Abstract
By impairing both function and survival, the severe reduction in oxygen availability associated with high-altitude environments is likely to act as an agent of natural selection. We used genomic and candidate gene approaches to search for evidence of such genetic selection. First, a genome-wide allelic differentiation scan (GWADS) comparing indigenous highlanders of the Tibetan Plateau (3,200–3,500 m) with closely related lowland Han revealed a genome-wide significant divergence across eight SNPs located near EPAS1. This gene encodes the transcription factor HIF2α, which stimulates production of red blood cells and thus increases the concentration of hemoglobin in blood. Second, in a separate cohort of Tibetans residing at 4,200 m, we identified 31 EPAS1 SNPs in high linkage disequilibrium that correlated significantly with hemoglobin concentration. The sex-adjusted hemoglobin concentration was, on average, 0.8 g/dL lower in the major allele homozygotes compared with the heterozygotes. These findings were replicated in a third cohort of Tibetans residing at 4,300 m. The alleles associating with lower hemoglobin concentrations were correlated with the signal from the GWADS study and were observed at greatly elevated frequencies in the Tibetan cohorts compared with the Han. High hemoglobin concentrations are a cardinal feature of chronic mountain sickness offering one plausible mechanism for selection. Alternatively, as EPAS1 is pleiotropic in its effects, selection may have operated on some other aspect of the phenotype. Whichever of these explanations is correct, the evidence for genetic selection at the EPAS1 locus from the GWADS study is supported by the replicated studies associating function with the allelic variants.
Keywords: chronic mountain sickness, high altitude, human genome variation, hypoxia, hypoxia-inducible factor
The high plateaus of Central Asia and the Andes were among the last areas occupied as Homo sapiens spread across the globe during the past 100,000–200,000 y. In the case of the Tibetan plateau, early visitors appeared more than 30,000 y ago, and the plateau has been colonized for more than 10,000 y (1, 2). The low partial pressure of oxygen resulting from the extreme altitude would have presented a formidable biological challenge to such colonists. Individuals from low-altitude populations (European and Han) who move to live at high altitude suffer from a number of potentially lethal diseases specifically related to the low levels of oxygen (3–5) and struggle to reproduce at these altitudes (6–9). The hypoxia of altitude (hypobaric hypoxia) would thus have exerted substantial evolutionary selection pressure.
The classic disease associated with long term residence at high altitude is chronic mountain sickness, or Monge's disease, after Carlos Monge-Medrano who first identified the condition among Andean highlanders (10). The cardinal feature is a very high concentration of the oxygen-carrying pigment, hemoglobin, in the blood, caused by an overproduction of red blood cells (excessive erythrocytosis). Tibetan highlanders are particularly resistant to developing chronic mountain sickness (4, 11), and exhibit little or no increase in hemoglobin concentration with increasing altitude, even at 4,000 m (13,200 ft) and only moderate increases beyond (12, 13). Typically, Tibetans average at least 1 g/dL and as much as approximately 3.5 gm/dL (i.e. approximately 10–20%) lower hemoglobin concentration in comparison with their Andean counterparts (14–16) or acclimatized lowlanders, such as the Han who have moved to altitudes above 2,500 m (4, 17–23). This suggests that Tibetans have evolved a blunted erythropoietic response to high-altitude hypoxia. The induction of erythrocytosis by hypoxia involves the hypoxia-inducible factor (HIF) family of transcription factors and, in particular, EPAS1 (or HIF2α) (24, 25).
Three independent studies, but with mutually reinforcing results, were performed by groups that have since come together to form a consortium with the aim of reporting on the findings. The first study was a genome-wide allelic differentiation scan that compared SNP frequencies of a Yunnan Tibetan population residing at 3,200–3,500 m with the HapMap Phase III Han sample. Mitochondrial, Y chromosome, and autosomal DNA evidence all suggest a north or east Asian origin for modern Tibetans (1, 26–28). Thus, the Han comprise a lowland population that is closely related to the Tibetans but which has not undergone selection for high-altitude adaptation. From this study, a signal of selection close to EPAS1 was identified at a genome-wide level of significance. The second study comprised a candidate gene analysis of EPAS1 in a separate sample of Tibetans from 4,200 m on the Tibetan plateau and identified a significant association between genotype and hemoglobin concentration, with the major alleles associating with the lower hemoglobin levels. These alleles were present at low frequency in the Han. The third study replicated the hemoglobin association in an independent sample of Tibetans from 4,300 m.
Results
Genome-Wide Allelic Differentiation Scan.
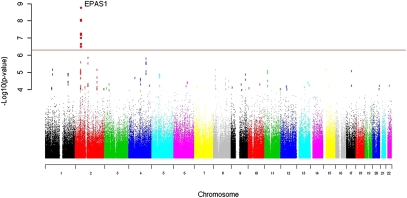
A genome-wide allelic differentiation scan (GWADS) was used to compare a cohort of Tibetan residents (n = 35) sampled from four townships at altitudes of 3,200–3,500 m in Yunnan Province, China, with HapMap Phase III Han individuals (n = 84). We postulated that any marked differences in SNP frequencies between the Yunnan Tibetan and the HapMap Han populations could reflect a history of divergent selection on functional variation that contributes to increased survival at high altitude. (See SI Materials and Methods for detailed methodology.) Of the 502,722 SNPs that were included in the analysis, eight SNPs emerged as having genome-wide significance (P values ranging from 2.81 × 10−7 to 1.49 × 10−9), all located within 235 kb on chromosome 2 (Fig. 1 and Table S1).
Fig. 1.
A genome-wide allelic differentiation scan that compares Tibetan residents at 3,200–3,500 m in Yunnan Province, China with HapMap Han samples. Eight SNPs near one another and EPAS1 have genome-wide significance. The horizontal axis is genomic position with colors indicating chromosomes. The vertical axis is the negative log of SNP-by-SNP P values generated from the Yunnan Tibetan vs. HapMap Han comparison. The red line indicates the threshold for genome-wide significance used (P = 5 × 10−7). Values are shown after correction for background population stratification using genomic control.
All eight GWADS significant SNPs were in high pairwise linkage disequilibrium in the Yunnan sample (0.23< r2 < 0.82), forming an extended haplotype with a frequency of 46% in the Yunnan Tibetan sample but only 2% in the Han sample [estimated via an expectation-maximization algorithm using Haploview software (29, 30)]. The SNPs lie between 366 bp and 235 kb downstream of EPAS1 but, as we show below, the region of high linkage disequilibrium extends into EPAS1 itself. In addition to this genome-wide significant finding relating to EPAS1, evidence for other signals of selection was also found. Regions of subgenome wide significance were in close proximity to other genes of the HIF pathway and present intriguing targets for follow-up studies (see SI Text for further details).
Candidate Gene Study for EPAS1.
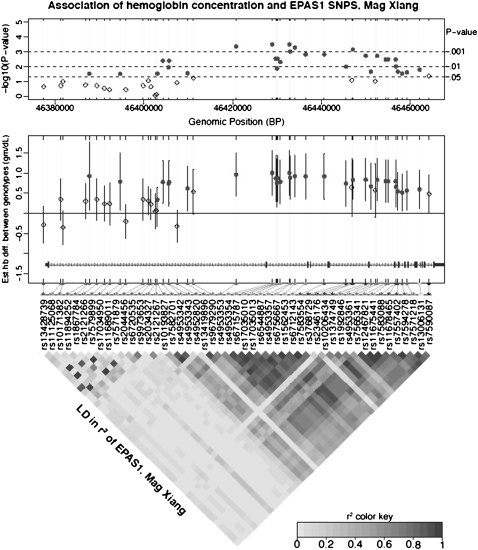
Independent of the GWADS study, a candidate gene study (based on the pathway linking hypoxia, EPAS1, and erythropoietin) addressed the functional consequence of EPAS1 variants by testing for association with hemoglobin concentration in a sample of 70 Tibetans residing at 4,200 m in Mag Xiang, Xigatse Prefecture in the Tibet Autonomous Region, China (Table S2). One hundred and three noncoding SNPs across the EPAS1 gene were selected for genotyping. Of these, 49 had a minor allele frequency ≥5%, and were thus amenable to regression analysis (Materials and Methods) that identified 31 SNP sites significantly associated with hemoglobin concentration (Fig. 2 and Table S3). The major (most frequent) allele of every significant SNP was associated with lower hemoglobin concentration. After adjusting for sex differences, individuals who were homozygous for the major allele had an average hemoglobin concentration that was 0.8 ± 0.15 g/dL (range from 0.3 to 1.0 g/dL) lower than individuals who were heterozygous for the major allele. Conditional linear regression analyses showed that once the most significant SNP (rs4953354) was included, no significant improvement in fit was obtained after Bonferroni correction by adding any other SNP, consistent with a single causal variant model. Many of the SNP sites were in high linkage disequilibrium (Fig. 2). Genotypes for the eight GWADS significant SNPs were available on 29 of the 70 individuals in the Mag Xiang cohort, too few to show statistical association with hemoglobin concentration. However, all eight GWADS SNPs were highly correlated (0.54 < r2 ≤ 1) with variants associating with hemoglobin concentration in the complete Mag Xiang cohort (Table S4). Thus, the genome-wide and the candidate-gene analyses can be linked, with the latter study demonstrating that there is a phenotype associated with the signal of selection.
Fig. 2.
Sex-adjusted hemoglobin concentrations and allelic variation in EPAS1 SNPs in a Tibetan sample from Mag Xiang (4,200 m), Tibet Autonomous Region, China. Values were, on average, 0.8 g/dL lower for individuals who were homozygous for the major alleles compared with those who were heterozygous. (Top) The results of testing variants at 49 SNPs with a minor allele frequency ≥5% for genotypic association with sex-adjusted hemoglobin concentration. (Middle) The estimated hemoglobin concentration difference (mean ± 95% confidence interval) between the major allele homozygote and heterozygote genotypes at each SNP. Filled circles represent SNPs detected as having a significant association with hemoglobin concentration, with the false discovery rate controlled at <0.05 across the EPAS1 locus. Open diamonds represent SNPs without significant association. (Bottom) The pairwise linkage disequilibrium measured as r2 between SNPs.
Replication of Candidate Gene Study for EPAS1.
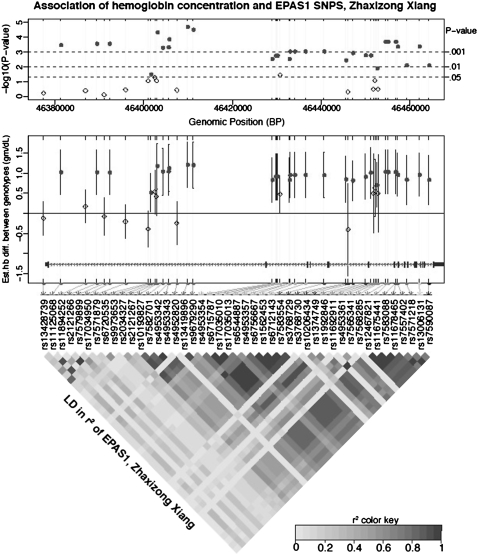
We replicated the association of EPAS1 SNPs with hemoglobin concentration in another sample of 91 Tibetans residing at 4,300 m in Zhaxizong Xiang, Xigatse Prefecture, China (Table S2). Of the 49 Mag Xiang SNPs with a minor allele frequency ≥5%, 48 were successfully genotyped in the Zhaxizong Xiang sample. Of these, 45 sites had a minor allele frequency ≥5% and 32 sites were significantly associated with hemoglobin concentration. After adjusting for sex differences, individuals who were homozygous for the major allele had an average hemoglobin concentration that was 1.0 ± 0.14 g/dL (range from 0.5 to 1.2 g/dL) lower than individuals who were heterozygous for the major allele (Fig. 3 and Table S3). Twenty-six SNPs were associated with hemoglobin concentration in both samples and the direction of the effect was the same. Conditional linear regression again found that, after including the most significant SNP (rs13419896), no further SNPs were significant after Bonferroni correction. This was also the case if the most significant SNP from the Mag Xiang sample (rs4953354) was used instead of rs13419896. Genotypes for the eight GWADS significant SNPs were available on 89 samples from the Zhaxizong Xiang cohort. Three of these SNPs correlated significantly with hemoglobin concentration (Table S4), thus supporting the association of a phenotype with the signal of selection that has been obtained from this area of the genome.
Fig. 3.
Sex-adjusted hemoglobin concentrations and allelic variation in EPAS1 SNPs in a Tibetan sample from Zhaxizong Xiang (4,300 m), Tibet Autonomous Region, China. Values were, on average, 1.0 g/dL lower for individuals who were homozygous for the major alleles compared with those who were heterozygous (Top) The results of testing variants at 45 SNPs with a minor allele frequency ≥5% for genotypic association with sex-adjusted hemoglobin concentration. (Middle) The estimated hemoglobin concentration difference (mean ± 95% confidence interval) between the major allele homozygote and heterozygote genotypes at each SNP. Filled circles represent SNP detected as having a significant association with hemoglobin concentration with the false discovery rate controlled at <0.05 across the EPAS1 locus. Open diamonds represent SNP without significant association. (Bottom) The pairwise linkage disequilibrium measured as r2 between SNPs.
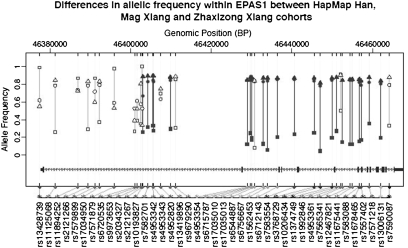
Comparing allelic frequencies between the two Tibetan samples and the HapMap Han sample, we note that the largest allele frequency differences occur at the EPAS1 SNP sites that are associated with low hemoglobin concentration (Fig. 4). Linkage disequilibrium (LD) among these 26 SNP sites was also elevated in the two Tibetan cohorts compared with the HapMap Han (SI Text).
Fig. 4.
Differences in allelic frequency at SNPs within EPAS1 between the HapMap Han, Mag Xiang and Zhaxizong Xiang cohorts. The horizontal axis is SNP position according to build 36.1. The vertical axis is allelic frequency, with the allele selected for display as the one occurring most frequently in the Mag Xiang cohort. Squares denote data for HapMap Han; circles denote data for Mag Xiang Tibetans; triangles denote data for Zhaxizong Xiang Tibetans. Filled symbols denote those SNPs having significant associations with hemoglobin in both Mag Xiang and Zhaxizong Xiang cohorts; open symbols denote those SNPs without both such associations.
Discussion
The results from the GWADS study revealed a level of allele frequency differentiation near EPAS1 that far exceeds the genome-wide average (Fig. 1). The association studies demonstrated that the SNP variants that were at higher frequencies within the Tibetan population were associated with lower hemoglobin concentrations. As large genome-wide association studies of the determinants of hemoglobin concentration in other populations at low altitude have failed to detect a signal associated with EPAS1 (31–34), our results suggest either that there is a genetic variant that is quite specific to the Tibetan population or that the variant is quite specific to moderating hemoglobin concentration only under conditions of high altitude. Such specificity of effect in relation to Tibetan highlanders is in keeping with a model of selection pressure on EPAS1 under the stress of high-altitude hypoxia. Interestingly, a comparison between the HapMap Han and Andean highlanders—both of whom have a vigorous erythropoietic response (15)—did not detect selection at the EPAS1 locus (35). It should be noted, however, that the Andean study (35) applied a different array of methodologies to detect selection and overlapping results are not necessarily expected, given the differing nature of the selection signals that particular techniques are powered to detect. It is also possible that the Andean and Tibetan populations have developed different genetic adaptations to the hypoxia of high altitude given the differences in physiology that are known to exist between these populations (13).
The association studies revealed that genetic variation across EPAS1 accounts for a large proportion of the variation in hemoglobin concentration in these populations. After controlling for sex, the average difference in hemoglobin concentration between major allele homozygotes and heterozygotes was 53% of one SD in the Mag Xiang sample and 50% in the Zhaxizong Xiang sample. In absolute terms, these differences were several fold larger than for any of the determinants of hemoglobin concentration in populations residing at low altitude (31–34). Our findings are, however, consistent with previous high estimates of heritability (h2) for hemoglobin concentration of 0.66 among Tibetans at 4,850–5,450 m (36) and of 0.86 among Tibetans at 3,800–4,065 m (15). These values estimate the proportion of additive genetic variation relative to total phenotypic variance. The combined findings of our association and conditional linear regression analysis are consistent with a model in which a single causal variant at the EPAS1 locus accounts for a substantial proportion of the heritability. Under this model, hemoglobin-associating SNPs should be interpreted as markers and are presumed to have differentiated because they are closely linked to an as yet to be identified causal variant. Functional studies will be required to identify how this variation works to restrain the hematopoietic response.
We have described a signal of natural selection on or near EPAS1 that is associated with a blunting of the normal erythropoietic response to hypoxia. As EPAS1 is pleiotropic, other responses to hypoxia may be similarly affected. Some insight into these may be given by studies of a few individuals/families, living at low altitudes, who have been reported to have gain of function mutations in EPAS1 (37–40). As expected, these individuals exhibit excessive erythrocytosis, but they also appear to be particularly susceptible to thrombotic events and to developing pulmonary hypertension—although the total number of cases reported is small. A larger number of cases have been reported for the slightly less specific genetic disorder of Chuvash Polycythemia, where homozygosity for hypomorphic alleles for VHL results in elevated levels of both HIF1α and EPAS1/HIF2α (41). The phenotype for Chuvash Polycythemia appears very similar to that for the specific EPAS1 gain of function mutations, with excessive erythrocytosis, an excess risk of thrombotic events at a young age, and pulmonary hypertension (42–45). In both conditions, the excessive erythrocytosis is generally managed by venesection in the belief that this may reduce the incidence of thrombotic events.
The clinical similarity between the phenotypes of these genetic disorders and chronic mountain sickness is striking. Indeed, it caused one group of investigators to observe in respect of the EPAS1 gain of function mutations that “it raises the possibility that polymorphic variation in HIF2α [EPAS1] contributes to the marked differential susceptibility to erythrocytosis, reduced plasma volume and pulmonary hypertension in humans at high altitude” (39). Chronic mountain sickness occurs among Tibetans at a lower prevalence than Han lowlanders (1.2% compared with 5.6%) living in the Tibet Autonomous Region (4, 46). Chronic mountain sickness remains at that low level throughout adulthood among Tibetans but, in Peruvians, prevalence increases with age from approximately 13% in 20 to 39 y olds to approximately 36% in 55 to 69 y olds at 4,300 m (47). In Andeans, excessive erythrocytosis at high altitude has been associated with significant pulmonary hypertension (48), an increased risk of stroke (49), and also an increased risk of poor outcome in pregnancy (stillbirth, preterm birth, or small for gestational age at birth) (50). These findings provide insight into some of the sources of elevated morbidity and mortality on which selection may have operated to influence allelic frequencies for EPAS1 among the early colonizers of the Tibetan plateau.
Although the similarity between chronic mountain sickness and the EPAS1 gain of function phenotype in lowlanders is striking, there nevertheless may be other aspects to the phenotype that are not revealed at low altitude but are only revealed at high altitude, when oxygen availability is also limited. In particular, EPAS1 plays very important, if still poorly understood, roles in both placental and embryonic development (51–54) and possibly also in the pathogenesis of fetal growth restriction (55). It is well recognized that reproductive success is more difficult at high altitude than at low altitude, and more difficult for nonnatives than natives (6). For example, pre- and postnatal mortality are threefold higher in the Han than in the Tibetans, and birth weight is significantly lower (56). This may relate in part to the presence of greater uterine arterial blood flow and lower hemoglobin concentration in the Tibetans (9, 57). As such, natural selection on EPAS1 may also have operated via effects during pregnancy that affect both pre- and postnatal mortality.
In conclusion, this study provides evidence for natural selection in Tibetan highlanders at a specific human gene locus. The finding is further supported by a demonstration, in two independent samples, that genetic variation at this locus has an associated phenotype. The known physiological roles associated with this gene locus provide insight into some of the factors that are likely to have influenced human adaptation and survival following colonization of the Tibetan Plateau.
Materials and Methods
Human Volunteers.
Ethics and consent.
This study was approved by the ethics committees of the Yunnan Population and Family Planning Institute (Kunming, China); the Beijing Genomics Institute at Shenzhen (Shenzhen, China); the Beijing Institute of Genomics, Chinese Academy of Sciences (Beijing, China)and Case Western University (Cleveland, OH). All work was conducted in accordance with the principles of the Declaration of Helsinki. All participants were recruited after obtaining informed consent.
Sample collection.
Sampling was conducted in three geographic regions of China approximately 2,400 kilometers apart. They were (i) the North Western region of Yunnan province (28°26’N 98°52'E), (ii) Mag Xiang, Xigatse Prefecture, Tibet Autonomous Region (29°15’N 88°53'E), and (iii) Zhaxizong Xiang, Xigatse Prefecture, Tibet Autonomous Region (28°34’N 86°38’E). Genotypic data from the HapMap Phase III Han population were also included in this analysis. Further details on sample collection are given in the SI Materials and Methods.
Genotyping.
All genotyping was conducted at the Beijing Institute of Genomics. The whole genome genotyping was conducted using the Illumina Veracode platform and 610-Quad high throughput genotyping chips. Genotyping within EPAS1 was conducted using a customer-designed Illumina GoldenGate assay (384 SNP plex) for all samples from Mag Xiang and some of the samples from Zhaxizong Xiang. The remainder of the samples from Zhaxizong Xiang were genotyped using MassARRAY assays. Further details of these and the quality control procedures are given in the SI Materials and Methods.
Phenotyping.
Hemoglobin concentration was measured in duplicate immediately after provision of a venipuncture blood sample by individuals in the Mag Xiang sample (58). Individuals were screened with the aim of including only healthy, native residents. Excluded were individuals who had anemia (men and women with hemoglobin concentrations below 13.7 g/dL and 12 g/dL, respectively), hypertension, fever, poor lung function, extreme hypoxemia, or who were currently or recently pregnant, or who had symptoms or medication indicative of heart or lung disease. The Zhaxizong Xiang sample was obtained in the course of a health survey and included all volunteers who were native residents.
Statistical Analysis.
GWADS.
To identify variation between the Yunnan Tibetan and the HapMap Han populations, we calculated SNP-by-SNP χ2 statistics for allele frequencies and corrected for background population stratification through a genomic control procedure (30). This approach allows genome-wide significant signals of allele frequency differentiation to be readily declared by examining genomic distributions of χ2 values in the sample of approximately 500,000 SNPs. A threshold of genome-wide significance was set at 5 × 10−7 (59). A full description of the method, including a simulation for two populations with a degree of genomic divergence equal to that between the Yunnan Tibetan and HapMap Han populations, is given in the SI Text.
Candidate gene studies.
Candidate gene association analysis of EPAS1 SNP genotype with hemoglobin concentration phenotype was performed separately in the two Tibet Autonomous Region samples. Mean characteristics for these populations are given in Table S2. For each SNP, a linear additive genetic model was fitted with hemoglobin concentration as the response variable, the SNP as the predictive variable (entered as a numerical variable—1, 2, 3—corresponding to the three genotypes sorted by descending allelic frequency) and with gender as a covariate. The P values of the likelihood ratio test were obtained from a comparison with the null model (i.e., only gender in the model). The estimated difference stands for the increase in the sex-adjusted mean with the addition of one copy of the minor allele taking the most frequent homozygous genotypes as the reference. Unless otherwise stated, an adjustment for multiple comparisons was implemented by controlling the false discovery rate at less than 0.05 across the EPAS1 gene. The R language and environment (R Project for Statistical Computing, http://www.r-project.org) was used for all related analysis and graphics. Conditional linear analyses were undertaken by including a specified SNP as an additional covariate in the model and were implemented using plink (http://pngu.mgh.harvard.edu/~purcell/plink/).
Supplementary Material
Acknowledgments
We thank Wei Chen, Jian Bai, and Feng Cheng of Beijing Institute of Genomics for their contribution in genotyping and data processing and three anonymous reviewers for their critical and constructive comments. We also thank the people of Shangri-La and De Qin Xians, Yunnan Province; Mag Xiang and Zhaxizong Xiang, Tibet Autonomous Region, for their cooperation and hospitality during data collection. We are grateful to the Tibet Academy of Social Sciences for their collaboration and enabling permission to collect data in Mag Xiang. This work was supported by the National Science Foundation; National Institutes of Health National Center for Research Resources, National Institute of General Medical Sciences, National Cancer Institute, National Heart, Lung, and Blood Institute; National Natural Science Foundation of China Grant 30890031 and Ministry of Science and Technology Grant 2006DFA31850 (to C.Z.); Chinese Academy of Sciences Grant KSCX2-YW-R-76 and Science and Technology Plan of the Tibet Autonomous Region Grant 2007-2-18 to Beijing Genomics Institute at Shenzhen; and an International Joint Project award from the Royal Society. This consortium grew from a catalysis meeting sponsored by the National Science Foundation-supported National Evolutionary Synthesis Center (http://www.NESCent.org).
Footnotes
The authors are listed alphabetically and declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002443107/-/DCSupplemental.
References
- 1.Zhao M, et al. Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau. Proc Natl Acad Sci USA. 2009;106:21230–21235. doi: 10.1073/pnas.0907844106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldenderfer M. Moving up in the world: Archaeologists seek to understand how and when people came to occupy the Andean and Tibetan plateaus. Am Sci. 2003;91:542–549. [Google Scholar]
- 3.Wu T, Miao C. High altitude heart disease in children in Tibet. High Alt Med Biol. 2002;3:323–325. doi: 10.1089/152702902320604340. [DOI] [PubMed] [Google Scholar]
- 4.León-Velarde F, et al. Consensus statement on chronic and subacute high altitude diseases. High Alt Med Biol. 2005;6:147–157. doi: 10.1089/ham.2005.6.147. [DOI] [PubMed] [Google Scholar]
- 5.Wu T. Life on the high Tibetan plateau. High Alt Med Biol. 2004;5:1–2. doi: 10.1089/152702904322963609. [DOI] [PubMed] [Google Scholar]
- 6.Julian CG, Wilson MJ, Moore LG. Evolutionary adaptation to high altitude: A view from in utero. Am J Hum Biol. 2009;21:614–622. doi: 10.1002/ajhb.20900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monge C. Acclimatization in the Andes. Baltimore: Johns Hopkins University Press; 1973. [Google Scholar]
- 8.Moore LG, Young D, McCullough RE, Droma T, Zamudio S. Tibetan protection from intrauterine growth restriction (IUGR) and reproductive loss at high altitude. Am J Hum Biol. 2001;13:635–644. doi: 10.1002/ajhb.1102. [DOI] [PubMed] [Google Scholar]
- 9.Moore LG, Zamudio S, Zhuang J, Sun S, Droma T. Oxygen transport in tibetan women during pregnancy at 3,658 m. Am J Phys Anthropol. 2001;114:42–53. doi: 10.1002/1096-8644(200101)114:1<42::AID-AJPA1004>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 10.Monge-M C, Encinas E, Heraud C, Hurtado A. La Enfermedad de los Andes (Sindromes Eritemicos) Lima: Imprenta Americana; 1928. [Google Scholar]
- 11.Pei SX, et al. Chronic mountain sickness in Tibet. Q J Med. 1989;71:555–574. [PubMed] [Google Scholar]
- 12.Beall CM. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8655–8660. doi: 10.1073/pnas.0701985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beall CM. Biodiversity of Human Populations in Mountain Environments. In: Korner C, Spehn EM, editors. Mountain Biodiversity: A Global Assessment. New York: The Parthenon Publishing Group; 2002. pp. 199–210. [Google Scholar]
- 14.Winslow RM, et al. Different hematologic responses to hypoxia in Sherpas and Quechua Indians. J Appl Physiol. 1989;66:1561–1569. doi: 10.1152/jappl.1989.66.4.1561. [DOI] [PubMed] [Google Scholar]
- 15.Beall CM, et al. Hemoglobin concentration of high-altitude Tibetans and Bolivian Aymara. Am J Phys Anthropol. 1998;106:385–400. doi: 10.1002/(SICI)1096-8644(199807)106:3<385::AID-AJPA10>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 16.Beall CM. Biodiversity of Human Populations in Moutain Environments. In: Korner C, Spehn EM, editors. Mountain Biodiversity: A Global Assessment. New York: The Parthenon Publishing Group; 2002. pp. 199–210. [Google Scholar]
- 17.Garruto RM, et al. Hematological differences during growth among Tibetans and Han Chinese born and raised at high altitude in Qinghai, China. Am J Phys Anthropol. 2003;122:171–183. doi: 10.1002/ajpa.10283. [DOI] [PubMed] [Google Scholar]
- 18.Ward MP, Milledge JS, West JB. High Altitude Medicine and Physiology. London: Chapman and Hall Medical; 1995. [Google Scholar]
- 19.Zhuang J, et al. Smaller alveolar-arterial O2 gradients in Tibetan than Han residents of Lhasa (3658 m) Respir Physiol. 1996;103:75–82. doi: 10.1016/0034-5687(95)00041-0. [DOI] [PubMed] [Google Scholar]
- 20.Moore LG, et al. Are Tibetans better adapted? Int J Sports Med. 1992;13(Suppl 1):S86–S88. doi: 10.1055/s-2007-1024605. [DOI] [PubMed] [Google Scholar]
- 21.Droma T, et al. Increased vital and total lung capacities in Tibetan compared to Han residents of Lhasa (3,658 m) Am J Phys Anthropol. 1991;86:341–351. doi: 10.1002/ajpa.1330860303. [DOI] [PubMed] [Google Scholar]
- 22.Sun SF, et al. Greater maximal O2 uptakes and vital capacities in Tibetan than Han residents of Lhasa. Respir Physiol. 1990;79:151–161. doi: 10.1016/0034-5687(90)90015-q. [DOI] [PubMed] [Google Scholar]
- 23.Zhuang J, et al. Hypoxic ventilatory responsiveness in Tibetan compared with Han residents of 3,658 m. J Appl Physiol. 1993;74:303–311. doi: 10.1152/jappl.1993.74.1.303. [DOI] [PubMed] [Google Scholar]
- 24.Chi JT, et al. Gene expression programs in response to hypoxia: Cell type specificity and prognostic significance in human cancers. PLoS Med. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rankin EB, et al. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest. 2007;117:1068–1077. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su B, et al. Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet. 2000;107:582–590. doi: 10.1007/s004390000406. [DOI] [PubMed] [Google Scholar]
- 27.Qian Y, et al. Multiple origins of Tibetan Y chromosomes. Hum Genet. 2000;106:453–454. doi: 10.1007/s004390000259. [DOI] [PubMed] [Google Scholar]
- 28.Gayden T, et al. Genetic insights into the origins of Tibeto-Burman populations in the Himalayas. J Hum Genet. 2009;54:216–223. doi: 10.1038/jhg.2009.14. [DOI] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 31.Benyamin B, et al. Common variants in TMPRSS6 are associated with iron status and erythrocyte volume. Nat Genet. 2009;41:1173–1175. doi: 10.1038/ng.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chambers JC, et al. Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. Nat Genet. 2009;41:1170–1172. doi: 10.1038/ng.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganesh SK, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soranzo N, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bigham AW, et al. Identifying positive selection candidate loci for high-altitude adaptation in Andean populations. Hum Genomics. 2009;4:79–90. doi: 10.1186/1479-7364-4-2-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beall CM, Blangero J, Williams-Blangero S, Goldstein MC. Major gene for percent of oxygen saturation of arterial hemoglobin in Tibetan highlanders. Am J Phys Anthropol. 1994;95:271–276. doi: 10.1002/ajpa.1330950303. [DOI] [PubMed] [Google Scholar]
- 37.Percy MJ, et al. A gain-of-function mutation in the HIF2A gene in familial erythrocytosis. N Engl J Med. 2008;358:162–168. doi: 10.1056/NEJMoa073123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Percy MJ, et al. Novel exon 12 mutations in the HIF2A gene associated with erythrocytosis. Blood. 2008;111:5400–5402. doi: 10.1182/blood-2008-02-137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gale DP, Harten SK, Reid CDL, Tuddenham EGD, Maxwell PH. Autosomal dominant erythrocytosis and pulmonary arterial hypertension associated with an activating HIF2 α mutation. Blood. 2008;112:919–921. doi: 10.1182/blood-2008-04-153718. [DOI] [PubMed] [Google Scholar]
- 40.Martini M, et al. A novel heterozygous HIF2AM535I mutation reinforces the role of oxygen sensing pathway disturbances in the pathogenesis of familial erythrocytosis. Haematologica. 2008;93:1068–1071. doi: 10.3324/haematol.13210. [DOI] [PubMed] [Google Scholar]
- 41.Ang SO, et al. Disruption of oxygen homeostasis underlies congenital Chuvash polycythemia. Nat Genet. 2002;32:614–621. doi: 10.1038/ng1019. [DOI] [PubMed] [Google Scholar]
- 42.Gordeuk VR, et al. Congenital disorder of oxygen sensing: Association of the homozygous Chuvash polycythemia VHL mutation with thrombosis and vascular abnormalities but not tumors. Blood. 2004;103:3924–3932. doi: 10.1182/blood-2003-07-2535. [DOI] [PubMed] [Google Scholar]
- 43.Sergeyeva A, et al. Congenital polycythemia in Chuvashia. Blood. 1997;89:2148–2154. [PubMed] [Google Scholar]
- 44.Smith TG, et al. Mutation of von Hippel-Lindau tumour suppressor and human cardiopulmonary physiology. PLoS Med. 2006;3:e290. doi: 10.1371/journal.pmed.0030290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bushuev VI, et al. Endothelin-1, vascular endothelial growth factor and systolic pulmonary artery pressure in patients with Chuvash polycythemia. Haematologica. 2006;91:744–749. [PubMed] [Google Scholar]
- 46.Ren YH, et al. Incidence of high altitude illnesses among unacclimatized persons who acutely ascended to Tibet. High Alt Med Biol. 2010;11:39–42. doi: 10.1089/ham.2009.1049. [DOI] [PubMed] [Google Scholar]
- 47.Monge-C C, Arregui A, León-Velarde F. Pathophysiology and epidemiology of chronic mountain sickness. Int J Sports Med. 1992;13(Suppl 1):S79–S81. doi: 10.1055/s-2007-1024603. [DOI] [PubMed] [Google Scholar]
- 48.Penaloza D, Arias-Stella J. The heart and pulmonary circulation at high altitudes: Healthy highlanders and chronic mountain sickness. Circulation. 2007;115:1132–1146. doi: 10.1161/CIRCULATIONAHA.106.624544. [DOI] [PubMed] [Google Scholar]
- 49.Jaillard AS, Hommel M, Mazetti P. Prevalence of stroke at high altitude (3380 m) in Cuzco, a town of Peru. A population-based study. Stroke. 1995;26:562–568. doi: 10.1161/01.str.26.4.562. [DOI] [PubMed] [Google Scholar]
- 50.Gonzales GF, Steenland K, Tapia V. Maternal hemoglobin level and fetal outcome at low and high altitudes. Am J Physiol Regul Integr Comp Physiol. 2009;297:R1477–R1485. doi: 10.1152/ajpregu.00275.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pringle KG, Kind KL, Sferruzzi-Perri AN, Thompson JG, Roberts CT. Beyond oxygen: Complex regulation and activity of hypoxia inducible factors in pregnancy. Hum Reprod Update. 2009 doi: 10.1093/humupd/dmp046. 10.1093/humupd/dmp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunwoodie SL. The role of hypoxia in development of the Mammalian embryo. Dev Cell. 2009;17:755–773. doi: 10.1016/j.devcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 53.Meade ES, Ma YY, Guller S. Role of hypoxia-inducible transcription factors 1alpha and 2alpha in the regulation of plasminogen activator inhibitor-1 expression in a human trophoblast cell line. Placenta. 2007;28:1012–1019. doi: 10.1016/j.placenta.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cowden Dahl KD, et al. Hypoxia-inducible factors 1alpha and 2alpha regulate trophoblast differentiation. Mol Cell Biol. 2005;25:10479–10491. doi: 10.1128/MCB.25.23.10479-10491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai SY, Kanenishi K, Ueno M, Sakamoto H, Hata T. Hypoxia-inducible factor-2alpha is involved in enhanced apoptosis in the placenta from pregnancies with fetal growth restriction. Pathol Int. 2004;54:843–849. doi: 10.1111/j.1440-1827.2004.01750.x. [DOI] [PubMed] [Google Scholar]
- 56.Moore LG, Young D, McCullough RE, Droma T, Zamudio S. Tibetan protection from intrauterine growth restriction (IUGR) and reproductive loss at high altitude. Am J Hum Biol. 2001;13:635–644. doi: 10.1002/ajhb.1102. [DOI] [PubMed] [Google Scholar]
- 57.Xing Y, et al. Hemoglobin levels and anemia evaluation during pregnancy in the highlands of Tibet: A hospital-based study. BMC Public Health. 2009;9:336. doi: 10.1186/1471-2458-9-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoit BD, et al. Nitric oxide and cardiopulmonary hemodynamics in Tibetan highlanders. J Appl Physiol. 2005;99:1796–1801. doi: 10.1152/japplphysiol.00205.2005. [DOI] [PubMed] [Google Scholar]
- 59.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.