in an Avanti Series centrifuge (Beckman-Coulter), the supernatant removed, and the pellet resuspended in 1 mL H2O in a microcentrifuge tube. This was centrifuged at 12,000 × g for 10 min, the supernatant removed, and 1 mL H2O added to wash. After five washes, the pellet was resuspended in 1 mL H2O and stored at 4 °C until use.
Cell Culture.
MDA-MB-435 cells (American Type Tissue Culture) were maintained in RPMI medium 1640 (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Sigma-Aldrich), and 1% penicillin/streptomycin (Sigma-Aldrich). Cultures were maintained at 37 °C and 4% CO2 atmosphere.
Tumor Xenograft.
The animal use protocol (protocol no. 2000-7675) was approved by the University of Toronto Animal Care Committee and met the requirements of animal use legislation in Ontario. MDA-MB-435 growth medium was replaced 24 h before seeding tumors. Cells were removed from culture flasks using a cell scraper, concentrated by centrifugation, and quantified using a hemocytometer. Cells were resuspended in conditioned media and Matrigel (BD Biosciences) at a 1∶1 ratio. A 200 μL volume containing 5 × 106 cells was injected subcutaneously onto the back of 6–8 week old CD1 athymic nude mice (Charles River Laboratories). Tumor growth was monitored until the mass reached 0.75 cm in length in any direction. Tumor-bearing mice were randomized before being used in either the biodistribution study or for imaging. GNP and streptavidin-Alexa fluor was administered to the mice in saline by intravenous injection of 150 μL into the tail vein. Mice were euthanized by CO2 asphyxiation, followed by cervical dislocation, or cervical dislocation while anesthetized. Tissues harvested for analysis were rinsed with PBS and blotted dry on clean filter paper.
Biodistribution Study and ICP-MS.
Mice bearing MDA-MB-435 tumors were injected with a single bolus of 1 × 1012 particles in 150 μL saline. At 6 and 24 HPI, mice were euthanized and tissue was collected for analysis. Two-hundred-fifty microliters of blood was obtained by cardiac puncture with a 25 gauge needle tip and 1 mL syringe. Organs were rinsed with PBS, blotted dry on clean filter paper, and weighed before storing at -20 °C until digestion. Organ samples were then digested in 10% HCl (Fluka), and Au content was quantified with ICP-MS to determine the total Au in the original organ.
Förster Resonance Energy Transfer.
To produce FRET on biotin-GNP, we used a variant of the pegylated GNP synthesis method described above. We synthesized 24 nm GNP using the citrate method, stabilized these with 500 biotin-PEG (Mr = 10,000 g/mol) per nanoparticle, then saturated the remaining surface area with mPEG-SH (Mr = 10,000 g/mol). These particles were found to produce a high energy transfer between the streptavidin-Alexa fluor 568 and 750 conjugates. After synthesis, the biotin-GNP were washed five times with PBS + 0.05% Tween20 before in vitro and in vivo experiments.
For in vitro characterization, we determined the concentration of the biotin-GNP, then prepared solutions of the particles and each of the fluorophore conjugates. These were then mixed with biotin-GNP either individually, to produce GNP-A568 and GNP-A750, or simultaneously, to produce GNP-A568/750. The products were then loaded into wells of a 0.7% agarose gel and separated by electrophoresis for 45 min at 100 V. The gel was then imaged on a Kodak In Vivo Multispectral Imaging System (Carestream Health) with the following filter combinations: A, excitation at 570 and emission 600; B, excitation 750 and emission 790; and C, excitation 570 emission 790. Fluorescence data were acquired for 5 min on a 4 megapixel camera, using 4 × 4 binning. Sixteen-bit images were then exported for processing in ImageJ. We performed a background subtraction, then applied a color table to each image before producing a montage of the three channels.
For in vivo measurements of FRET in harvested tissue, mice were injected with 1 × 1012 biotin-GNP 24 h before injection of fluorophores. A solution of the two streptavidin-Alexa fluor conjugates was prepared in saline, with a 1∶1 molar ratio and each at 2 μM. One-hundred-fifty microliters were then injected intravenously by tail vein. After a 2-h incubation, the animals were killed by cervical dislocation while anesthetized, a blood sample was collected by cardiac puncture, and organs were harvested. These were imaged on the Kodak system using the same settings described above for the in vitro analysis. The 16-bit images were exported and analyzed in ImageJ identically to gel images.
Imaging Data Collection and Analysis.
For the biotin-GNP and mPEG-GNP treatment groups, mice were injected with 1 × 1012 GNP 24 h prior to injection with streptavidin-Alexa fluor 750 (Invitrogen). Mice in the preassembled GNP-A750 group were injected with assembled construct immediately prior to imaging. Competitive inhibition controls were injected with 150 μL of a 5 uM bolus of streptavidin 30 min before injection of streptavidin-Alexa fluor 750. In all cases, imaging commenced as soon as possible (within 5 min). Mice were anesthetized with 5% isofluorane, transferred to a Kodak In Vivo Multispectral Imaging System (Carestream Health), and maintained with 3.5% isofluorane. Ten image stacks were acquired using Kodak Molecular Imaging Software version 5.0.0.86, by excitation at 710, 720, 730, 750, and 770 nm and emission at 830 nm. Data were acquired for 2 min at each filter combination, approximately 11 min per image stack, on a 4 megapixel camera with 4 × 4 binning and applying a field normalizer. After completion of imaging experiments, the animals were killed by cervical dislocation while anesthetized, a blood sample was obtained by cardiac puncture, and organs were harvested for imaging using the same settings as for in vivo imaging. Organ fluorescence data were expressed as a percent of the total fluorescence measured in all tissues, normalizing the signal in the volume of blood measured (50 uL) to total blood volume in a mouse (assumed to be 2 mL).
Image stacks were spectrally unmixed using Kodak Multispectral software version 1.1 (Carestream Health). Unmixed 16-bit images were then background-subtracted in ImageJ using a Rolling Ball radius of 150 pixels (50 for images of harvested organs). Image intensity data were then obtained by ROI analysis, tracing either the entire animal minus the tumor, the tumor outline on the animal, or the outer edge of the harvested organs using the “Freehand Selection” tool, and determining the mean intensity. Color tables were applied to mouse (“Smart”) and organ (“Red Hot”) images in ImageJ, and brightness was calibrated appropriately for presentation.
Histology, Silver Enhancement, and Fluorescence Microscopy.
Histological samples obtained after animal euthanization were immediately fixed in 10% buffered formalin (Sigma-Aldrich). Sample processing, mounting, and hematoxylin staining were carried out by the University Health Network laboratories using standard procedures. Silver enhancement was carried out on dewaxed mounted sections prior to hematoxylin staining, using the Silver Enhancement Kit for Light and Electron Microscopy (Ted Pella). Tissue sections were rinsed with double-distilled (dd) H2O, an equal volume of the kit’s two reagents were mixed in a microcentrifuge tube, and 50 μL was deposited on top of the section. Enhancement was allowed to proceed for 20–30 min before being stopped by rinsing with dd H2O.
Fluorescence histology images were obtained on an Olympus IX51 (Olympus) with excitation by a X-Cite120 light source (Exfo), and collection by a Retiga EXi CCD camera (QImaging). Brightfield color histology images were collected on a Axiovert 135 (Zeiss) and Coolsnap-Procf camera (Media Cybernetics).
Statistics.
Values of percent injected dose for biodistribution of GNP were compared between 6 h (n = 3) and 24 h (n = 3) after injection using a two-way Student’s t-test. An increase in signal from tumors of biotin-GNP treated mice were compared against mPEG-GNP controls (n = 3) using a two-way Student t test. Differences in contrast agent biodistribution within harvested organs 2 HPI were tested by ANOVA and post hoc Dunnet’s test, comparing tissues from in vivo assembly group animals against the three controls (n = 3). Differences in tissues harvested at 24 HPI were examined with a two-way Student t test. Signal-over-noise was calculated by [(target ROI—nontarget ROI)/(standard deviation of nontarget ROI)]. We tested for a higher signal-over-noise and larger area-under-the-curve from in vivo assembly vs. GNP-A750 passive targeting nanoparticles using a two-way Student t test (n = 3).
Supplementary Material
Acknowledgments.
The authors thank Kelvin So at the University Health Network, Toronto for his technical support, and Dr. Vladimir Baranov, University of Toronto, for providing tissue processing and ICP-MS services. This study was funded by the Canadian Institutes of Health Research (MOP:193110; RMF-7255), the Natural Sciences and Engineering Research Council of Canada (RGPIN 288231-09), the Canadian Foundation for Innovation, and the Ontario Innovation Trust Fund. S.D.P. acknowledges the National Sciences and Engineering Research Council of Canada (Postgraduate Scholarship D) and the Ontario Ministry of Training, Colleges, and Universities (Ontario Graduate Scholarship) for support through graduate student research scholarships.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001367107/-/DCSupplemental.
References
- 1.Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature. 2008;452:580–589. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt GP, Reiser MF, Baur-Melnyk A. Whole-body MRI for the staging and follow-up of patients with metastasis. Eur J Radiol. 2009;70:393–400. doi: 10.1016/j.ejrad.2009.03.045. [DOI] [PubMed] [Google Scholar]
- 3.Davis ME, Chen Z, Shin DM. Nanoparticle therapeutics: An emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 4.Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WCW. Mediating tumor targeting efficiency of nanoparticles through design. Nano Lett. 2009;9:1909–1915. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- 5.Larsen EK, et al. Size-dependent accumulation of PEGylated silane-coated magnetic iron oxide nanoparticles in murine tumors. ACS Nano. 2009;3:1947–1951. doi: 10.1021/nn900330m. [DOI] [PubMed] [Google Scholar]
- 6.Yuan F, et al. Vascular-permeability in a human tumor xenograft—molecular-size dependence and cutoff size. Cancer Res. 1995;55:3752–3756. [PubMed] [Google Scholar]
- 7.Fang C, et al. In vivo tumor targeting of tumor necrosis factor-alpha-loaded stealth nanoparticles: Effect of MePEG molecular weight and particle size. Eur J Pharm Sci. 2006;27:27–36. doi: 10.1016/j.ejps.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Jain RK. Transport of molecules, particles, and cells in solid tumors. Annu Rev Biomed Eng. 1999;1:241–263. doi: 10.1146/annurev.bioeng.1.1.241. [DOI] [PubMed] [Google Scholar]
- 9.Hobbs SK, et al. Regulation of transport pathways in tumor vessels: Role of tumor type and microenvironment. Proc Natl Acad Sci USA. 1998;95:4607–4612. doi: 10.1073/pnas.95.8.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayyagari AL, et al. Long-circulating liposomal contrast agents for magnetic resonance imaging. Magn Reson Med. 2006;55:1023–1029. doi: 10.1002/mrm.20846. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, et al. RGD peptide functionalized and reconstituted high-density lipoprotein nanoparticles as a versatile and multimodal tumor targeting molecular imaging probe. FASEB J. 2010 doi: 10.1096/fj.09-139865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao XH, et al. In vivo molecular and cellular imaging with quantum dots. Curr Opin Biotechnol. 2005;16:63–72. doi: 10.1016/j.copbio.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Akerman ME, Chan WCW, Laakkonen P, Bhatia SN, Ruoslahti E. Nanocrystal targeting in vivo. Proc Natl Acad Sci USA. 2002;99:12617–12621. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park J, et al. Cooperative nanoparticles for tumor detection and photothermally triggered drug delivery. Adv Mater. 2010;22:880–885. doi: 10.1002/adma.200902895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park J, et al. Cooperative nanomaterial system to sensitize, target, and treat tumors. Proc Natl Acad Sci USA. 2010;107:981–986. doi: 10.1073/pnas.0909565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simberg D, et al. Biomimetic amplification of nanoparticle homing to tumors. Proc Natl Acad Sci USA. 2007;104:932–936. doi: 10.1073/pnas.0610298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decuzzi P, Pasqualini R, Arap W, Ferrari M. Intravascular delivery of particulate systems: Does geometry really matter? Pharm Res. 2009;26:235–243. doi: 10.1007/s11095-008-9697-x. [DOI] [PubMed] [Google Scholar]
- 18.Tam F, Goodrich GP, Johnson BR, Halas NJ. Plasmonic enhancement of molecular fluorescence. Nano Lett. 2007;7:496–501. doi: 10.1021/nl062901x. [DOI] [PubMed] [Google Scholar]
- 19.Cheng D, Xu Q. Separation distance dependent fluorescence enhancement of fluorescein isothiocyanate by silver nanoparticles. Chem Commun. 2007:248–250. doi: 10.1039/b612401a. [DOI] [PubMed] [Google Scholar]
- 20.Chou LYT, Fischer HC, Perrault SD, Chan WCW. Visualizing quantum dots in biological samples using silver staining. Anal Chem. 2009;81:4560–4565. doi: 10.1021/ac900344a. [DOI] [PubMed] [Google Scholar]
- 21.Pimm MV, Fells HF, Perkins AC, Baldwin RW. I-131 and in-111 labeled avidin and streptavidin for pre-targeted immunoscintigraphy with biotinylated anti-tumor monoclonal-antibody. Nucl Med Commun. 1988;9:931–941. doi: 10.1097/00006231-198811000-00010. [DOI] [PubMed] [Google Scholar]
- 22.Vanosdol WW, Sung C, Dedrick RL, Weinstein JN. A distributed pharmacokinetic model of 2-step imaging and treatment protocols—application to streptavidin-conjugated monoclonal-antibodies and radiolabeled biotin. J Nucl Med. 1993;34:1552–1564. [PubMed] [Google Scholar]
- 23.Saga T, et al. 2-step targeting of experimental lung metastases with biotinylated antibody and radiolabeled streptavidin. Cancer Res. 1994;54:2160–2165. [PubMed] [Google Scholar]
- 24.AlvarezDiez TM, Polihronis J, Reilly RM. Pretargeted tumor imaging with streptavidin immunoconjugates of monoclonal antibody CC49 and in-111-DTPA-biocytin. Nucl Med Biol. 1996;23:459–466. doi: 10.1016/0969-8051(96)00022-4. [DOI] [PubMed] [Google Scholar]
- 25.Paganelli G, et al. Antibody-guided three-step therapy for high grade glioma with yttrium-90 biotin. Eur J Nucl Med. 1999;26:348–357. doi: 10.1007/s002590050397. [DOI] [PubMed] [Google Scholar]
- 26.Sakahara H, Saga T. Avidin-biotin system for delivery of diagnostic agents. Adv Drug Deliver Rev. 1999;37:89–101. doi: 10.1016/s0169-409x(98)00101-x. [DOI] [PubMed] [Google Scholar]
- 27.Ntziachristos V, Ripoll J, Wang LHV, Weissleder R. Looking and listening to light: The evolution of whole-body photonic imaging. Nat Biotechnol. 2005;23:313–320. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]
- 28.Gao XH, Cui YY, Levenson RM, Chung LWK, Nie SM. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 29.Choi HS, et al. Design considerations for tumor-targeted nanoparticles. Nat Nanotechnol. 2009;5:42–47. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruoslahti E, Bhatia SN, Sailor MJ. Targeting of drugs and nanoparticles to tumors. J Cell Biol. 2010;188:759–768. doi: 10.1083/jcb.200910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frens G. Controlled nucleation for regulation of particle-size in monodisperse gold suspensions. Nat Phys Sci. 1973;241:20–22. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.