Abstract
An orchestrated balance of pro- and antiinflammatory cytokine release is critical for an innate immune response sufficient for pathogen defense without excessive detriment to host tissues. By using an unbiased forward genetic approach, we previously reported that IL-1R–associated kinase 1 binding protein 1 (IRAK1BP1) down-modulates Toll-like receptor-mediated transcription of several proinflammatory cytokines. To gain insights into the physiological relevance of the inhibitory role of IRAK1BP1 in inflammation, we generated mutant mice lacking IRAK1BP1. Here we report that IRAK1BP1 does not inhibit signaling pathways generally but rather changes the transcriptional profile of activated cells, leading to an increase in IL-10 production and promoting LPS tolerance. This shift in cytokine transcription correlates with an increased ratio of functional NF-κB subunit dimers comprised of p50/p50 homodimers relative to p50/p65 heterodimers. The increase in nuclear p50/p50 was consistent with the ability of IRAK1BP1 to bind to the p50 precursor molecule and IκB family member p105. We conclude that IRAK1BP1 functions through its effects on NF-κB as a molecular switch to bias innate immune pathways toward the resolution of inflammation.
Keywords: inhibitor, innate immunity, p105, Toll-like receptor, tolerance
An effective host defense against fulminant pathogen dissemination and subsequent disease relies not only on the ability of the innate immune system to mount a robust and complete acute inflammatory response but also on control of these responses to avoid tissue injury and chronic inflammation (1–3). Thus, the necessary role of inflammation in resolution or containment of infections must be balanced carefully against its potent ability to induce immune-mediated disease.
Most of our knowledge of the molecular components regulating acute inflammation has arisen from studies of the Toll-like receptor (TLR), IL-1 receptor (IL-1R), and TNF receptor (TNFR) pathways. It is clear from these studies that the innate immune response is controlled at multiple levels within the cell, from receptor proximal events, such as the binding of adaptor proteins and activation of upstream kinases, to posttranslational events, such as processing and release of effector proteins (4). Furthermore, various cytokines and chemokines can exert antagonistic effects on target cells (5). For example, IL-1R antagonist has been shown to inhibit directly the receptor interaction of IL-1 and IL-1R (6, 7). The potent antiinflammatory molecule IL-10 exerts numerous effects, including the feedback inhibition of TLR-stimulated cytokine production in macrophages as well as the expansion of T-regulatory cells (8). As such, activated cells of the innate immune system can dictate the course of an inflammatory response through the levels of pro- and antiinflammatory cytokines they secrete.
The unique transcriptional responsiveness of individual genes is one mechanism by which the activation of common upstream pathways results in temporally coordinated and signal-specific cytokine production. For example, “first-wave” genes such as TNF-α have constitutively accessible promoter regions and are transcribed immediately upon activation, whereas “second-wave” genes such as IL-6 require chromatin remodeling and new protein synthesis for transcription (9, 10). This difference leads to distinct temporal patterns of TNF-α and IL-6 secretion. Chromatin structure also plays a role in LPS tolerance, such that recent TLR4 stimulation results in a subset of inflammatory genes becoming inaccessible upon reactivation and a new class of genes being primed for increased levels of transcription (11).
In addition to differences in chromatin accessibility, individual genes are optimally responsive to specific transcription factors, including the ubiquitous inflammatory regulator NF-κB. The NF-κB family is comprised of five proteins containing a Rel-homology domain: p65 (RelA), RelB, c-Rel, p50, and p52, which bind to target promoters as dimers (12). Three of these subunits—p65, RelB, and c-Rel—contain a transactivation domain (TD) and strongly activate transcription, either as homodimers or, more commonly, as heterodimers with a non-TD–containing subunit [e.g., p65/p50 (13)]. Homodimers of p50 molecules form readily and are thought to compete for promoter binding sites of inflammatory genes, where they block the binding of TD-containing dimers and consequently inhibit transcription (14). Although p50/p50 homodimers have been associated with an inhibitory effect on the transcription of inflammatory cytokines such as TNF-α, IL-12-p40, and IL-6, they also increase the production of the antiinflammatory cytokine IL-10 (15, 16). This differential effect has been linked to the ability of p50/p50 homodimers to recruit additional factors, including cAMP response element-binding (CREB)-binding protein (CBP), as well as promoter-specific preferences for different NF-κB dimers. The IL-10 promoter, in contrast to that of many proinflammatory genes, exhibits a strong preference for exclusively binding p50/p50 homodimers, which contribute to IL-10 transcription by complexing with CBP (15). Although the effect of various NF-κB complexes on the expression of pro- and antiinflammatory processes has been demonstrated using in vitro and gene-targeting studies, the extent to which naturally occurring cellular processes manipulate this system to regulate the immune response is not known.
We previously demonstrated that IL-1R–associated kinase 1 binding protein 1 (IRAK1BP1) is an inhibitory component of TLR-, IL-1–, and TNFR-mediated pathways (17). We now confirm the antiinflammatory role of IRAK1BP1 in physiologically relevant conditions and show that IRAK1BP1 promotes up-regulation of IL-10 in macrophages. We demonstrate that this effect is associated with IRAK1BP1 promoting nuclear translocation of inhibitory p50/p50 homodimers relative to p50/p65 heterodimers. As a result of this IRAK1BP1-dependent effect, the p50/p50 homodimers down-regulate transcription of proinflammatory cytokines and contribute to the state of LPS-induced tolerance. Thus, IRAK1BP1 modulates the balance of inflammatory responses and LPS-tolerance by affecting the relative ratio of endogenous NF-κB subunits.
Results
Increased IL-6 Production in IRAK1BP1-Knockout Mouse Cells.
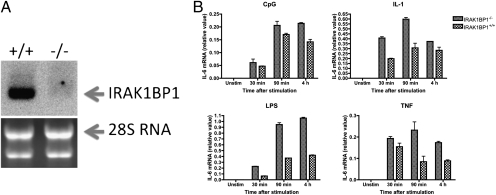
We previously reported that IRAK1BP1 down-regulates TLR-mediated transcription of several proinflammatory cytokines. To gain further insights into the inhibitory role of IRAK1BP1 in inflammation, we generated IRAK1BP1-knockout mice. A search of the Omni Bank database (http://www.lexgen.com) of ES clones with retrovirus insertions identified a clone containing a retroviral integration of a splice acceptor site fused to neomycin resistance cassette and a poly-A sequence between the first and the second exons of mouse IRAK1BP1 (Fig. S1). Using these ES cells, we obtained mice carrying the disrupted IRAK1BP1 gene on a C57BL/6Jx129SvEv genetic background. This “gene-trapped” allele resulted in a complete loss of IRAK1BP1 mRNA in mouse embryonic fibroblasts (MEFs) as measured by Northern blot analysis (Fig. 1A).
Fig. 1.
Increased IL-6 expression in IRAK1BP1 knockout mouse cells. (A) Northern blot analysis of IRAK1BP1 expression in knockout and wild-type fibroblasts. Ribosomal 28S RNA was used as loading control. (B) Embryonic fibroblasts from newly isolated IRAK1BP1-deficient or wild-type control embryos were activated with LPS (100 ng/mL), CpG (200 nm), IL-1 (20 ng/mL), and TNF (10 ng/mL) for different times followed by isolation of total RNA and quantitative PCR analysis of IL-6 mRNA accumulation. Data are representative of one (A) or three (B) independent experiments.
MEFs have constitutively high levels of IRAK1BP1 (17). Accordingly, we compared levels of up-regulation of IL-6 mRNA in MEFs from IRAK1BP1−/− embryos and littermate controls in response to TNF, IL-1, and TLR agonists (Fig. 1B). Stimulation with LPS, synthetic CpG-oligonucleotides (CpG), IL-1β, or TNF-α resulted in increased IL-6 mRNA accumulation in MEFs from IRAK1BP1−/− embryos relative to those from wild-type embryos (Fig. 1B). These data suggest that the effect of IRAK1BP1 occurs through a general inhibitory role in inflammatory gene transcription rather than acting at the level of receptor-proximal signaling events.
IRAK1BP1 Independently Activates IL-10 and Inhibits IL-6 Transcription.
Although we previously demonstrated an inhibitory role for IRAK1BP1 in the transcription of several proinflammatory cytokines from their endogenous loci following TLR, IL-1R, and TNF-R stimulation (17), another report suggested that overexpression of IRAK1BP1 increased activation of NF-κB reporter plasmids following TNF-α stimulation (18). To broaden the list of target genes for IRAK1BP1, we extended our analysis to include the antiinflammatory cytokine IL-10. The effect of IRAK1BP1 on IL-10 was analyzed first in RAW264.7 cells, because these macrophages, like MEFs, have a constitutively high level of IRAK1BP1.
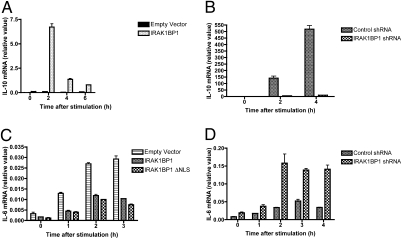
Quite surprisingly, overexpression of IRAK1BP1 in the murine macrophage cell line RAW264.7 led to a greater than 10-fold increase in IL-10 mRNA following stimulation with the TLR2 agonist lipoteichoic acid (LTA) (Fig. 2A). Conversely, cells stably expressing an IRAK1BP1-targeting shRNA construct were severely deficient in their ability to produce IL-10 mRNA in response to the TLR4 agonist LPS (Fig. 2B). A similar effect of the silencing of IRAK1BP1 on IL-10 levels also was observed for LTA-stimulated macrophages.
Fig. 2.
Differential regulation of cytokine transcription by IRAK1BP1. (A) RAW264.7 macrophages were lentivirally transduced with IRAK1BP1 or an empty vector control and stimulated with 2 μg/mL LTA. IL-10 mRNA was measured by real-time PCR. (B) RAW264.7 macrophages stably expressing IRAK1BP1-specific or control (nonhomologous) shRNA constructs (17) were stimulated with 100 ng/mL LPS. IL-10 mRNA was measured by real-time PCR. (C) L929 cells were lentivirally transduced with empty vector, IRAK1BP1, or an IRAK1BP1 mutant deficient in the putative nuclear localization sequence (IRAK1BP1 ΔNLS). Cells were stimulated with 100 ng/mL TNFα, and IL-6 mRNA was measured by real-time PCR. (D) L929 cells stably expressing the lentiviral constructs used in B were stimulated with 100 ng/mL IL-1β, and IL-6 mRNA was measured by real-time PCR. Error bars are ± range of duplicate wells. All data are representative of at least three independent experiments.
IL-10 has been shown to exert an autocrine effect on macrophages, impairing their ability to respond to TLR stimulation (19, 20). The ability of IRAK1BP1 to promote an increase in IL-10 mRNA raised the possibility that its inhibition of proinflammatory gene transcription occurs secondarily through an autocrine loop. To test whether the inhibition of IL-6 transcription by IRAK1BP1 relies on IL-10 secretion, we assessed its effect in the L929 murine fibroblast cell line, which is responsive to IL-1 and TNF-α stimulation but does not produce IL-10 (21). As shown in Fig. 2C and D, IL-6 mRNA produced by stimulating L929 cells with TNF-α or IL-1 was sensitive to IRAK1BP1 overexpression or knockdown, respectively. To dismiss further the autocrine effect of IL-10 on IL-6 expression in activated macrophages, we compared levels of IL-6 mRNA in the presence of neutralizing IL-10 or control antibodies. As anticipated, we did not observe that blocking the IL-10 receptor pathway had any noticeable effect of on IL-6 expression (Fig. S2). Thus, IRAK1BP1 inhibits IL-6 production independently from its ability to promote IL-10 synthesis.
Because previous work suggested that a putative nuclear localization sequence (NLS) was involved in the effect of IRAK1BP1 (22), we also transduced L929 cells with a construct coding for a truncated IRAK1BP1 that lacks the NLS. Overexpression of this mutant was equally capable of inhibiting TNF-α–stimulated IL-6 production (Fig. 2C), suggesting that the NLS of IRAK1BP1 is not required for its antiinflammatory effects.
IRAK1BP1 Promotes LPS Tolerance at Specific Cytokine Loci.
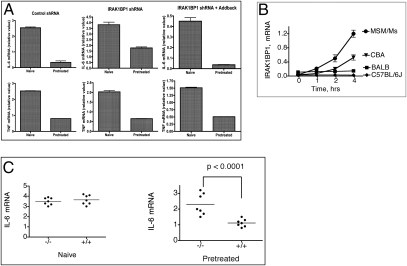
One of the important biological phenomena mechanistically linked to the independent and opposite effects on accumulation of IL-6 and IL-10 mRNAs is the state of LPS tolerance (23). To assess the effect of IRAK1BP1 on tolerance to LPS, we first silenced endogenous IRAK1BP1 in RAW macrophages and also generated an additional cell line expressing IRAK1BP1 with three silent mutations in the shRNA binding site as a control for specificity of the hairpin (Fig. S3). Next, we repeatedly activated with LPS both types of cells as well as control cells (Fig. 3A). Although pretreatment with LPS resulted in a greater than 5-fold reduction in IL-6 mRNA upon subsequent stimulation in control cells, IRAK1BP1 knockdown cells exhibited a reduction of only approximately 2-fold. Add-back of IRAK1BP1 to the knockdown cell line restored tolerance as measured by IL-6 production to the magnitude observed in control cells (Fig. 3A).
Fig. 3.
IRAK1BP1 promotes LPS-induced tolerance for IL-6 but not TNF-α transcription. (A) RAW264.7 cells stably expressing the indicated shRNA and IRAK1BP1 add-back constructs were pretreated with 100 ng/mL LPS for 18 h. After 1 h of recovery, cells were restimulated along with naïve cells with 100 ng/mL LPS for 4 h. IL-6 and TNF-α mRNA were measured using real-time PCR. (B) Peritoneal macrophages from different mouse strains were activated with LPS (10 ng/mL) and analyzed for IRAK1BP1 mRNA levels (relative units) by means of qualitative PCR. (C) Mice with heterozygosity for the targeted allele of Irak1bp were outcrossed to CBA mice for three generations and then intercrossed. Peritoneal macrophages from the resultant N3F1 progeny were pretreated with LPS for 18 h, allowed to recover, and treated again, along with naïve cells, with LPS for 2 h. IL-6 mRNA was measured using real-time PCR. Mice were categorized according to their haplotype for the Irak1bp1-knockout allele. Data are representative of three (A) or two (B and C) independent experiments.
To extend our studies to primary macrophages and to identify an Irak1bp1 allele that is responsive to TLR stimulation (17), we examined several mouse strains and found that the CBA allele of Irak1bp1 is responsive to activation by TLRs (Fig. 3B). Therefore, to examine the effect of IRAK1BP1 knockout in primary mouse macrophages, we outcrossed the knockout allele to the CBA strain for three generations. Mice heterozygous for the knockout allele were intercrossed, and littermates were examined in an LPS-tolerance test with respect to their haplotype for the Irak1bp1 allele. Fig. 3C shows that lower levels of IL-6 mRNA after prestimulation with LPS were significantly associated with the functional allele of Irak1bp1, a finding consistent with the role of IRAK1BP1 in establishing the state of tolerance in primary macrophages.
Interestingly, for both RAW cells (Fig. 3A) and primary macrophages (17), the effect of IRAK1BP1 on TNF mRNA was negligible, suggesting that tolerance at the level of transcription may be under unique control for individual cytokines as well in addition to the more broadly acting mechanisms such as receptor and signaling intermediate down-modulation. Furthermore, these findings suggest that, in addition to regulating primary responses, IRAK1BP1 may have an important role in the resolution phase of inflammation, promoting IL-10 production and preventing the subsequent release of proinflammatory mediators such as IL-6 in the hours and days after an acute response.
IRAK1BP1 Affects the Ratio of Nuclear NF-κB Subunits Downstream of Early Signaling Events.
One of several mechanisms involved in the complex cellular phenomenon of LPS tolerance (23, 24) is an antiinflammatory bias in NF-κB subunits resulting from nuclear accumulation of p50/p50 homodimers.
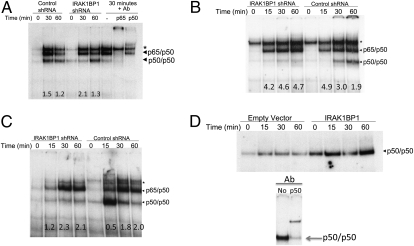
Accordingly, we assessed the effect of IRAK1BP1 knockdown on the TLR-stimulated nuclear translocation and DNA-binding ability of NF-κB. As shown in Fig. 4A, knockdown of IRAK1BP1 in RAW cells led to a delayed induction of p50/p50 homodimer binding despite a similar amount of active p65/p50 in the nucleus after 30 min of LTA stimulation. To assess whether IRAK1BP1 exerted a similar effect on NF-κB translocation resulting from other inflammatory pathways such as IL-1R and TNFR pathways, we performed a similar comparison in L929 fibroblasts stably expressing the same knockdown and in control shRNA constructs. As shown in Fig. 4 B and C, cells deficient in IRAK1BP1 exhibited a delayed and reduced accumulation of functional p50/p50 homodimers relative to p65/p50 molecules in the nucleus after stimulation with these acute-phase cytokines. Thus, physiologic levels of IRAK1BP1 are required in macrophages and fibroblasts to achieve normal ratios of nuclear NF-κB dimers.
Fig. 4.
IRAK1BP1 affects the ratio of p50/p50 to p65/p50. (A) RAW264.7 cells stably expressing the indicated shRNA constructs were stimulated with 2 μg/mL LTA, and the DNA binding activity of the indicated NFκB dimers was assessed using EMSA with a consensus probe. Identification of bands was performed using supershift of the IRAK1BP1 shRNA at the 30-min time point with anti-p50 and anti-p65 antibodies (Right). Values at the bottom represent the ratio of p65/p50 to p50/p50. *, nonspecific band. (B and C) L929 cells stably expressing the shRNA constructs shown were stimulated with 100 ng/mg IL-1β (B) or TNFα (C), and NFκB binding was assessed as in B. Values at the bottom represent the ratio of p65/p50 to p50/p50. (D) RAW264.7 cells expressing IRAK1BP1 or empty vector controls were stimulated with 2 μg/mL LTA. Nuclear extracts were prepared and used for EMSA analysis with an NFκB consensus (A) or an IL-10 promoter (B; position −59/−39) oligonucleotide probe. Arrow indicates the position of p50/p50 homodimers that are shifted with p50-specific antibodies. Data are representative of three (A) or two (B–D) independent experiments.
The IL-10 promoter has been shown to bind p50/p50 molecules with high specificity, leading to activation of IL-10 transcription through recruitment of CBP (15). Given the observed role of IRAK1BP1 in promoting IL-10 production (Fig. 2 A and B), we reasoned that it would induce a corresponding increase in NF-κB molecules bound to the promoter. We used an oligonucleotide probe corresponding to position −59/−39 relative to the IL-10 transcription start site, which has been implicated previously in NF-κB binding (15). As expected, overexpression of IRAK1BP1 correlated with an increased amount of NF-κB binding to the IL-10 promoter-specific probe (Fig. 4D).
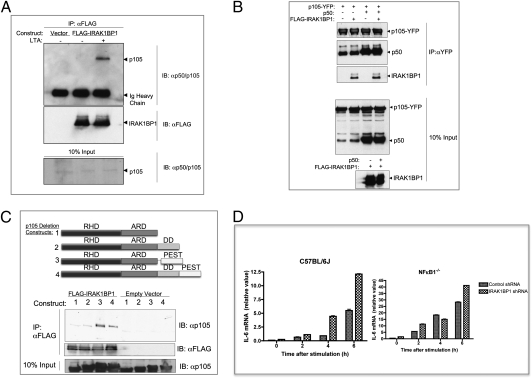
IRAK1BP1 Associates with and Stabilizes p105.
To characterize the upstream events associated with the increase in nuclear p50/p50 molecules, we assessed the ability of IRAK1BP1 to bind NF-κB family proteins. As shown in Fig. 5A, coimmunoprecipitation experiments demonstrated a stimulus-enhanced association between IRAK1BP1 and p105 in RAW macrophages. p105 plays a complex and central role in the synthesis and function of NF-κB complexes containing p50. In addition to the IκB function of the p105 C terminus, p105 is constitutively processed by the 20S proteasome to generate p50 molecules from its N terminus (25). Thus, the association of IRAK1BP1 with p105 places IRAK1BP1 at a major site of p50 regulation. Furthermore, p105 exhibits strong cytoplasmic localization and thus is a plausible target, given that IRAK1BP1 with a deletion in the putative NLS motif retains the ability to inhibit inflammatory gene transcription (Fig. 2C).
Fig. 5.
Characterization and functional outcome of IRAK1BP1’s association with p105. (A) RAW264.7 cells stably expressing FLAG-tagged IRAK1BP1 or empty vector controls were left untreated or stimulated for 30 min with 2 μg/mL LTA. Cytoplasmic protein extracts were prepared and used for coimmunoprecipitation with M2 anti-FLAG antibodies. Immunoprecipitation products were resolved on an SDS/PAGE gel and used for Western blot analysis with the indicated antibodies. (B) 293T cells stably expressing FLAG-tagged IRAK1BP1 or empty vector controls were transfected with a construct encoding p105 fused to YFP at the C terminus to avoid labeling of processed p50 and, where indicated, were transfected with another plasmid encoding unlabeled p50 (residues 1–435 in p105). After cell lysis, anti-YFP antibodies were used for immunoprecipitation of p105. Bound p50 and IRAK1BP1 were assessed using Western blotting with anti-p105/p50 or M2 anti-FLAG antibodies, respectively. (C) Plasmids coding for p105 with deletions in the domains shown or wild-type p105 (construct 4) were transfected into 293T cells expressing FLAG-tagged IRAK1BP1 or empty vector. Immunoprecipitation with anti-FLAG M2 antibodies was followed by Western blot analysis for bound p105. (D) MEFs were prepared from wild-type and NFκB1−/− mice (both from the C57BL/6J background) and infected with lentiviral constructs expressing control or IRAK1BP1-targeting shRNA. After stably selecting transducants, MEFs were stimulated with 2 μg/mL LTA for the indicated times, and IL-6 mRNA was assessed using real-time PCR. Error bars represent ± range from duplicate wells. Representative data from three independent experiments are shown.
To test the possibility that IRAK1BP1 competes directly with p50 for binding with p105, we introduced C-terminally YFP-labeled p105 into 293T cells along with IRAK1BP1 or empty vector controls. Immunoprecipitation using anti-YFP antibodies allowed the assessment of bound p50 and IRAK1BP1 molecules. As shown in Fig. 5B, overexpression of IRAK1BP1 was not sufficient to displace endogenous p50 molecules from p105. To assess further the possibility that IRAK1BP1 and p50 compete directly for binding to p105, we introduced ectopically expressed p50 into the same cells. Although the addition of plasmid-derived p50 did result in a greater occupancy of p105 molecules, IRAK1BP1 binding was not affected. Thus, the relative associations of IRAK1BP1 and p50 with p105 are independent of each other's concentrations.
Upon stimulation, p105 is phosphorylated in the C-terminal PEST domain by IκB kinase with subsequent ubiquitination and proteolysis by the β-TrCP (SCF-type ubiquitin E3 ligase) complex (26, 27). We assessed the role of these two regulatory domains in the association with IRAK1BP1 using the p105 deletion mutants shown in Fig. 5C. These experiments demonstrated that the PEST domain is essential for optimal IRAK1BP1 association. By contrast, the death domain was not required for binding. Rather, deletion of this region consistently enhanced the effect, suggesting that it may negatively regulate the association of IRAK1BP1 with p105.
Because the PEST and death domain regions of p105 affected its ability to interact with IRAK1BP1, we reasoned that the association of these proteins may have functional consequences for stimulus-induced p105 phosphorylation and/or degradation. Accordingly, we introduced YFP-labeled p105 into L929 fibroblasts stably expressing IRAK1BP1 or an empty vector control for pulse chase analysis. We observed an expected degradation of labeled p105 protein in control cells. Overexpression of IRAK1BP1, however, was sufficient to prevent any observable p105 degradation up to 150 min after IL-1β stimulation (Fig. S4). IRAK1BP1 also was stable over this time course. Collectively, these data demonstrate that IRAK1BP1 associates with regulatory regions of the p50 precursor and IκB family protein p105 and that overexpression of IRAK1BP1 prevents p105 degradation. By associating with p105, IRAK1BP is centrally positioned to affect the nuclear balance of NF-κB subunits from the cytoplasm.
The strong correlation between the levels of IRAK1BP1 and the ratio of p50/p50 to p65/p50 NF-κB molecules suggested that IRAK1BP1’s effect on cytokine production may depend heavily on the availability of particular NF-κB family proteins. If IRAK1BP1 exerts its effect through p105, then removal of p105 would eliminate the effect. We used a lentiviral shRNA construct to knock down IRAK1BP1 in MEFs from wild-type C57BL/6J and NFκB1−/− mice in which the p50 precursor and IκB family member p105 (NF-κB1) has been targeted by homologous recombination (28). As shown in Fig. 5D, depletion of IRAK1BP1 mRNA increased IL-6 production more than 2-fold in wild-type MEFs. Cells lacking NF-κB1, on the other hand, were less affected by the presence of IRAK1BP1. Although these results imply that IRAK1BP1 may have somewhat pleiotropic effects on inflammatory pathways, they demonstrate that NF-κB1 is part of the major axis by which its antiinflammatory outcome occurs.
Discussion
In this report, we extend our earlier forward genetic study demonstrating an antiinflammatory role for IRAK1BP1 and show that IRAK1BP1-deficient embryonic fibroblasts have higher responses to TLR agonists, IL-1, and TNF, thereby confirming that IRAK1BP1 exerts its antiinflammatory activity at a level common to all these pathways. In addition, we show that IRAK1BP1-deficient macrophages are significantly more responsive to a repeated activation with LPS, therefore suggesting that IRAK1BP1 not only is involved directly in down-regulation of inflammatory cytokines but also contributes to the state of LPS-induced tolerance. To reveal the inhibitory role of IRAK1BP1 in physiological conditions, it was important to identify mouse strains that carry an LPS-responsive allele of Irak1bp1. We previously found the Irak1bp1 allele to be responsive to activation of TLRs in wild-derived but not classical inbred mouse strains such as C57BL/6J, A/J, and DBA/J. In this study, we expanded the list of the strains and identified that macrophages from CBA mice were able to up-regulate IRAK1BP1 in response to TLR agonists. Accordingly, we looked at the effect of targeting the Irak1bp1 allele on the CBA background. These findings emphasize the importance of the mouse background in revealing gene function. They also show the importance of genetic studies in evolutionarily divergent mouse strains that developed unique and different gene networks when compared with classical inbred strains. Taken together, our findings provide a strong rationale for further investigation of innate immune responses in wild-derived mice.
Further investigation of the inhibitory effect of IRAK1BP1 showed that its overexpression in macrophages results in significantly more IL-10 in addition to decreased levels of IL-6. An independent effect on IL-6 production was observed in fibroblasts that do not produce detectable levels of IL-10, thus dismissing the possibility of IL-10-dependent down-regulation of IL-6. These results correlated with the ability of IRAK1BP1 to bind to the p50 precursor molecule p105 and induce a functional increase in the ratio of p50/p50 homodimers to p65/p50 heterodimers available for binding cytokine promoters.
The mechanism by which p50/p50 molecules differentially affect cytokine gene transcription has been well described (15). Our results expand upon the role of p50 homodimer-mediated NF-κB regulation to include interactions with the TLR-inducible protein IRAK1BP1 in macrophages. Although the mechanism by which IRAK1BP1 induces an increase in p50/p50 DNA binding remains to be elucidated, its ability to bind p105, along with the observed phenotypic activity of the NLS-deleted IRAK1BP1, offers some clues. These data indicate that IRAK1BP1 may induce a cytoplasmic modification leading to increased nuclear accumulation of p50, a bias in dimer formation, an increase in DNA-binding affinity, or a combination of these outcomes. The apparent stabilization of p105 in cells overexpressing IRAK1BP1 (Fig. S4) is counterintuitive—p105 degradation is thought to be required for nuclear translocation of its NF-κB substrates primarily consisting of p50/p50 homodimers (27). This stabilization may reflect a byproduct of other effects, such as posttranslational modifications of p50 molecules that are bound to p105, in which case p105 may serve an adaptor-like function, allowing IRAK1BP1 to access p50 molecules soon to be translocated to the nucleus. IRAK1BP1 may rely on a stable p105 scaffold to increase the prevalence of p50 homodimers over p65/p50 heterodimers. The requirement of the PEST region of p105 for optimal levels of IRAK1BP1 binding suggests that other proteins known to bind this domain in a stimulus-dependent manner, such as the β-TrCP complex, may be involved in the recruitment of IRAK1BP1, again possibly as a mechanism by which it contacts NF-κB subunits destined for nuclear translocation.
Unlike transformed cell lines, quiescent primary macrophages lack any detectable IRAK1BP1 transcript. However, here and in our previous report we showed that in some mouse strains, particularly those prone to robust early cytokine transcription, IRAK1BP1 is highly induced by 4 h after transcription (17). At this point, its ability to promote p50/p50 homodimer formation, leading to increased IL-10 production and inhibition of continued proinflammatory gene transcription, aids in dampening the burst of inflammatory mediators already released upon initial activation. Finally, its role in LPS tolerance assures that continued stimulation does not lead to unchecked cytokine release. In addition to its transcriptional profile in activated macrophages, previous work also has shown that IRAK1BP1 is constitutively expressed at high levels in the mouse brain and testis, two immune-privileged organs, suggesting that its expression also may be used to modulate the effect of inflammatory mediators such as IL-1 and TNF in specific tissues where their potentially harmful effects may outweigh their immune-activating benefits (18).
Inflammation is not a preprogrammed cascade of molecular events but rather is a dynamic process driven by constant feedback between the immune system, pathogens, and the surrounding host tissue. As such, our knowledge of how the innate immune response is regulated continues to incorporate a wider and more complex picture: Cytokine-producing cells undergo epigenetic and transcriptional reprogramming to produce specific effectors at various times, target cells modulate their own receptor and signaling pathways in response to unique stimuli, and cells of the adaptive immune system initiate new developmental fates based on their cytokine milieu. An antiinflammatory role for IRAK1BP1 fits perfectly in this scenario. By shifting the balance of NF-κB subunits available for promoter binding in both cytokine-producing and -sensing cell types, it changes the outcome of stimulation even as the cellular environment and upstream signaling processes remain active. Our increasing understanding of the initial events leading from the proinflammatory innate immune response to resolution of inflammation will shed light on the possible molecular pathology underlying diseases that result from prolonged or insufficiently sustained release of cytokines.
Experimental Procedures
Mice and Isolation of MEFs.
C57BL/6J and NFκB1−/− (B6.Cg-Nfkb1 < tm1Bal>/J) mice were purchased from Jackson Laboratories and maintained in a pathogen-free mouse facility at Tufts University School of Medicine with approval of the Tufts-New England Medical Center Institutional Animal Care and Use Committee. MEFs were isolated from 13.5-d mouse embryos.
Antibodies and Reagents.
Anti-FLAG M2 antibodies were from Sigma. Phospho and total ERK and glycogen synthase kinase antibodies and p65 and phospho p105 Ser933 antibodies were from Cell Signaling. Anti-total p105/p50 (SC114) was from Santa Cruz Biotechnology. Anti-YFP antibody (ab290-50) and anti-TBP (ab51841) were from Abcam. Purified LTA from Staphylococcus aureus was obtained from Invivogen, and Salmonella Minnesota Re595 LPS was purchased from Alexis. IL-1β was obtained from A. Ischenko (Research Institute of Highly Pure Biopreparates, St. Petersburg, Russia), and TNF-α from V. Kravchenko (The Scripps Research Institute, La Jolla, CA).
Plasmids and Cloning.
Silencing and overexpression of IRAK1BP1 were performed as described earlier (17). The constructs for N-terminally YFP-labeled p105 was from Dr. G. Gosh (UCSD, San Diego, CA), p50 from Dr. S. Smale (UCLA, Los Angeles, CA), and the p105 deletion mutants from Dr. S. Ley (National Institute of Medical Research, UK) have been described. C-terminally YFP-labeled p105 was cloned using PCR techniques into pYFP.c1 (a gift from S. Bunnell, Department of Pathology, Tufts, Boston, MA).
Generation of Cell Lines and Transfection.
Stably transduced shRNA and IRAK1BP1-overexpressing cell lines were generated using lentiviral infection that was performed as described earlier (17).
Real-Time PCR.
cDNA was made with 9-mer random primers and was analyzed by real-time PCR gene expression analysis using mouse gene-specific Taqman primer-probe sets (Applied Biosystems).
Protein Analysis.
Cells were incubated for 10 min in cold lysis buffer [50 mM Tris (pH 8), 150 mM NaCl, 2 mM EDTA, 1% Triton-X100, 1 mM NaVanadate, 10 mM NaF] supplemented with Halt Protease Inhibitor Mixture (Thermo Scientific) and spun at 13,000 rpm at 4 °C. Supernatants were used for protein analysis.
For immunoprecipitations, 2 μg anti-FLAG M2 or 0.5 μL anti-GFP serum were conjugated to 30 μL of protein G Sepharose slurry (Thermo Scientific) for 2 h and immunoprecipitated with samples at 4 °C overnight. Proteins eluted from washed beads were resolved in SDS/PAGE gels (Bio-Rad), transferred to a nitrocellulose membrane, blocked in TBS, 5% BSA, and 1% Tween, and used for immunoblotting.
EMSA Analysis.
Cells were stimulated with 2 μg/mL LTA in DMEM for various time intervals after which nuclear proteins were extracted using the Nucbuster kit (Novagen) and incubated with Klenow-labeled, double-stranded oligonucleotide containing an NF-κB binding site. The DNA–protein complexes were incubated for 20 min, resolved on a 5% native polyacrylamide gel, and visualized by autoradiography. The sequences for the NF-κB consensus site and IL-10 promoter were as given in ref. 15.
Pulse-Chase Analysis.
Cells were labeled with 35S-methionine for 30 min and then chased for various times in the presence of 200 mg/mL methionine and resuspended in lysis buffer (see above). Proteins bound to anti-GFP were resolved by SDS–PAGE and analyzed by autoradiography.
Supplementary Material
Acknowledgments
This study was supported by Grant R01AI056234 from the National Institute of Health (to A.P.). J.C. is recipient of a Dean's Fellowship award from the Sackler School of Graduate Biomedical Sciences, Tufts University, Department of Pathology, Tufts, Boston, MA.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006894107/-/DCSupplemental.
References
- 1.Altare F, et al. Inherited interleukin 12 deficiency in a child with bacille Calmette-Guerin and Salmonella enteritidis disseminated infection. J Clin Invest. 1998;102(12):2035–2040. doi: 10.1172/JCI4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter S, O'Neill LA. How important are Toll-like receptors for antimicrobial responses? Cell Microbiol. 2007;9:1891–1901. doi: 10.1111/j.1462-5822.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 3.Picard C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 4.Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of Toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 5.Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 6.Hannum CH, et al. Interleukin-1 receptor antagonist activity of a human interleukin-1 inhibitor. Nature. 1990;343:336–340. doi: 10.1038/343336a0. [DOI] [PubMed] [Google Scholar]
- 7.Carter DB, et al. Purification, cloning, expression and biological characterization of an interleukin-1 receptor antagonist protein. Nature. 1990;344:633–638. doi: 10.1038/344633a0. [DOI] [PubMed] [Google Scholar]
- 8.Couper KN, Blount DG, Riley EM. IL-10: The master regulator of immunity to infection. J Immunol. 2008;180:5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez-Carrozzi VR, et al. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saccani S, Pantano S, Natoli G. Two waves of nuclear factor kappaB recruitment to target promoters. J Exp Med. 2001;193:1351–1359. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 12.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 14.Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 15.Cao S, Zhang X, Edwards JP, Mosser DM. NF-kappaB1 (p50) homodimers differentially regulate pro- and anti-inflammatory cytokines in macrophages. J Biol Chem. 2006;281:26041–26050. doi: 10.1074/jbc.M602222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirotani T, et al. The nuclear IkappaB protein IkappaBNS selectively inhibits lipopolysaccharide-induced IL-6 production in macrophages of the colonic lamina propria. J Immunol. 2005;174:3650–3657. doi: 10.4049/jimmunol.174.6.3650. [DOI] [PubMed] [Google Scholar]
- 17.Conner JR, Smirnova II, Poltorak A. Forward genetic analysis of Toll-like receptor responses in wild-derived mice reveals a novel antiinflammatory role for IRAK1BP1. J Exp Med. 2008;205:305–314. doi: 10.1084/jem.20071499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vig E, et al. SIMPL is a tumor necrosis factor-specific regulator of nuclear factor-kappaB activity. J Biol Chem. 2001;276:7859–7866. doi: 10.1074/jbc.M010399200. [DOI] [PubMed] [Google Scholar]
- 19.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O'Garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815–3822. [PubMed] [Google Scholar]
- 20.de Waal Malefyt R, Abrams J, Bennett B, Figdor CG, de Vries JE. Interleukin 10 (IL-10) inhibits cytokine synthesis by human monocytes: An autoregulatory role of IL-10 produced by monocytes. J Exp Med. 1991;174:1209–1220. doi: 10.1084/jem.174.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones EA, Flavell RA. Distal enhancer elements transcribe intergenic RNA in the IL-10 family gene cluster. J Immunol. 2005;175:7437–7446. doi: 10.4049/jimmunol.175.11.7437. [DOI] [PubMed] [Google Scholar]
- 22.Kwon HJ, et al. Tumor necrosis factor alpha induction of NF-kappaB requires the novel coactivator SIMPL. Mol Cell Biol. 2004;24:9317–9326. doi: 10.1128/MCB.24.21.9317-9326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bohuslav J, et al. Regulation of an essential innate immune response by the p50 subunit of NF-kappaB. J Clin Invest. 1998;102:1645–1652. doi: 10.1172/JCI3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziegler-Heitbrock L. The p50-homodimer mechanism in tolerance to LPS. J Endotoxin Res. 2001;7(3):219–222. [PubMed] [Google Scholar]
- 25.Moorthy AK, et al. The 20S proteasome processes NF-kappaB1 p105 into p50 in a translation-independent manner. EMBO J. 2006;25:1945–1956. doi: 10.1038/sj.emboj.7601081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beinke S, et al. NF-kappaB1 p105 negatively regulates TPL-2 MEK kinase activity. Mol Cell Biol. 2003;23:4739–4752. doi: 10.1128/MCB.23.14.4739-4752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beinke S, Ley SC. Functions of NF-kappaB1 and NF-kappaB2 in immune cell biology. Biochem J. 2004;382:393–409. doi: 10.1042/BJ20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.