Abstract
Hepatitis C virus (HCV) infection is associated with dysregulation of both lipid and glucose metabolism. As well as contributing to viral replication, these perturbations influence the pathogenesis associated with the virus, including steatosis, insulin resistance, and type 2 diabetes. AMP-activated protein kinase (AMPK) plays a key role in regulation of both lipid and glucose metabolism. We show here that, in cells either infected with HCV or harboring an HCV subgenomic replicon, phosphorylation of AMPK at threonine 172 and concomitant AMPK activity are dramatically reduced. We demonstrate that this effect is mediated by activation of the serine/threonine kinase, protein kinase B, which inhibits AMPK by phosphorylating serine 485. The physiological significance of this inhibition is demonstrated by the observation that pharmacological restoration of AMPK activity not only abrogates the lipid accumulation observed in virus-infected and subgenomic replicon-harboring cells but also efficiently inhibits viral replication. These data demonstrate that inhibition of AMPK is required for HCV replication and that the restoration of AMPK activity may present a target for much needed anti-HCV therapies.
Hepatitis C virus (HCV) infection is a major cause of chronic liver disease affecting an estimated 3% of the world's population (1). HCV is a positive-stranded RNA virus with a 9.6-kb genome that encodes a large polyprotein, translated in a cap-independent fashion and processed by cellular and viral proteases to produce 10 mature proteins: core, envelope proteins E1/E2, a cation channel p7, and six nonstructural proteins, NS2 to NS5B (2). HCV infection is associated with accumulation of intracellular lipid, which manifests as steatosis (fatty liver) in patients and is a predictor of serious liver disease (3). Furthermore, data from cell-culture studies have shown that inhibition of cellular lipid biosynthesis is detrimental to virus replication (4, 5), consistent with a role for lipid droplets in both viral genome replication and assembly of infectious particles (6). Although HCV infection has been shown to activate genes such as peroxisome proliferator-activated receptors (PPARα/γ/δ) and sterol regulatory element-binding protein-1 (SREBP-1) (7–9) that increase lipid biogenesis and inhibit mitochondrial β-oxidation, the mechanisms underpinning this regulation remain obscure. Intriguingly, HCV infection also is associated with the development of insulin resistance and type 2 diabetes. This association may result in part from dysregulation of lipid metabolism, but recent data also have pointed to direct effects of HCV on hepatic glucose uptake, again by uncharacterized mechanisms (10).
A key regulator of both lipid and glucose metabolism is AMP-activated protein kinase (AMPK). AMPK is a heterotrimeric complex consisting of α, β, and γ subunits and has been referred to as a metabolic “master switch,” because its activity is regulated by the energy status of the cell. AMPK responds to ATP depletion by detecting changes in the AMP:ATP ratio (11). AMPK is active only after phosphorylation of the α subunit at a threonine residue within the kinase domain (T172) by upstream kinases, most important of which is a heterotrimeric complex between serine/threonine kinase 11 (LKB1), sterile 20 protein-related adaptor, and mouse protein 25 (12) (Fig. S1). T172 also can be phosphorylated by the Ca2+ sensing kinase, CAMKKβ (13, 14). AMP promotes T172 phosphorylation by inhibiting dephosphorylation at this residue, an effect antagonized by ATP (15, 16). Once activated, AMPK phosphorylates multiple substrate proteins to effect general inhibition of ATP-consuming metabolic pathways and simultaneous activation of ATP-generating pathways, restoring ATP levels (11). These effects are particularly important in regulating liver metabolism, where activation of AMPK augments fatty acid oxidation and decreases glucose output and cholesterol and triglyceride synthesis (11, 17).
Here we demonstrate that in cells harboring HCV subgenomic replicons or infected with HCV, AMPK T172 phosphorylation was inhibited. This inhibition corresponded with an increase in phosphorylation by the serine/threonine kinase, AKT, at an inhibitory serine (S485). Furthermore, treatment with AMPK agonists effectively inhibited both viral genome replication and lipid accumulation, suggesting that restoration of AMPK activity may provide a target for much needed anti-HCV therapies.
Results
AMPK Activity Is Inhibited in HCV Subgenomic Replicon-Harboring Cells.
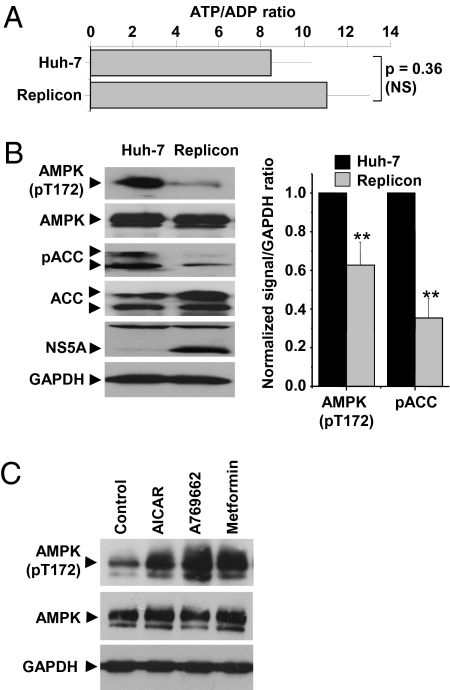
Viral infection might be expected to lead to an increase in AMP concentration because of increased energy demands upon the host cell. Cellular AMP concentrations are very low and therefore difficult to measure, but the ADP:ATP ratio changes in concert with AMP:ATP ratio (because of the adenylate kinase reaction) and can be used as a surrogate for AMP:ATP (17). Surprisingly, when we compared the ATP/ADP ratios in Huh-7 cells and in cells stably harboring an HCV genotype 1b culture-adapted subgenomic replicon (18) (hereafter termed “replicon cells”), we observed no significant difference between the two cell populations (Fig. 1A). We speculated that this observation might result from the activation of AMPK in replicon cells, thereby restoring the energy balance in these cells. Active AMPK phosphorylates a large number of targets, including the two isoforms of acetyl-CoA carboxylase (ACC1/2). Importantly, phosphorylation of ACC by AMPK inhibits enzymatic activity (17), decreasing cellular fatty acid synthesis. We therefore assessed AMPK activity in replicon cells by immunoblot analysis with antibodies directed against either the activated T172 phosphorylated form of AMPK or phosphorylated ACC. This analysis revealed that replicon cells displayed a 40% decrease in AMPK T172 phosphorylation and a 70% reduction in ACC phosphorylation (Fig. 1B) with no change in total levels of AMPK or ACC. The two bands detected by the phospho-ACC antibody correspond to the ACC1/2 isoforms (ACC1 is phosphorylated on S79; ACC2 is phosphorylated on S220). Contrary to our predictions, these data imply that the phosphorylation and activation of AMPK are inhibited in replicon cells. To confirm that AMPK still was able to be activated in replicon cells, we treated them with three AMPK agonists. Two of the agonists, aminoimidazole carboxamide ribonucleotide (AICAR) (19) and metformin (20), have been well characterized, and the latter is used for treatment of type II diabetes; the third, thienopyridone A769662, selectively activates AMPK heterotrimers containing the β1 isoform (21). All three compounds could override the AMPK inhibition, as shown by elevated levels of AMPK T172 phosphorylation (Fig. 1C) in replicon cells.
Fig. 1.
Inhibition of AMPK activity in cells harboring the HCV subgenomic replicon. (A) ATP/ADP ratio was measured as described to assess AMP/ATP indirectly (n = 3). NS, not significant. (B) Cell lysates resolved by SDS/PAGE were immunoblotted with the indicated antibodies (Left). Levels of phosphorylated AMPK or ACC were quantified by densitometry in comparison with GAPDH (n = 3) (Right). All error bars indicate mean ± SEM. **Significant difference from Huh-7 (P < 0.05). (C) Replicon cells were left untreated (Control) or treated with AICAR (1 mM), A769662 (100 μM), or metformin (1 mM) for 4 h before immunoblot analysis with the indicated antibodies.
AMPK Activation Inhibits HCV Genome Replication and Abrogates Lipid Accumulation in Replicon Cells.
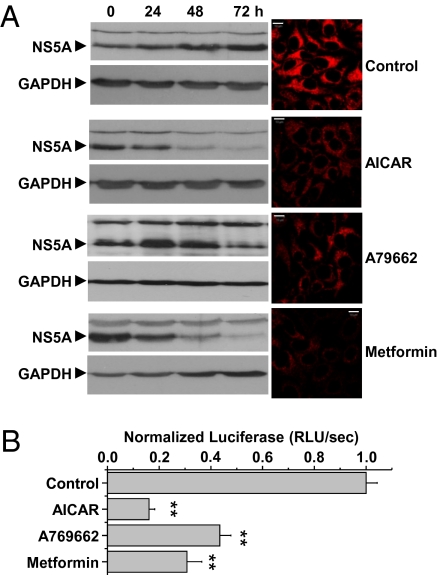
Because HCV mediated an inhibition of AMPK activity, we asked whether this effect might be important for virus replication. We treated replicon cells with AICAR, metformin, and A769662 and analyzed levels of the viral nonstructural protein NS5A at various time points following treatment. Treatment of replicon cells for 72 h dramatically reduced NS5A levels, most effectively in AICAR-treated cells (Fig. 2A). NS5A levels in replicon cells are an accepted indirect measure of genome replication, but it was possible that the decrease in NS5A abundance was caused by degradation or inhibition of translation. Therefore, to investigate directly if AMPK activation inhibited genome replication, we transiently transfected Huh-7 cells with luciferase-based genotype 1b or 2a (JFH-1) replicons, allowing direct correlation of HCV replication to luciferase activity. AICAR, A769662, and metformin treatment significantly decreased luciferase activity (Fig. 2B and Fig. S2A) in a dose-dependent manner (Fig S2B) as compared with untreated controls, suggesting that inhibition of AMPK is required for HCV genome replication. None of these compounds had any effect on cell viability at the highest concentrations used (Fig S2C).
Fig. 2.
AMPK activation inhibits HCV replication. (A) Replicon cells were left untreated (Control) or treated with AICAR (1 mM), A769662 (100 μM), or metformin (1 mM) for 0, 24, 48, and 72 h, and replicon replication was assessed by immunoblotting for NS5A (Left). NS5A immunofluorescence was performed at 72 h posttransfection (Right). Identical settings were maintained for image capture. Representative confocal images are shown. (Scale bars, 10 μm.) (B) Cells transfected with in vitro transcripts of a luciferase subgenomic replicon (genotype 1b) were treated with the indicated AMPK agonists overnight. Luciferase activity was used as a measure of replication. Error bars indicate mean ± SEM. **Significant difference from control (P < 0.05).
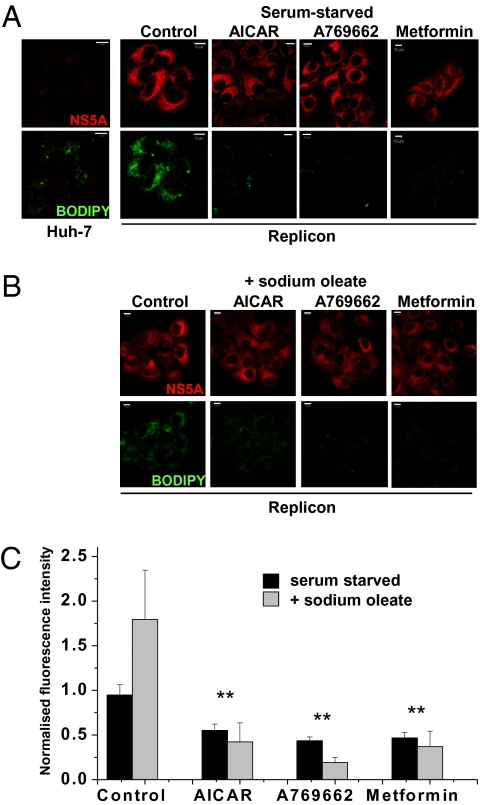
HCV replication is associated with an intracellular accumulation of lipid, and drugs that block cholesterol and fatty acid biosynthesis have been shown to regulate replicon replication in Huh-7 cells (4, 5). AMPK inhibits lipogenesis by modulating the activity of transcription factors required for lipogenic gene expression (e.g., PPARγ/δ and SREBP-1). We postulated that the lipid accumulation induced by HCV might be mediated by inhibition of AMPK. To test this possibility, we treated replicon cells with AICAR, A769662, or metformin and visualized cellular lipid content using BODIPY. This analysis revealed that replicon cells displayed higher levels of BODIPY fluorescence than parental Huh-7 cells (Fig. 3A and Fig. S2D). Upon short-term (4-h) treatment with AMPK agonists, cellular lipid content was reduced rapidly and dramatically (Fig. 3A), implying that inhibition of AMPK activity was in part responsible for the HCV-induced increase in cellular lipid abundance. This short-term treatment did not affect NS5A levels (compared with 72-h treatment; Fig. 2A), suggesting that the loss of lipid accumulation preceded the disruption of viral genome replication.
Fig. 3.
Effect of AMPK activators on lipid abundance. (A and B) Abundance of NS5A or cellular lipids (BODIPY) was evaluated in Huh-7 or replicon cells incubated in serum-free medium (A) or supplemented with sodium oleate (B). Cells were stained for lipid content with BODIPY dye for 1 h after NS5A labeling. Identical settings were maintained for image capture. Representative confocal images are shown. (Scale bars, 10 μm.) Replicon cells were treated with AMPK agonists for 4 h before processing. (C) For quantification of lipid abundance, images were captured and analyzed using Imaris software. Values were normalized to the control BODIPY levels minus oleate. Error bars indicate mean ± SEM. **Significant difference from Huh-7 (P < 0.05).
In vivo, hepatocytes derive fatty acids from the bloodstream, whereas Huh-7 cells derive intracellular lipids mainly via cellular lipogenesis from glucose. Because the latter process is regulated by AMPK, we investigated whether the loss of intracellular lipids following AMPK activation occurred when Huh-7 cells were provided with an exogenous source of fatty acids, by supplementing the culture media with oleate (Fig. 3B). Quantification of BODIPY fluorescence (Fig. 3C) confirmed that the presence of oleate increased lipid levels 2-fold in replicon cells; however, in AICAR-, A769662-, or metformin-treated cells, lipid levels were reduced compared with untreated cells, independent of the presence of exogenous fatty acid. These data suggest that AMPK activation can maintain an inhibitory effect on HCV lipid accumulation even when extracellular lipid is available and would be predicted to override HCV-mediated lipid accumulation in an infected liver.
HCV Mediates AMPK Inhibition via AKT Phosphorylation of S485.
We next addressed the mechanism by which the HCV replicon mediated inhibition of AMPK activity. Both NS4B and NS5A have been shown to activate the protein kinase AKT (9, 22). Because S485 phosphorylation of the AMPKα subunit by AKT has been reported to prevent AMPK activation in the presence of increased AMP (23), we asked whether HCV suppressed AMPK activity by activating AKT-dependent S485 phosphorylation of AMPKα.
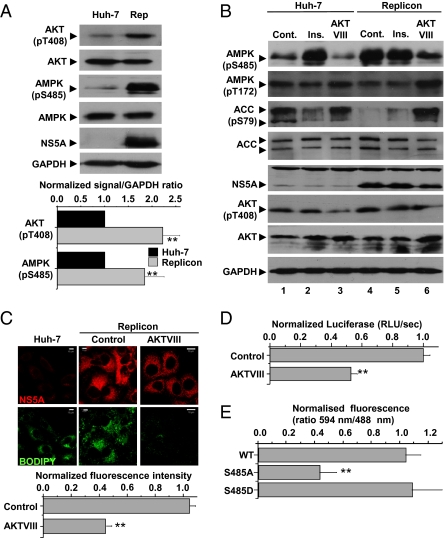
As previously shown (22), levels of active, phosphorylated AKT were increased 2.3-fold in replicon cells compared with Huh-7 cells (Fig. 4A), and concomitantly levels of S485 phosphorylated AMPK were enhanced 1.8-fold (Fig. 4A), providing a potential mechanism for the observed inhibition of AMPK.
Fig. 4.
Activation of AKT in replicon cells. (A) Cell lysates were analyzed by immunoblotting with the indicated antibodies. Levels of phosphorylated AKT or AMPK were quantified by densitometry in comparison with GAPDH (n = 3). **Significant difference from Huh-7 (P < 0.05). (B) Cell lysates were analyzed by immunoblotting with the indicated antibodies. Lanes 2 and 5: insulin (Ins) treatment (100 nM) for 4 h before harvest; lanes 3 and 6: AKTVIII treatment (5 μM) for 4 h before harvest. Cont, control. (C) Abundance of NS5A or lipids (BODIPY) was evaluated in untreated Huh-7 or replicon cells following treatment with AKTVIII (5 μM) for 4 h. For quantification of lipid abundance, images were captured and analyzed using Imaris software. **Significant difference from Huh-7 (P < 0.05). (D) Cells transfected with in vitro transcripts of a luciferase subgenomic replicon (genotype 1b) were treated with AKTVIII (5 μM) overnight. Luciferase activity [relative luminescence units (RLU)/sec] was used as a measure of replication. **Significant difference from control (P < 0.05). (E) Replicon cells were transiently transfected with plasmids expressing Myc-tagged wild-type, S485A or S485D AMPKα. At 48 h posttransfection, expression of both exogenous AMPKα and NS5A was detected by immunofluorescence with the appropriate antibodies. The ratio of fluorescence at 594 nm (AMPKα) to 494 nm (NS5A) was quantitated for 15 cells per condition using ImageJ software. All error bars indicate mean ± SEM. **Significant difference from wild-type (WT) AMPK (P < 0.05).
We reasoned that if AMPK inhibition was mediated by AKT, this inhibition could be prevented by inhibiting AKT. We therefore assessed AMPK activation in replicon cells treated with either insulin (an activator of AKT via upstream PI3K activation) or AKTVIII (a selective AKT inhibitor). Fig. 4B shows that insulin treatment of Huh-7 cells stimulated AMPK S485 phosphorylation with a concomitant loss of both AMPK T172 and ACC phosphorylation (compare lanes 1 and 2), confirming that AKT activation inhibits AMPK activation (23). This inhibition could be reversed by blocking AKT activity, because both AMPK T172 and ACC phosphorylation were restored following AKTVIII treatment (compare lanes 1 and 3). As expected, AKTVIII treatment also resulted in a loss of AMPK S485 phosphorylation. These data confirmed that AMPK activation is inhibited by AKT in Huh-7 cells. By contrast, replicon cells exhibited high levels of AMPK S485 phosphorylation which were unaffected by insulin treatment (compare lanes 4 and 5), although they were reduced after AKTVIII treatment (lane 6). Treatment of replicon cells with AKTVIII resulted in concomitant restoration of AMPK activity, as shown by increased AMPK T172 and ACC phosphorylation (compare lanes 4 and 6). Furthermore, AKT inhibition (in common with AMPK activation; Fig. 3) resulted in a rapid reduction in cellular lipid content, as indicated by a loss of BODIPY staining (Fig. 4C), further confirming that inhibition of AMPK activation via AKT was responsible for the HCV-induced increase in cellular lipid abundance. Consistent with the AKT dependence of AMPK inhibition, AKTVIII treatment also significantly inhibited replicon luciferase expression and, thus, genome replication (Fig. 4D).
To confirm further the role of S485 phosphorylation in the HCV-mediated inhibition of AMPK, we overexpressed wild-type or mutated forms of the AMPKα subunit (which will form heterotrimers with endogenous β/γ subunits, displacing the endogenous α subunit). Because the exogenously expressed AMPKα was Myc-tagged, we were able to assess NS5A abundance (and therefore the levels of genome replication) by quantifying anti-NS5A fluorescence in cells that were positive for the Myc tag. This analysis revealed that NS5A abundance was unaffected by overexpression of either wild-type AMPKα or a phosphomimetic mutant (S485D) (Fig. 4E and Fig. S3). By contrast, overexpression of a nonphosphorylatable mutant (S485A) dramatically reduced the abundance of NS5A, confirming that phosphorylation of AMPKα at residue S485 is required for genome replication.
HCV-Infected Cells Exhibit AMPK Inhibition.
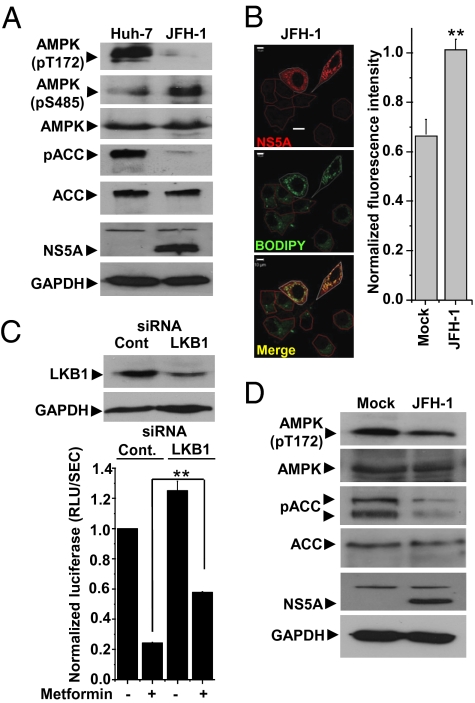
Although the presence of the replicon was both necessary and sufficient to mediate AMPK inhibition, it was important to determine whether AMPK activity was perturbed in the context of virus infection. Huh-7 cells were transfected with in vitro transcribed full-length RNA of the cell-culture permissive genotype 2a HCV isolate, JFH-1 (24). Consistent with the replicon data, AMPK T172 and ACC phosphorylation were abrogated, whereas AMPK S485 phosphorylation was elevated in JFH-1 RNA-transfected cells (Fig. 5A). Importantly, as for replicon cells, levels of cellular lipids were elevated compared with mock-transfected cells (Fig. 5B and Fig. S4A). To demonstrate further the effects of AMPK activation on viral replication, we showed that metformin treatment reduced replication of a modified virus that expressed luciferase (J6/JFH-1Luc) (25) by 75% (Fig. 5C). To confirm that this reduction was AMPK mediated, we transfected cells with LKB1 siRNA. Although this siRNA was able to ablate LKB1 expression only partially (Fig. 5C), the inhibitory effect of metformin on virus replication was significantly reduced compared with control cells (Fig. 5C). To verify that these data were not a consequence of electroporation of viral RNA into Huh-7 cells, we directly infected cells with JFH-1 virus and again observed a reduction of both AMPK T172 and ACC phosphorylation at 48 h postinfection, confirming that AMPK activity was inhibited (Fig. 5D). We also assessed the effects of AMPK agonists on virus production in cells infected with JFH-1 virus, because expected levels of both intracellular and released infectious virus were reduced 3- to 10-fold by all three compounds (Fig S4B). Taken together, these data confirm that the inhibition of AMPK is not restricted to HCV replicons but also occurs in the context of virus infection, where it is required for efficient HCV genome replication.
Fig. 5.
Inhibition of AMPK activity is observed in cells transfected with full-length JFH-1 RNA or infected with JFH-1 virus. (A) Cells were electroporated with full-length JFH-1 RNA and analyzed for AMPK activation status as described in Fig 1B. (B) Abundance of NS5A or cellular lipids (BODIPY) was analyzed as described in Fig. 3. JFH-1 transfected cells are outlined in white; untransfected cells in the same field are outlined in red. For quantification of lipid abundance, images were captured and analyzed using Imaris software. **Significant difference from untransfected (Mock) cells (P < 0.05). (C) Cells were electroporated with either control (Cont) or LKB-1–specific siRNA, incubated for 72 h to allow silencing of the target gene, and then transfected with J6/JFH-1Luc RNA. Samples at 48 h posttransfection were analyzed for luciferase activity. Metformin was added to cells at 24 h posttransfection. Cell lysates were analyzed by immunoblotting with the indicated antibodies to confirm LKB1 silencing. Results are expressed as mean ± SEM (n = 3). **Significant difference from untreated (P < 0.05). (D) Huh-7 cells were mock-infected or infected with JFH-1 virus at a multiplicity of infection of 0.5 focus-forming units per cell. Cell lysates were prepared at 48 h posttransfection and analyzed by immunoblotting with the indicated antibodies.
Discussion
Our data clearly demonstrate that HCV inhibits the activity of AMPK. Superficially, this observation seems counterintuitive, because active viral replication probably will place high energy demands upon the cell, increasing both ATP consumption and the AMP/ATP ratio. The concomitant increase in AMPK activity would switch on processes that generate ATP while switching off those that consume ATP. In the long term, this effect might benefit the virus by prolonging the life of the host cell. Why, then, does HCV mediate AMPK inhibition? Our data suggest that one answer is that loss of function of AMPK increases hepatic lipid accumulation. An increasing body of evidence shows that HCV is critically dependent on cellular lipids throughout the virus life cycle. By blocking AMPK activity, the virus can ensure that lipid biosynthesis can continue at a high level, permitting the accumulation of lipid that is required for virus replication. By overriding AMPK inhibition with AMPK agonists, not only is lipid accumulation in replicon cells abrogated, but virus genome replication also is inhibited, thereby reducing the production of infectious virus (Fig. 2 and Figs. S2B and S4B). This possibility raises exciting prospects for therapeutic approaches to HCV treatment using well-characterized AMPK agonists such as metformin, a safe and well-tolerated drug that already is used extensively for treatment of diabetes. Indeed, a recent study (26) demonstrated that inclusion of metformin with IFN/ribavirin therapy had a beneficial antiviral outcome, albeit for HCV patients who also had type II diabetes.
What might be the effect of AMPK inhibition in the HCV-infected liver? In addition to a role in fatty acid synthesis, liver AMPK controls glucose homeostasis, mainly through the inhibition of gluconeogenic gene expression and glucose production (11). In primary cultured hepatocytes, glucose production is suppressed by constitutively active forms of AMPK, and thus the glucose-lowering effect of metformin in the treatment of diabetes can be attributed partly to its ability to suppress gluconeogenesis through AMPK activation (20, 27, 28). This effect is pertinent, because HCV infection has been associated with a higher prevalence of type II diabetes (29). It is interesting to speculate that the HCV-mediated AMPK inhibition in infected liver may contribute to this phenotype.
We demonstrate that AMPK is inhibited in replicon cells, implying that this function can be ascribed to one or more of the nonstructural proteins. Because AMPK inhibition is mediated via AKT, this inhibition suggests a role for NS4B and/or NS5A, both of which have been shown previously to stimulate PI3K/AKT (9, 17). Consistent with this finding, it has been reported that both NS4B and NS5A independently can lead to accumulation of cholesterol and fatty acids in hepatic cells (9, 30). The NS5A effect was shown to be mediated by inhibition of PPARs, and for NS4B lipid accumulation was mediated via activation of SREBP-1c, whose expression is negatively regulated by AMPK (20). The observation that NS4B-mediated AKT signaling was required for this activation further implies a role of AMPK inhibition in NS4B-induced lipid accumulation.
Activation of the PI3K/AKT pathway is a common theme in viruses that establish chronic infections; for example, the HIV-1 Nef protein binds to and activates PI3K, stimulating AKT signaling (31). Intriguingly, Nef also induces increases in cholesterol biosynthesis and modulates the lipid content of nascent virions (32), although whether this effect is mediated via AMPK is undetermined. By contrast, our data suggest a role of AMPK inhibition in facilitating HCV genome replication which, in common with other positive-strand RNA viruses, occurs within a “membranous web” derived from intracellular vesicles (33). It is tempting to speculate that lipid biogenesis induced by AMPK inhibition may be required for the establishment of the correct architecture of this complex. This possibility is particularly pertinent because it has been shown that blocking fatty acid synthesis through the inhibition of ACC decreases HCV replication (34).
A number of recent reports have described other interactions between viruses and AMPK: The HIV-1 Tat protein has been reported to inhibit AMPK, and, consistent with this effect, AMPK agonists inhibited Tat transactivation of the HIV-1 LTR (35). Infection with human cytomegalovirus (36) also resulted in an inhibition of AMPK T172 phosphorylation. Contrastingly, the SV40 small T antigen maintains energy homeostasis during glucose deprivation by activating AMPK (37), and avian reovirus infection also has been shown to stimulate AMPK T172 phosphorylation (38). The challenge will be to dissect the precise role that AMPK activity plays in the life cycle of these viruses, allowing a better rationale for the use of AMPK agonists in antiviral therapy. In this regard, we demonstrate that reversing AMPK inhibition in HCV culture systems can reduce the accumulation of lipids, suggesting that HCV may affect one or more steps in cholesterol and/or fatty acid biosynthesis directly through AMPK inactivation. The key issue that remains to be addressed is to dissect the precise physiological interplay between HCV proteins and the control of AMPK activity.
Materials and Methods
Cell Culture.
Huh-7 cells were cultured in DMEM with 10% FCS, 1% nonessential amino acids, 2 mM L-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin at 37 °C in a humidified 5% CO2 incubator. Subgenomic replicon-harboring cell lines (genotype 1b FK5.1) (18) were maintained in DMEM with 250 μg/mL G418.
Adenine Nucleotide Quantification.
Cells were scraped into a minimal volume of 5% perchloric acid and centrifuged (13,800 × g at 4 °C) to remove debris. The perchloric acid was neutralized, and nucleotides were extracted using an equal volume of a 1:1 1,1,2-trichlorotrifluororethane:trioctylamine mix. The ATP:ADP ratio was measured using capillary electrophoresis and was used as a surrogate for AMP:ATP (17).
Immunoblotting.
Cells were lysed in Glasgow lysis buffer [GLB;10 mM Pipes-KOH (pH 7.2), 120 mM KCl, 30 mM NaCl, 5 mM MgCl2, 1% Triton X-100 (Sigma), 10% glycerol] (Promega) plus protease and phosphatase inhibitors (2 mM Na3VO4, 5 mM NaF, 5 mM Na4P2O7). Fifty micrograms of protein were resolved by SDS/PAGE, transferred to a PVDF membrane using a semidry transfer apparatus, and probed with appropriate primary and secondary antibodies. ImageJ (National Institutes of Health) densitometry was used for quantification.
Immunofluorescence.
Cells on glass coverslips were fixed with 3% paraformaldehyde, permeabilized in 0.1% Triton X-100, and blocked in PBS/1% BSA for 30 min. Cells were labeled with a polyclonal sheep anti-NS5A serum followed by Alexa Fluor 594 (Invitrogen) anti-sheep secondary. Lipid content was detected with the BODIPY 493/503 dye (Invitrogen) for 1 h after NS5A staining. Cells were viewed on a Zeiss 510-META laser scanning confocal microscope under an oil-immersion 63× objective lens (NA = 1.40). Representative images are displayed as single optical sections of 50-μm thickness. For detection of exogenous AMPKα, cells were transfected with plasmids expressing Myc-AMPKα, fixed 48 h posttransfection, permeabilized, and probed with a mouse monoclonal anti-Myc antibody (1 μg/mL) followed by an Alexa Fluor 488 anti-mouse secondary. For quantification cells were serum-starved overnight ± 20 μg/mL sodium oleate and AMPK agonists as described. Images were captured and analyzed using Imaris (Bitplane AG) or ImageJ software. Thresholds of each channel were set at 10% of the maximum intensity. Vesicles of a diameter of 0.5 μm were counted and divided by the cell number for each image.
Transient Subgenomic Replicon Luciferase Assays.
T7 transcripts were generated from linearized DNA templates of JFH1 (SGR-Luc-JFH-1) (40) or genotype 1b (FK5.1Luc) luciferase subgenomic replicons. Then 4 × 106 cells were washed in diethylpyrocarbonate (DEPC)-treated PBS, resuspended in 400 μL PBS, and electroporated with replicon RNA (5 μg) in 0.4-cm cuvettes at 950 μF, 270 V. Then 1× 104 cells were seeded into each well of 96-well plates. Cells were lysed directly in 96-well plates at 4 and 24 (JFH-1) or 4 and 48 h posttransfection (FK5.1) in 1× passive lysis buffer (PLB) (Promega). Luciferase activity was measured using luciferase assay reagent (LAR; Promega) on a BMG plate reader. AMPK agonists were added at 8 or 32 h posttransfection for JFH-1 and FK5.1, respectively. Statistical significance of differences was determined using the paired Student's t test. P < 0.05 was accepted as significant.
Virus Assays.
Cells were washed in DEPC-treated PBS and resuspended at 2 × 107 cells/mL. Then 8 × 106 cells were electroporated with 10 μg of JFH-1 RNA. For replication assays, cells electroporated with siRNA were incubated for 72 h to allow silencing of the target gene before transfection with 1 μg J6/JFH-1Luc RNA using Lipofectin (Invitrogen) following the manufacturer's instructions. Samples were harvested in 100 μL PLB at 4/48 h posttransfection. For infection experiments, virus inoculum was titrated by focus-forming assay (41) and used to infect Huh-7 cells at a multiplicity of infection of 0.5 in complete medium. For Western blotting, cells were lysed in GLB as described above at 48 h posttransfection. Virus harvest and titrations were performed as described (41).
Supplementary Material
Acknowledgments
We thank R. Bartenschlager and V. Lohmann (University of Heidelberg, Heidelberg, Germany) for FK5.1 constructs, J. McLauchlan (MRC Virology Unit, Glasgow, United Kingdom) for SGR-Luc-JFH-1, T. Wakita (National Institute of Infectious Diseases, Tokyo) for JFH-1, C. Rice (The Rockefeller University, New York) for J6/JFH-1Luc, and A. Macdonald and D. Rowlands (University of Leeds, Leeds, UK) for critical reading of this manuscript. This work was supported by Grants G0401577 from the Medical Research Council (to M.H.) and 082812 from the Wellcome Trust (to M.H. and S.D.C.G.) Work in the Peers and Hardie laboratories also is supported by the Wellcome Trust. M.E.H. is supported by a Cooperative Awards in Science and Engineering (CASE) PhD Studentship from the Biotechnology and Biological Sciences Research Council and Arrow Therapeutics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0912426107/-/DCSupplemental.
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 3.McLauchlan J. Lipid droplets and hepatitis C virus infection. Biochim Biophys Acta. 2009;1791:552–559. doi: 10.1016/j.bbalip.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Gastaminza P, et al. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82:2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapadia SB, Barth H, Baumert T, McKeating JA, Chisari FV. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J Virol. 2007;81:374–383. doi: 10.1128/JVI.01134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyanari Y, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka N, et al. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J Clin Invest. 2008;118:683–694. doi: 10.1172/JCI33594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oem JK, et al. Activation of sterol regulatory element-binding protein 1c and fatty acid synthase transcription by hepatitis C virus non-structural protein 2. J Gen Virol. 2008;89:1225–1230. doi: 10.1099/vir.0.83491-0. [DOI] [PubMed] [Google Scholar]
- 9.Park CY, Jun HJ, Wakita T, Cheong JH, Hwang SB. Hepatitis C virus nonstructural 4B protein modulates sterol regulatory element-binding protein signaling via the AKT pathway. J Biol Chem. 2009;284:9237–9246. doi: 10.1074/jbc.M808773200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasai D, et al. HCV replication suppresses cellular glucose uptake through down-regulation of cell surface expression of glucose transporters. J Hepatol. 2009;50:883–894. doi: 10.1016/j.jhep.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 11.Hardie DG. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 12.Hawley SA, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003 doi: 10.1186/1475-4924-2-28. 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawley SA, et al. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Woods A, et al. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2005;2:21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 15.Davies SP, Helps NR, Cohen PTW, Hardie DG. 5′-AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2C alpha and native bovine protein phosphatase-2AC. FEBS Lett. 1995;377:421–425. doi: 10.1016/0014-5793(95)01368-7. [DOI] [PubMed] [Google Scholar]
- 16.Sanders MJ, Grondin PO, Hegarty BD, Snowden MA, Carling D. Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem J. 2007;403:139–148. doi: 10.1042/BJ20061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardie DG, Hawley SA. AMP-activated protein kinase: The energy charge hypothesis revisited. Bioessays. 2001;23:1112–1119. doi: 10.1002/bies.10009. [DOI] [PubMed] [Google Scholar]
- 18.Krieger N, Lohmann V, Bartenschlager R. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J Virol. 2001;75:4614–4624. doi: 10.1128/JVI.75.10.4614-4624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corton JM, Gillespie JG, Hawley SA, Hardie DG. 5-aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur J Biochem. 1995;229:558–565. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhou G, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott JW, et al. Thienopyridone drugs are selective activators of AMP-activated protein kinase beta1-containing complexes. Chem Biol. 2008;15:1220–1230. doi: 10.1016/j.chembiol.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Street A, Macdonald A, Crowder K, Harris M. The hepatitis C virus NS5A protein activates a phosphoinositide 3-kinase-dependent survival signaling cascade. J Biol Chem. 2004;279:12232–12241. doi: 10.1074/jbc.M312245200. [DOI] [PubMed] [Google Scholar]
- 23.Horman S, et al. Insulin antagonizes ischemia-induced Thr172 phosphorylation of AMP-activated protein kinase alpha-subunits in heart via hierarchical phosphorylation of Ser485/491. J Biol Chem. 2006;281:5335–5340. doi: 10.1074/jbc.M506850200. [DOI] [PubMed] [Google Scholar]
- 24.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tscherne DM, et al. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80:1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romero-Gómez M, et al. Spanish Treatment of Resistance to Insulin in Hepatitis C Genotype 1 Group Treatment of insulin resistance with metformin in naïve genotype 1 chronic hepatitis C patients receiving peginterferon alfa-2a plus ribavirin. Hepatology. 2009;50:1702–1708. doi: 10.1002/hep.23206. [DOI] [PubMed] [Google Scholar]
- 27.Lochhead PA, Salt IP, Walker KS, Hardie DG, Sutherland C. 5-aminoimidazole-4-carboxamide riboside mimics the effects of insulin on the expression of the 2 key gluconeogenic genes PEPCK and glucose-6-phosphatase. Diabetes. 2000;49:896–903. doi: 10.2337/diabetes.49.6.896. [DOI] [PubMed] [Google Scholar]
- 28.Shaw RJ, et al. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Machado MV, Cortez-Pinto H. Insulin resistance and steatosis in chronic hepatitis C. Ann Hepatol. 2009;8(Suppl 1):S67–S75. [PubMed] [Google Scholar]
- 30.Kim K, et al. Hepatitis C virus NS5A protein increases hepatic lipid accumulation via induction of activation and expression of PPARgamma. FEBS Lett. 2009;583:2720–2726. doi: 10.1016/j.febslet.2009.07.034. [DOI] [PubMed] [Google Scholar]
- 31.Wolf D, et al. HIV-1 Nef associated PAK and PI3-kinases stimulate Akt-independent Bad-phosphorylation to induce anti-apoptotic signals. Nat Med. 2001;7:1217–1224. doi: 10.1038/nm1101-1217. [DOI] [PubMed] [Google Scholar]
- 32.Zheng YH, Plemenitas A, Fielding CJ, Peterlin BM. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV-1 progeny virions. Proc Natl Acad Sci USA. 2003;100:8460–8465. doi: 10.1073/pnas.1437453100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egger D, et al. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci USA. 2005;102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang HS, Wu MR. SIRT1 regulates Tat-induced HIV-1 transactivation through activating AMP-activated protein kinase. Virus Res. 2009;146:51–57. doi: 10.1016/j.virusres.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Kudchodkar SB, Del Prete GQ, Maguire TG, Alwine JC. AMPK-mediated inhibition of mTOR kinase is circumvented during immediate-early times of human cytomegalovirus infection. J Virol. 2007;81:3649–3651. doi: 10.1128/JVI.02079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar SH, Rangarajan A. Simian virus 40 small T antigen activates AMPK and triggers autophagy to protect cancer cells from nutrient deprivation. J Virol. 2009;83:8565–8574. doi: 10.1128/JVI.00603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji WT, Lee LH, Lin FL, Wang L, Liu HJ. AMP-activated protein kinase facilitates avian reovirus to induce mitogen-activated protein kinase (MAPK) p38 and MAPK kinase 3/6 signalling that is beneficial for virus replication. J Gen Virol. 2009;90:3002–3009. doi: 10.1099/vir.0.013953-0. [DOI] [PubMed] [Google Scholar]
- 40.Targett-Adams P, McLauchlan J. Development and characterization of a transient-replication assay for the genotype 2a hepatitis C virus subgenomic replicon. J Gen Virol. 2005;86:3075–3080. doi: 10.1099/vir.0.81334-0. [DOI] [PubMed] [Google Scholar]
- 41.Hughes M, et al. A conserved proline between domains II and III of hepatitis C virus NS5A influences both RNA replication and virus assembly. J Virol. 2009;83:10788–10796. doi: 10.1128/JVI.02406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.