Abstract
Meiosis generates four haploid daughters from a diploid parental cell. Central steps of meiosis are the pairing and recombination of homologous chromosomes followed by their segregation in two rounds of cell division. Meiotic recombination is monitored by a specialized DNA damage checkpoint pathway and is guided by a unique chromosomal structure called synaptonemal complex (SC), but how these events are coordinated is unclear. Here, we identify the SC protein Red1 as a crucial regulator of early meiosis. Red1 interacts with two subunits of the 9-1-1 checkpoint complex via two distinct 9-1-1 subunit-specific motifs. Association of 9-1-1 with Red1 is essential not only for meiotic checkpoint activation but for SC formation. Moreover, Red1 becomes SUMO-modified, which fosters interaction of Red1 with the central SC element Zip1, thereby securing timely SC formation. Thus, Red1, in addition to its structural role in the SC, is a crucial coordinator of meiosis by coupling checkpoint signaling to SC formation.
Keywords: checkpoint, meiosis, SUMO
The central activity of meiosis is the equal distribution of the genetic material of a diploid cell into four daughter cells. A key step of meiosis occurs in pachytene, in which the homologous chromosomes (i.e., the parental chromosomes, each containing two sister chromatids) align (synapsis). This alignment facilitates the exchange of parental information by homologous recombination and is crucial for chromosome segregation during the first meiotic division.
The juxtaposition of the homologs in pachytene involves a unique chromosome structure known as the synaptonemal complex (SC), which assembles along the entire length of the bundled chromosomes (1, 2). The SC-induced arrangement of chromosomes appears to favor genetic exchange between homologs rather than sister chromatids. SC formation starts along each pair of homologous sister chromatids with the assembly of 50-nm-thick fibers, termed axial elements, which later become the so-called “lateral elements” of the SC. The axial element-coated parental homologs finally associate via components of the central element, which zip the axial elements together, forming the SC. In Saccharomyces cerevisiae, the proteins Hop1 and Red1 are structural components of the lateral element, whereas the coiled-coil protein Zip1, which homo (oligo)-dimerizes during SC formation, forms the “steps” of the ladder-like central region. SC “zipping” is thought to take place by serial Zip1-Red1 interactions along the entire SC axis (1, 2).
Early meiosis is under the control of the meiotic recombination checkpoint (called the meiotic checkpoint here; it is also known as the pachytene checkpoint), which prevents meiotic progression in the presence of unrepaired recombination intermediates. A key element of the meiotic surveillance pathway is the ring-shaped, heterotrimeric 9-1-1 complex (the proteins Rad9, Hus1, and Rad1 in mammals; the proteins Ddc1, Mec3, and Rad17 in S. cerevisiae), which is structurally related to the homotrimeric DNA-encircling DNA polymerase sliding clamp proliferating cell nuclear antigen (PCNA) (3). Unlike PCNA, 9-1-1 is specifically loaded close to damaged sites and, presumably, also close to resected meiotic DNA double-strand breaks (DSBs) (4, 5). In meiosis, these sites are DSBs purposely induced by the topoisomerase-related enzyme Spo11 to trigger meiotic recombination (6, 7). Binding of 9-1-1 to damaged sites recruits downstream effectors, which usually activate checkpoint kinases involved in cell cycle arrest. The archetypal mediator for checkpoint signaling is the conserved protein TopBP1 (in humans, Dpb11 in S. cerevisiae, and Cut5/Rad4 in Schizosaccharomyces pombe), which physically interacts with 9-1-1 via its Ddc1 subunit (8, 9). Notably, however, Dpb11-dependent signaling via (S. cerevisiae) Rad9 and Rad53 does not seem to play a major role in the checkpoint response to programmed Spo11-induced meiotic DSBs (10–14), raising the question of how 9-1-1 transduces meiotic DSB-induced checkpoint signaling in meiosis.
In this study, we show that Red1, a lateral element protein of the SC, connects central events of early meiosis and lies at the heart of meiotic checkpoint signaling. In particular, our studies revealed that Red1 functions as a downstream effector of 9-1-1 in the meiotic DNA damage surveillance pathway. We found that Red1 interacts with two subunits of the 9-1-1 checkpoint complex, involving two subunit-specific domains of Red1. This association is not only essential for meiotic checkpoint signaling but, surprisingly, for SC formation. Interestingly, a fraction of Red1 becomes SUMO-modified during meiosis. We found that this modification, apparently by fostering the interaction of Red1 with the central element protein Zip1, promotes the timing of SC assembly. Thus, Red1 functions as a key regulator of meiosis by connecting meiotic checkpoint signaling to SC formation through physical interactions.
Results
The 9-1-1 Checkpoint Complex Binds the SC Protein Red1.
A key step of meiosis is the formation of DSBs catalyzed by Spo11 (6), which initiate meiotic recombination. Such DSBs are believed to be marked with the ring-shaped 9-1-1 complex (10), but how checkpoint signaling is transduced from 9 to 1-1 in the meiotic pathway remains unclear. Because Dpb11 (TopBP1), the normal downstream effector of 9-1-1, does not seem to play a major role (14), we speculated that perhaps a meiosis-specific protein might cooperate with 9-1-1 in the meiotic surveillance pathway. Among the meiosis-specific proteins that are known to act downstream of 9-1-1 are the SC proteins Red1 and Hop1 and the protein kinase Mek1, a putative homolog of the checkpoint kinase Rad53/CHK2 (15–18). Moreover, Red1 and Hop1 are phosphorylated in response to DSB formation (or 9-1-1 activity) (5, 17), and Hop1 phosphorylation by Mec1 is required for activation of Mek1 (17).
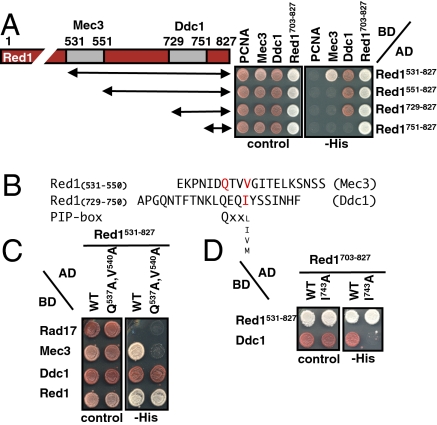
Because of these findings, we considered the proteins Hop1, Red1, and Mek1 as candidates for the direct meiotic downstream effector of 9-1-1. Specifically, we asked whether 9-1-1 physically interacts with these proteins. To this end, all three subunits of 9-1-1 were expressed as two-hybrid fusions and assayed for interaction with the three candidate proteins. Although we found no interaction of Hop1 and Mek1 with any of the three 9-1-1 subunits in this assay (Fig. S1A), we observed strong interaction of 9-1-1 with Red1 (Fig. 1A). Surprisingly, 9-1-1–Red1 interaction involves two subunits of the heterotrimeric complex, Mec3 and Ddc1, and both subunits interact with the C-terminal domain of Red1 (Red1531–827). Thus, the DNA-encircling checkpoint complex 9-1-1 appears to be connected to the SC through Red1, suggesting that Red1 is the missing downstream factor for meiotic checkpoint signaling.
Fig. 1.
Red1 interacts with two 9-1-1 subunits. (A) Mapping of Red1 domains interacting with Mec3 and Ddc1 in two-hybrid assays. Mec3 binds Red1 at a region from amino acids 531–551 and Ddc1 at a region from amino acids 729–751. (Right) Two-hybrid interactions identified on selective media (−His) of fusions with an activating domain (AD) or DNA binding domain (BD) are shown. The white colony color is indicative of better growth. Images were taken after 3 d at 30 °C. (Left) Mec3 and Ddc1 binding sites are shown in gray in the diagram. Growth on complete media is shown as a control. (B) Comparison of the 9-1-1 binding domains in Red1 with the bona fide PIP box consensus core motif. Amino acids altered to alanine are shown in red. (C) Amino acid replacements in the Mec3 binding site of Red1 abolish Mec3 interaction, as indicated by two-hybrid assays (3 d at 30 °C). (D) Similarly, the amino acid replacement in the Ddc1 binding site of Red1 abolishes Ddc1 interaction.
Red1 Bears Two 9-1-1 Subunit-Specific Binding Motifs.
9-1-1 associates tightly with chromatin, and perhaps because the 9-1-1–Red1 interaction is transient or requires a fully assembled DNA-bound 9-1-1 complex, we could not detect a significant association of 9-1-1 with the Red1 full-length version or truncated Red1 fragments in coimmunoprecipitation or GST pull-down experiments. Indeed, previous studies indicated that only a small fraction of Red1 colocalizes with 9-1-1, as judged by immunofluorescence microscopy of chromosome spreads (5). To circumvent this problem, we decided to map the binding sites by two-hybrid assays and to confirm the interaction in vivo by mutational studies. Interestingly, we identified two 9-1-1 interaction sites in Red1, which are specific for individual 9-1-1 subunits. Whereas the Mec3 interaction site is located between residues 531 and 551, the Ddc1 interaction site resides between residues 729 and 751 of Red1 (Fig. 1 A and B). Two-hybrid assays also confirmed Red1 oligomerization involving Red1’s C-terminal tail (residues 703–827) (19) (Fig. 1A), suggesting that the C-terminal region of Red1 is particularly crucial for Red1 function.
Previous studies indicated that 9-1-1 interacts with partner proteins via hydrophobic residues (20–22), but no consensus element has been identified so far. Given the overall similarity of 9-1-1 to PCNA (3), we speculated that 9-1-1 might interact with Red1 similar to how PCNA associates with its alternative partners. Most PCNA-interacting proteins (PIPs) use a hydrophobic so-called “PIP box” that fits into a hydrophobic pocket of a PCNA subunit (23). The core element of a PIP box is the sequence QxxΨ (with Ψ being the residues L, M, V, and I), but additional residues flanking this element often crucially contribute to PCNA binding. Because we identified QxxΨ motifs in both Red1 segments (Fig. 1B), we focused on these sequences for further analysis. Notably, when we altered residues Q537 and V540 of Red1 to alanines, two-hybrid interaction to Mec3 was completely lost but Ddc1 interaction was unaltered (Fig. 1C and Fig. S1B). Conversely, changing Q740 and I743 (or I743 alone) to alanine abolished interaction of Ddc1 but not of Mec3 (Fig. 1D and Fig. S1C). However, because Q740 was not essential for Ddc1 interaction and both 9-1-1 binding elements did not bind PCNA (Fig. 1A), these sites, despite their similarity to PIP boxes, do not function as bona fide PIP boxes (i.e., they are specific to 9-1-1). Importantly, although Red1 dimerization involves a similar region, the Red1 mutant variant defective in Ddc1 association (I743A) was still proficient in Red1-Red1 binding (Fig. 1D and Fig. S1C), demonstrating that this mutant variant is specifically defective in Ddc1 interaction and that overall protein expression and folding are unaltered. Similarly, the interaction with Hop1 is unlikely to be affected by this alteration because Hop1 binding involves a more N-terminal domain of Red1 (19).
9-1-1–Red1 Interaction Is Critical for Meiosis.
To confirm the 9-1-1–Red1 interaction in vivo and to study its functional significance, we expressed the Red1 variants defective in interaction with Mec3 (Q537A, V540A; termed Red1−Mec3), Ddc1 (I743A; termed Red1−Ddc1), or both (Red1−Mec3,−Ddc1) as the only source of Red1 from the diploid genome. Interestingly, when we scored for the viability of the respective spores as a measure for meiosis competence, we found a dramatic reduction with mutants expressing the Red1 variants that are defective in Ddc1 interaction (Red1−Ddc1 and Red1−Mec3,−Ddc1) (Table S1). By contrast, spore viability was only moderately affected if Red1 fails to associate exclusively with the Mec3 subunit of the 9-1-1 complex. These data suggest that successful meiosis requires a physical association of the 9-1-1 checkpoint complex (particularly its Ddc1 subunit) with the SC protein Red1. Because even deletion of RED1 does not prevent genome-wide meiotic DSB production (24), disruption of the 9-1-1–Red1 interaction may interfere with later functions in the meiotic pathway. In fact, because the spore viability of Red1−Ddc1-expressing cells was as low as for cells in which checkpoint activation is absent [e.g., 9-1-1–deficient cells (10), Table S1; hop1SCD (17)] and even lower than for cells that are unable to form SCs (e.g., Δzip1 mutants, Table S1), we hypothesized that 9-1-1–Red1 interaction might be crucial for checkpoint activation, SC formation, or both.
9-1-1–Red1 Interaction Is Essential for Meiotic Checkpoint Signaling.
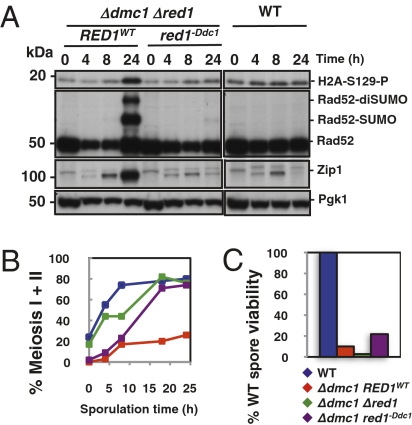
We next asked whether 9-1-1–Red1 interaction plays a direct role in the meiotic checkpoint. To this end, we additionally deleted the gene for the meiotic recombinase Dmc1 because cells deficient in this enzyme accumulate resected DSBs and recombination intermediates, which, in turn (when WT Red1 is expressed), leads to checkpoint activation (25, 26). We have shown previously that meiotic checkpoint activation can be followed not only by monitoring the induction of histone H2A serine-129 phosphorylation (equivalent to mammalian γ-H2AX) but by monitoring that of the SUMOylated form of Rad52, which accumulates dramatically in Δdmc1 cells (26). When we used these assays, we found no significant defects with mutants expressing the Red1 variant deficient in Mec3 binding (red1−Mec3) (Fig. S2A). By contrast, red1−Ddc1 mutants completely failed to induce the meiotic checkpoint (Fig. 2A), comparable to mutants that are deficient in 9-1-1 (10). We also monitored Zip1 levels because pachytene-arrested cells are known to accumulate SCs and its proteins because they essentially do not progress further in the meiotic cell cycle (4). Indeed, Zip1 accumulated to high levels in Δdmc1 cells that express WT RED1 (Fig. 2A). By contrast, if the red1−Ddc1 allele was expressed in these cells, Zip1 expression was induced normally but the protein did not accumulate further (Fig. 2A). Thus, we conclude that Zip1 does not accumulate, because the checkpoint signaling cascade is inactive in red1−Ddc1 cells.
Fig. 2.
9-1-1–Red1 interaction is essential for meiotic checkpoint signaling. (A) Ddc1 binding-deficient Red1 variant (Red1−Ddc1) reverts meiotic checkpoint arrest of Δdmc1 deletion strains. Extracts of synchronously sporulating cells were made at the indicated times and probed by Western blot analysis for Zip1 and Pgk1 expression and in parallel for phosphorylated H2A (equivalent to mammalian γ-H2AX) and Rad52 SUMOylation as measures for checkpoint activation. The Y1083, CE571, and CE579 strains were used. (B) Meiotic (nuclear) divisions monitored by DAPI staining of chromosomal DNA. Samples from synchronously growing cultures were harvested at the indicated times and fixed in ethanol for subsequent DAPI staining. Symbols are identical to those used in C. (C) For germination assays, spores were isolated and zymolase-treated after 72 h in sporulation media and then plated on YDP plates at 30 °C, as described previously (43). For B and C, the Y1083, Y2031, CE571, and CE579 strains were used.
A distinctive phenotype of meiotic checkpoint mutants (e.g., deletions in 9-1-1 genes) is that they are able to progress through meiosis even when meiotic recombination is defective (e.g., in Δdmc1 cells) but with the consequence of severe deficiencies in spore viability (10). In fact, we found that Δdmc1 red1−Ddc1 cells (in contrast to Δdmc1 RED1 cells) also fail to arrest and undergo nuclear divisions, as monitored by DAPI staining (Fig. 2B). Moreover, although red1−Ddc1 cells (proficient in DMC1) progressed through meiosis similar to WT cells and showed no defects in meiotic nuclear divisions (Fig. S2B), they exhibited dramatically reduced spore viability, as indicated by a germination assay (Table S1 and Fig. 2C). Together, these data indicate that Red1 is essential for the meiotic checkpoint pathway, apparently because checkpoint signaling is directly transmitted through interaction of 9-1-1 (Ddc1) with the SC protein Red1. This finding puts Red1 directly downstream of 9-1-1 in the meiotic checkpoint cascade, suggesting that Red1 fulfills a TopBP1-like function during meiosis.
9-1-1–Red1 Interaction Is Essential for SC Formation.
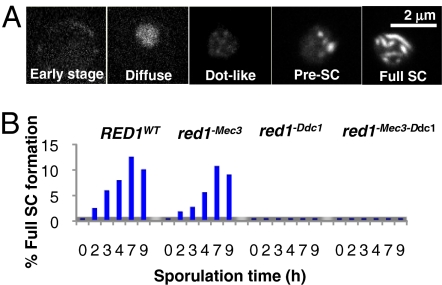
Because the red1−Ddc1 mutation has a very strong negative effect on spore viability (Table S1) and Red1 is a structural component of the SC, we speculated that the interaction of Red1 with 9-1-1 might also be important for SC formation. To test this hypothesis, we monitored the different stages of SC formation by spinning disk microscopy using strains that express a GFP-tagged Zip1 variant. Because GFP tagging to either end of Zip1 significantly inactivates the protein, we used a variant that harbors GFP embedded within the protein's central coiled-coil region (27). Judging by the spore viability of this strain, this Zip1 variant is almost as functional as the WT protein (Table S1), and full SCs are formed already 2 h after sporulation induction (Fig. 3A and B and Fig. S3). When we assayed for SC formation using GFP-tagged Zip1 (in a DMC1WT background), we noticed a moderate defect in red1−Mec3 mutants but a virtually complete loss of complete SCs in red1−Ddc1 mutants and red1 mutants defective in interaction with both 9-1-1 subunits (red1−Mec3,−Ddc1) (Fig. 3B). Interestingly, although preassemblies of SCs (pre-SCs) were detectable at similar levels, fully formed SCs were not assembled if Red1 failed to bind 9-1-1 via Ddc1. Notably, the failure to form SCs was not attributable to the slightly reduced Red1 levels (Fig. S4) because even higher expression of the respective mutant (red1−Ddc1) could not trigger SC formation and checkpoint signaling. Thus, we conclude that the interaction of Red1 with the Ddc1 subunit of 9-1-1 is essential not only for meiotic checkpoint signaling but for SC formation. The data also suggest that this interaction ensures that the two processes are directly coupled.
Fig. 3.
9-1-1–Red1 interaction is essential for SC formation. (A) SC formation assay. SCs were visualized using GFP-tagged Zip1 and spinning disk microscopy, as described in Fig. S3. Maturation of SCs was categorized in the indicated classes (early stage, diffuse, dot-like, pre-SCs, and full SCs). In the quantified assay, only full SCs and pre-SCs were distinguished and counted. (B) Full SCs and pre-SCs in WT and 9-1-1 binding-deficient Red1 variants (red1−Mec3, red1−Ddc1, and red1−Mec3,−Ddc1) were monitored, as described in A. Shown are mean values from five independent experiments. For each time point, more than 100 cells were analyzed. The strains used were CE774, CE775, CE776, and CE777.
Red1 SUMOylation in Meiosis.
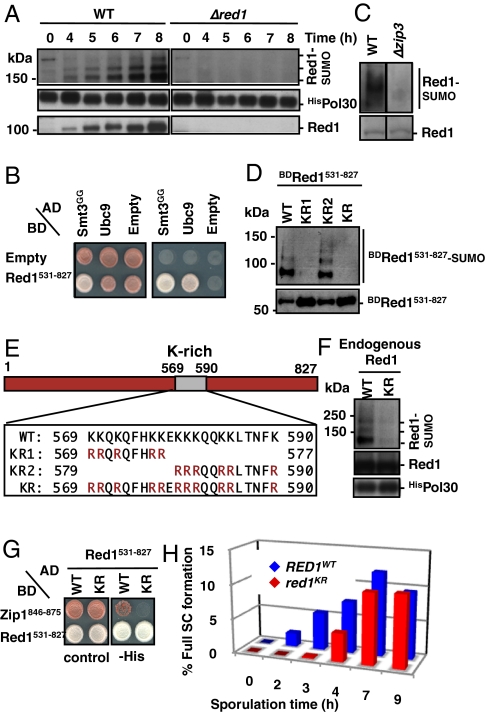
Several lines of evidence indicate that protein modification by the ubiquitin-related protein SUMO plays a role in meiosis (28–34). In particular, the SC itself seems to contain SUMO because the SC can be stained with SUMO-specific antibodies along its entire axis (28). In fact, recent findings suggest that SC elements may be sandwiched together by SUMO chains, which seem to bind both Zip1 and Red1 (34). When we isolated SUMO conjugates by a previously established protocol (26, 35), we detected Red1 as a SUMOylation substrate, suggesting another layer of control by SUMO (29). By using Red1-specific antibodies, we followed the level of Red1 and its SUMOylated form after synchronous meiosis induction. Corresponding to the structural role of Red1 in SCs, Red1 levels strongly rose on sporulation induction, when SCs are known to form (Fig. 4A). Concomitantly, SUMOylated Red1 species also accumulated (Fig. 4A), indicating that the SUMO modification may be directly linked to SC formation. As expected for this reversible modification, only a small fraction of Red1 was SUMO-modified at steady state, suggesting that SUMOylation of Red1 may be transient. Additionally, we found that the carboxyl (C)-terminal domain of Red1 (e.g., residues 531–827) particularly binds SUMO (yeast Smt3) and the SUMO-conjugating enzyme Ubc9 in two-hybrid assays (Fig. 4B). Interestingly, SUMOylation of Red1 was almost absent in Δzip3 null mutants (Fig. 4C), indicating that Zip3, a meiosis-specific potential SUMO ligase (29), might be directly responsible for the bulk of Red1 SUMOylation. Moreover, the ordered ladder-like pattern of SUMO-modified Red1 species suggests that Red1 might be modified by a poly-SUMO chain.
Fig. 4.
Red1 SUMOylation during meiosis mediates timing of SC formation. (A) SUMOylation of endogenous Red1. Diploid homozygous SK1 WT (CE382) or Δred1 (CE387) cells with an integrated version of HisSUMO were released into synchronous sporulation, and cell extracts were harvested after the indicated times. HisSUMO conjugates (monitored by Ni-NTA pull-down, followed by Western blotting using an anti-Red1 antibody) detect Red1 species carrying one, two, or more SUMO moieties. To control for pull-down efficiency, HisPol30 (PCNA)-expressing cultures were mixed with the meiotic cultures before lysis and pull-down, and HisPol30 was detected by Western blot analysis using an anti-Pol30 antibody (Bottom, Red1 input levels). (B) Red1 interacts with SUMO (Smt3) and Ubc9. For the two-hybrid assay, cells were transformed with respective activation domain (AD) and binding domain (BD) fusions and spotted on selective media and were grown for 3 d at 30 °C. (C) Red1 SUMOylation depends in vivo on the SUMO E3 ligase Zip3 (zip3 strain, CE566; done as in A; Lower, Red1 input levels). (D) Identification of SUMO-acceptor sites in Red1 using DF5 cells expressing HisSUMO and BDRed1531–827, in which Red1 sequences were derived from WT Red1 or Red1 variants carrying KR exchanges (KR1, KR2, KR; defined in E). SUMO conjugates were isolated by Ni-NTA pull-down from lysates and detected by Western blotting using anti-BD monoclonal antibodies (Santa Cruz) (Lower, Red1 input levels). (E) Diagram of Red1 indicating a lysine-rich (K-rich) region (amino acids 569–590). Red1 variants harboring KR replacements of all lysines within regions of amino acids 569–577 (designated KR1), amino acids 579–590 (designated KR2), and amino acids 569–590 (designated KR) are indicated. (F) SUMOylation of endogenous Red1KR. Ni-NTA pull-down of SUMO conjugates from lysates of SK1 strains (8 h after induction of sporulation) expressing Red1 WT (CE659) or the Red1KR (CE662) variant from the genome using RED1 promoter and terminator elements. Western blot analysis using an anti-Red1 antibody. Control of pull-down efficiency as in A (HisPol30). (Middle) Red1 input levels are shown. (G) SIM-containing C-terminal region of Zip1 specifically binds SUMOylation-proficient WT Red1 but not Red1KR. Two-hybrid interactions of a C-terminal fragment of Zip1 (amino acids 846–875) with a Red1 fragment (amino acids 531–827) derived from WT Red1 or Red1KR identified on selective media (−His). (H) Full SC formation in WT (CE774) and red1KR mutant (CE778) was monitored as described in Fig. 3A and Fig. S3. Shown are mean values from six independent experiments. For each time point, more than 100 cells were analyzed.
Red1 SUMOylation Sustains Normal Meiosis.
To address the significance of the SUMO modification, we searched for the SUMO acceptor lysine residues in Red1. We assumed that cells expressing a Red1 variant that cannot be modified by SUMO would allow us to test the previously suggested hypothesis that the interaction of specifically SUMOylated Red1 with Zip1 is essential for SC assembly (29). Because the steady-state levels of SUMOylated Red1 are very low during meiosis (about 1–5% of Red1 protein is modified), a direct analysis of SUMOylation sites in endogenous Red1 was not practical. However, we noticed that a two-hybrid construct bearing the C-terminal domain of Red1 (residues 531–827, termed Red1531–827) was strongly SUMOylated (apparently independent of a ligase) in yeast cells if His-tagged SUMO (HisSUMO/Smt3) was overexpressed (Fig. 4D). Taking advantage of this finding, we introduced lysine-to-arginine (KR) replacements in this region and assayed for Red1 SUMOylation after isolation of HisSUMO-conjugates by Ni-NTA pull-downs. A particular lysine-rich (K-rich) region is located between residues 569 and 590 of Red1 (Fig. 4E), and several lysine residues within this domain indeed seem to serve as acceptor sites for SUMOylation in vivo. Additional mutational analysis revealed that the most frequently used SUMOylation sites are located between residues 569 and 577 and lack consensus ψKXE SUMOylation motifs but that other lysines further C-terminal to this region also contribute to Red1 SUMOylation in vivo (Fig. 4 D and E). In fact, changing all lysine residues to arginines in this region, although potentially interfering with any other type of modification, dramatically reduced the SUMOylation level of this Red1-fusion protein to less than 10% compared with modification of the WT protein (Fig. 4D, lane KR).
Importantly, when we introduced these changes into full-length Red1 and expressed the protein from its endogenous genetic locus (designated Red1KR), SUMOylation was equally strongly reduced (Fig. 4F). This indicates that the major SUMOylation sites of endogenous Red1 also reside within the K-rich region of Red1’s C-terminal tail and that the modifications can occur at multiple, perhaps alternative, sites. To address the physiological relevance of Red1 SUMOylation during meiosis, we first determined the viability of spores of the SUMOylation-defective red1KR mutants. Notably, the deficiency in SUMO modification had no significant effect on the protein level (Fig. 4F and Fig. S4), did not result in a checkpoint arrest (Fig. S2B), and maintained a checkpoint arrest in a Δdmc1 deletion background similar to RED1WT (Fig. S2A). By contrast, only about 40% (compared with 89% in the relevant WT strain) of the mutant spore population was able to germinate and give rise to viable cells (Table S1). Thus, we conclude that SUMOylation of Red1 is crucial for normal meiosis.
Red1 SUMOylation Mediates SC Formation Timing.
Because SCs contain SUMO and the SC protein Red1 is a SUMOylation substrate, we speculated that the modification might directly affect SC formation. Notably, Zip1 harbors a SUMO-interacting motif (SIM); therefore, it has been suggested that SC formation may directly arise through binding of Zip1 to SUMOylated lateral element proteins or to poly-SUMO chains (29, 34). Using two-hybrid assays, we confirmed that Zip1 can dimerize (oligomerize) and bind SUMO and that it also binds Red1 (Fig. S5 A–C). Importantly, the Zip1-Red1 interaction involved the C-terminal domain of Zip1 that harbors the SIM, and binding of this Zip1 domain to Red1 did not occur to significant levels if the SUMOylation-defective Red1 variant was used (Fig. 4G). Moreover, Red1 binds the de-SUMOylation enzyme Ulp2, but it only does so when the SUMOylation acceptor lysines in Red1 are present (Fig. S5D). Together, these findings suggest that Red1-SUMOylation may indeed be critical for promoting Zip1-Red1 interaction, and thus also for SC formation, as has been suggested previously (29). To test this model, we monitored the progressing steps of SC formation again by spinning disk microscopy of strains that express GFP-tagged Zip1 (Fig. 3A and Fig. S3). By comparing the SUMO-deficient mutant red1KR with Red1 WT strains, we found that although the red1KR strain is capable of forming full SCs, their formation is significantly delayed by several hours (Fig. 4H). This indicates that Red1 SUMOylation is not essential for the assembly of SCs, as has been suggested previously (29), but that it rather promotes the initiation of SC formation, thereby securing timely SC assembly. Apparently, this delay has a profound influence on meiosis progression, as indicated by the reduced spore viability of the same strain (Table S1). Moreover, the Red1 variant that is completely deficient in 9-1-1 interaction (red1−Mec3,−Ddc1) is still modified by SUMO (Fig. S5E), demonstrating that 9-1-1 binding is not a prerequisite for Red1 SUMOylation. Importantly, however, the grade of the SC deficiencies of the respective red1 mutants (Fig. 3B) corresponded well with their deficiencies in spore viability (Table S1). Together, these data indicate that Red1 might be the direct downstream partner of 9-1-1 and that meiotic checkpoint signaling, SC formation, and the correct timing of SC formation, are all directly linked to the SC protein Red1.
Discussion
In this study, we uncovered detailed molecular mechanisms for two major functions of meiosis: meiotic checkpoint signaling and SC formation. Both processes are directly functionally linked to the lateral element protein Red1 through covalent and noncovalent protein-protein interactions. Whereas the bulk of Red1 appears to play a structural role within the SC, only a small subfraction of Red1 is apparently involved in signaling. A central finding of this work is that Red1 acts as a downstream effector of 9-1-1 in meiotic checkpoint signaling. Intriguingly, Red1 appears to associate with two subunits of the ring-shaped heterotrimeric complex, by which means undesired interactions of 9-1-1 (e.g., with Dpb11/TopBP1) are perhaps effectively blocked. Also interesting is the finding that 9-1-1 binding involves sequences reminiscent of PIP boxes, which are usually found in PIPs. Importantly, however, although PCNA and 9-1-1 are structurally highly related (3), these regions of Red1 bind 9-1-1 but not PCNA, thereby confirming that the two related clamps operate in distinct pathways. Moreover, the two binding sites in Red1 are 9-1-1 subunit-specific, suggesting that different 9-1-1 subunits may perhaps play distinct roles in DNA damage pathways. Although we identified the binding of Red1 with 9-1-1 by two-hybrid assays, we assume that this interaction is direct and occurs in vivo, given the subunit-specific phenotypes of the 9-1-1–deficient Red1 mutants and the presence of the two 9-1-1 subunit-specific PIP box-like sequences in Red1.
Because we found that intact 9-1-1 binding sites in Red1 are essential for 9-1-1–dependent meiotic checkpoint signaling, we propose that the SC protein Red1 may take the position of TopBP1/Dpb11 in the meiotic checkpoint pathway (Fig. S6). Interestingly, the interaction essential for checkpoint signaling for both Red1 and TopBP1/Dpb11 is the Ddc1 subunit of 9-1-1, suggesting that the two proteins might indeed be alternative 9-1-1 binding partners. This model has important implications. Linking 9-1-1 to Red1 may further trigger downstream signaling events that take place at the SC, such as the phosphorylation of Red1 and Hop1, which, in turn, may recruit the kinase Mek1, which might recognize the phosphorylated forms of the proteins by means of its forkhead-associated domain. Notably, Mek1 plays a major role in ensuring interhomolog vs. intersister recombination (36), perhaps partially through its coupling to the SC. This phenomenon, often called “barrier-to-sister chromatid recombination,” leads to high crossover rates and high spore viabilities. Our finding that experimental disruption of 9-1-1–Red1 interaction leads to a high number of inviable spores supports this model.
Another advantage of a 9-1-1–Red1 interaction is the possibility of connecting checkpoint signaling to SC formation. Mutants defective in checkpoint signaling are known to be at least partially defective in SC formation (17, 37, 38). Our data now show that this functional interconnection is brought about through the physical 9-1-1–Red1 interaction. In fact, the finding that the very same mutant (red1−Ddc1) is completely defective in both processes shows how exquisitely these two essential meiotic events are coupled.
We also discovered that Red1 is subject to another layer of control. We found that yeast Red1 is SUMOylated during early meiosis concomitantly with the appearance of SCs. Interestingly, when we looked in human 293T cells that express tagged versions of either SUMO1 or SUMO2 (humans possess three SUMO variants), we found that Sycp3 (alias Scp3, Cor1), a possible functional analogue of yeast Red1 (39), which is known to bind human Ubc9 (40), is modified specifically by SUMO2 (Fig. S7). Thus, SUMOylation of lateral element SC proteins (Red1 and Sycp3) appears to be a conserved feature. Interestingly, a SIM-containing C-terminal fragment of the central element protein Zip1 from yeast specifically interacts with the SUMO-modified form of Red1, suggesting that SC zipping is stimulated by Red1 SUMOylation. This finding draws interesting parallels to other SUMO-stimulated interactions, for example, the binding of PCNA to the SIM-containing Srs2 helicase, an interaction that is greatly stimulated by PCNA SUMOylation (41). Notably, strongly reduced Red1 SUMOylation (in red1KR cells) causes a delay in complete SC formation by several hours (without the occurrence of any polycomplexes), which, in turn, causes reduced spore viability. Interestingly, this effect on SC assembly by the red1KR mutant is much milder than the effect observed in zip3 deletion strains or in zip1 mutants defective in SUMO binding (29). This suggests that the putative SUMO ligase Zip3 and the SIM motif of Zip1 may have additional roles in meiosis.
An interesting open question is whether SUMO-modified Red1 recruits Zip1 along the entire chromosome. The staining of whole chromosomes with anti-SUMO antibodies indeed suggests a broad role for SUMO in these complexes (28), perhaps as a “zipping glue.” However, the low steady-state level of modified Red1 and our finding that Red1 SUMOylation triggers SC formation timing (rather than SC formation itself) argue for a regulatory role instead. In our model, Red1-SUMO initiates zipping by helping to recruit the initial Red1-Zip1 associations, perhaps analogous to the role of the ends of zippers in clothing. Firm interaction between the SC elements may then be completed by poly-SUMO chains that seem to bind both Zip1 and Red1 (34). Given the recent finding that Red1 binds SUMO chains via two SIMs in the C-terminal domain between residues 710 and 770 (34), SC assembly or disassembly might involve an intramolecular interaction between SUMO chains added to Red1’s lysine-rich region and the two SIMs at its C terminus. Notably, Red1 SUMOylation is stimulated by the putative meiotic ligase Zip3, which is known to associate with the MRX complex and concentrates at the first meiotic DSBs (42). Zip3 also binds the central element SC protein Zip1 (42), and these proteins, together with SUMO, first localize to recombination sites before Zip1 and SUMO spread along the entire chromosome (28). Thus, Zip3 may SUMOylate only Red1 molecules that are close to the DSBs, thereby perhaps stimulating the first specific Red1-Zip1 interactions. Because Zip3 also associates with the recombinase Rad51 (42), the SUMO ligase might be guided by homology search to the homologous chromosome and initiate SUMOylation of Red1 also at the recipient homologous chromosome. This model seems particularly attractive because it suggests a mechanism of how the information for timely zipping is transmitted from one chromosome to its homolog.
Materials and Methods
Yeast strains used are listed in Table S2. Strains are isogenic to strain SK1 (Y1083, for all meiotic assays), DF5 (Fig. 4D), or PJ69-7A (for all yeast two-hybrid assays). Detailed methods for the construction of mutants and strains, synchronous meiotic time course experiments, spore viability, germination and meiotic nuclear division assays, live cell microscopy, and protein techniques can be found in SI Text.
Supplementary Material
Acknowledgments
We thank J. Wächter, A. Strasser, and U. Cramer for technical assistance; G. Karras, S. Müller (Max-Planck Institute of Biochemistry), and I. Psakhye for comments on the manuscript; D. Kaback (University of Medicine and Dentistry of New Jersey), S. Müller, and M. Knop (European Molecular Biology Laboratory) for materials; and T. Walther for help with microscopy. This work was supported by the Max Planck Society (S.J.), Deutsche Forschungsgemeinschaft, Fonds der chemischen Industrie, Center for Integrated Protein Science Munich, and Role of Ubiquitin and Ubiquitin-like Modifiers in Cellular Regulation European Union Network of Excellence. C.S.E. was supported by a Ph.D. fellowship of the Studienstiftung des Deutschen Volkes.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004248107/-/DCSupplemental.
References
- 1.Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 2.Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- 3.Doré AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex—Implications for clamp loading and regulation. Mol Cell. 2009;34:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 4.Roeder GS, Bailis JM. The pachytene checkpoint. Trends Genet. 2000;16:395–403. doi: 10.1016/s0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- 5.Hong EJ, Roeder GS. A role for Ddc1 in signaling meiotic double-strand breaks at the pachytene checkpoint. Genes Dev. 2002;16:363–376. doi: 10.1101/gad.938102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 7.Prieler S, Penkner A, Borde V, Klein F. The control of Spo11’s interaction with meiotic recombination hotspots. Genes Dev. 2005;19:255–269. doi: 10.1101/gad.321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puddu F, et al. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lydall D, Nikolsky Y, Bishop DK, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- 11.Cartagena-Lirola H, Guerini I, Manfrini N, Lucchini G, Longhese MP. Role of the Saccharomyces cerevisiae Rad53 checkpoint kinase in signaling double-strand breaks during the meiotic cell cycle. Mol Cell Biol. 2008;28:4480–4493. doi: 10.1128/MCB.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowling EL, Maloney DH, Fogel S. Meiotic recombination and sporulation in repair-deficient strains of yeast. Genetics. 1985;109:283–302. doi: 10.1093/genetics/109.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber L, Byers B. A RAD9-dependent checkpoint blocks meiosis of cdc13 yeast cells. Genetics. 1992;131:55–63. doi: 10.1093/genetics/131.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perera D, et al. TopBP1 and ATR colocalization at meiotic chromosomes: Role of TopBP1/Cut5 in the meiotic recombination checkpoint. Mol Biol Cell. 2004;15:1568–1579. doi: 10.1091/mbc.E03-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu L, Weiner BM, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- 16.Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 17.Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132:758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 18.Wan L, de los Santos T, Zhang C, Shokat K, Hollingsworth NM. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol Biol Cell. 2004;15:11–23. doi: 10.1091/mbc.E03-07-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woltering D, et al. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol Cell Biol. 2000;20:6646–6658. doi: 10.1128/mcb.20.18.6646-6658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo Z, et al. Comprehensive mapping of the C-terminus of flap endonuclease-1 reveals distinct interaction sites for five proteins that represent different DNA replication and repair pathways. J Mol Biol. 2008;377:679–690. doi: 10.1016/j.jmb.2007.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi G, et al. Physical and functional interactions between MutY glycosylase homologue (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan X, et al. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res. 2007;35:2463–2472. doi: 10.1093/nar/gkm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 25.Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 26.Sacher M, Pfander B, Hoege C, Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat Cell Biol. 2006;8:1284–1290. doi: 10.1038/ncb1488. [DOI] [PubMed] [Google Scholar]
- 27.Scherthan H, et al. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:16934–16939. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hooker GW, Roeder GS. A Role for SUMO in meiotic chromosome synapsis. Curr Biol. 2006;16:1238–1243. doi: 10.1016/j.cub.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 29.Cheng CH, et al. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Carvalho CE, Colaiácovo MP. SUMO-mediated regulation of synaptonemal complex formation during meiosis. Genes Dev. 2006;20:1986–1992. doi: 10.1101/gad.1457806. [DOI] [PubMed] [Google Scholar]
- 31.Cheng CH, Lin FM, Lo YH, Wang TF. Tying SUMO modifications to dynamic behaviors of chromosomes during meiotic prophase of Saccharomyces cerevisiae. J Biomed Sci. 2007;14:481–490. doi: 10.1007/s11373-007-9176-0. [DOI] [PubMed] [Google Scholar]
- 32.Spirek M, et al. SUMOylation is required for normal development of linear elements and wild-type meiotic recombination in Schizosaccharomyces pombe. Chromosoma. 2010;119:59–72. doi: 10.1007/s00412-009-0241-5. [DOI] [PubMed] [Google Scholar]
- 33.Macqueen AJ, Roeder GS. Fpr3 and Zip3 ensure that initiation of meiotic recombination precedes chromosome synapsis in budding yeast. Curr Biol. 2009;19:1519–1526. doi: 10.1016/j.cub.2009.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin FM, Lai YJ, Shen HJ, Cheng YH, Wang TF. Yeast axial-element protein, Red1, binds SUMO chains to promote meiotic interhomologue recombination and chromosome synapsis. EMBO J. 2010;29:586–596. doi: 10.1038/emboj.2009.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 36.Niu H, et al. Mek1 kinase is regulated to suppress double-strand break repair between sister chromatids during budding yeast meiosis. Mol Cell Biol. 2007;27:5456–5467. doi: 10.1128/MCB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grushcow JM, et al. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics. 1999;153:607–620. doi: 10.1093/genetics/153.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailis JM, Roeder GS. Synaptonemal complex morphogenesis and sister-chromatid cohesion require Mek1-dependent phosphorylation of a meiotic chromosomal protein. Genes Dev. 1998;12:3551–3563. doi: 10.1101/gad.12.22.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schalk JA, et al. Localization of SCP2 and SCP3 protein molecules within synaptonemal complexes of the rat. Chromosoma. 1998;107:540–548. doi: 10.1007/s004120050340. [DOI] [PubMed] [Google Scholar]
- 40.Tarsounas M, Pearlman RE, Gasser PJ, Park MS, Moens PB. Protein-protein interactions in the synaptonemal complex. Mol Biol Cell. 1997;8:1405–1414. doi: 10.1091/mbc.8.8.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 42.Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102:245–255. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- 43.Herman PK, Rine J. Yeast spore germination: A requirement for Ras protein activity during re-entry into the cell cycle. EMBO J. 1997;16:6171–6181. doi: 10.1093/emboj/16.20.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.