Abstract
High light can be lethal for photosynthetic organisms. Similar to plants, most cyanobacteria protect themselves from high irradiance by increasing thermal dissipation of excess absorbed energy. The photoactive soluble orange carotenoid protein (OCP) is essential for the triggering of this photoprotective mechanism. Light induces structural changes in the carotenoid and the protein, leading to the formation of a red active form. Through targeted gene interruption we have now identified a protein that mediates the recovery of the full antenna capacity when irradiance decreases. In Synechocystis PCC 6803, this protein, which we called the fluorescence recovery protein (FRP), is encoded by the slr1964 gene. Homologues of this gene are present in all of the OCP-containing strains. The FRP is a 14-kDa protein, strongly attached to the membrane, which interacts with the active red form of the OCP. In vitro this interaction greatly accelerates the conversion of the red OCP form to the orange form. We propose that in vivo, FRP plays a key role in removing the red OCP from the phycobilisome and in the conversion of the free red OCP to the orange inactive form. The discovery of FRP and its characterization are essential elements in the understanding of the OCP-related photoprotective mechanism in cyanobacteria.
Keywords: carotenoid, nonphotochemical quenching, phycobilisome, photoreceptor, Synechocystis
Because too much light can be lethal for photosynthetic organisms, photoprotection against excess absorbed light energy is an essential and universal attribute of oxygenic photosynthetic organisms. Survival, productivity, and habitat preference are largely determined by development of photoprotective mechanisms. One of these mechanisms, which is induced by high irradiance, dissipates the excess harmful absorbed energy into heat at the levels of the antennae.
In all photosynthetic organisms, the function of the light harvesting antenna is similar: to collect and concentrate light energy in the photochemical reaction centers in which light energy is converted into chemical energy. In plants and green algae, light is principally harvested by membrane-embedded complexes noncovalently binding chlorophyll and carotenoid molecules. These complexes show a large structural and functional flexibility. They are reversibly switched from a very efficient energy collecting state into a photoprotected state. This state allows the conversion of excess energy into heat, decreasing the energy arriving at the reaction centers under high light conditions (1–4). Cyanobacteria are oxygenic photosynthetic prokaryotes that play a key role in global carbon cycling. To harvest light. most cyanobacteria use a large, membrane-extrinsic complex called the phycobilisome (PB), which is composed of several types of chromophorylated phycobiliproteins and of linker peptides (for reviews, see refs. 5–7).
It has been recently shown that, in cyanobacteria, like in plants, there exists a photoprotective process that decreases the energy transfer between the antenna and the reaction centers by increasing thermal dissipation (8–13). However, this mechanism is not triggered by a lowering of the luminal pH, as occurs in plants; instead, it is induced by the light activation of a soluble carotenoid protein, the orange carotenoid protein (OCP), which binds the keto-carotenoid, 3′ hydroxyechinenone (12, 13). In darkness or dim light the OCP appears orange (OCPo). Upon illumination with blue-green light (400–550 nm), structural changes in the carotenoid and the protein occur and photoconvert it into a relatively unstable red form, OCPr (14). The accumulation of this red form is essential for the induction of the photoprotective mechanism (14, 15). It results in an increase of the thermal dissipation of the energy absorbed by the PBs (14). A concomitant decrease of the PB fluorescence emission and of the energy transfer from the PB to the reaction centers also occurs (9, 14). In mutants in which the OCP was absent (12) or unable to form or stabilize the red OCP form (14, 15), blue-green light did not induce fluorescence quenching or the photoprotective mechanism. Conversely, the presence of high concentrations of OCP increases fluorescence quenching (13, 14).
In darkness, OCPr spontaneously reverts into the orange form and, in vivo, a recovery of the lost fluorescence is observed. However, the fluorescence recovery kinetics in vivo are slower than the OCPr-to-OCPo dark conversion in vitro, suggesting that the OCPr form is more stable in vivo than in vitro (14). This OCPr stabilization could be explained by a specific interaction of OCPr with the PBs. A possible direct interaction between the OCP and the PBs was suggested by PB reconstitution experiments in the presence of the OCP (16). In vivo, an unknown protein might be involved in the detachment of the OCPr from the PB and in the destabilization of OCPr. Here, we describe a protein that plays this role, being essential for fluorescence recovery after high irradiance exposure.
Results
In Synechocystis PCC6803 (hereafter called Synechocystis), the strain used for almost all studies about the OCP-related photoprotective mechanism, the OCP is encoded by the slr1963 gene (17). It is constitutively expressed (13), although under stress conditions (e.g., high light, salt stress, iron starvation) the levels of OCP transcripts and proteins are increased (13, 18, 19). Highly conserved (80–50% identity) homologues of the OCP gene are found in most PB-containing cyanobacteria (26 of 39 strains) for which genomic data are available (20). These strains are able to perform blue-green light–induced fluorescence quenching indicating that the OCP-related photoprotective mechanism is widespread (20).
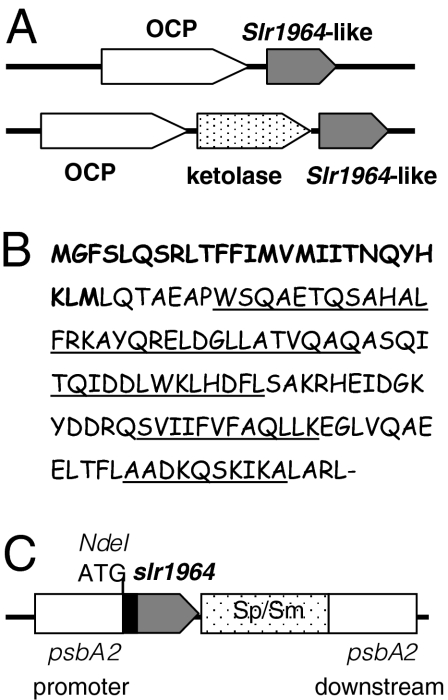
The genomic context of the OCP gene varies considerably, however. In most of the freshwater strains the OCP encoding gene is followed by a gene coding for a conserved hypothetical protein (slr1964 in Synechocystis; Fig. 1A and Table S1). The function of this small hypothetical protein was not known until this work. In all of the marine Synechococci (except Synechococcus sp. PCC7335), including Synechococcus WH5701 and Cyanobium PCC 7001 (which can live in marine or fresh water), a gene encoding a putative β-carotene ketolase is found between the OCP-like gene and the conserved slr1964-like gene (Fig. 1A and Table S1). Only three OCP-containing strains seem not to have homologues for the slr1964 gene, or the homology of these genes is too low to be detected by the BLAST programs.
Fig. 1.
The slr1964 gene. (A) Gene arrangement of the slr1963-like (OCP-like) genes and slr1964-like genes in freshwater and marine cyanobacteria strains. (B) The sequence of the slr1964 Synechocystis gene. The additional N-terminal sequence existing only in Synechocystis and Microcystis is in bold. The amino acids forming the α-helices are underlined. (C) Gene arrangement in the psbA2 region of the mutant overexpressing slr1964. The slr1964 gene is under the control of the psbA2 promoter.
The predicted Slr1964-like proteins have 106 to 111 amino acids with the exception of Synechocystis and Mycrocystis NIES-843 proteins, which contain an N-terminal prolongation of 22 to 25 amino acids (Fig. S1). In Synechocystis, this additional sequence contains four ATG or GTG codons. The first GTG is considered as the first Met of the protein but the fourth codon coding for a Met (Met26) coincides with the first Met of the other homologues (Fig. 1B and Fig. S1). The protein is predicted to be a soluble protein (with a molecular mass of 15.38 kDa in Synechocystis) without any known pattern. The structure includes a long α-helix in the N-terminal moiety and three shorter ones in the rest of the protein but none of them is predicted to be a transmembrane helix (Fig. 1B).
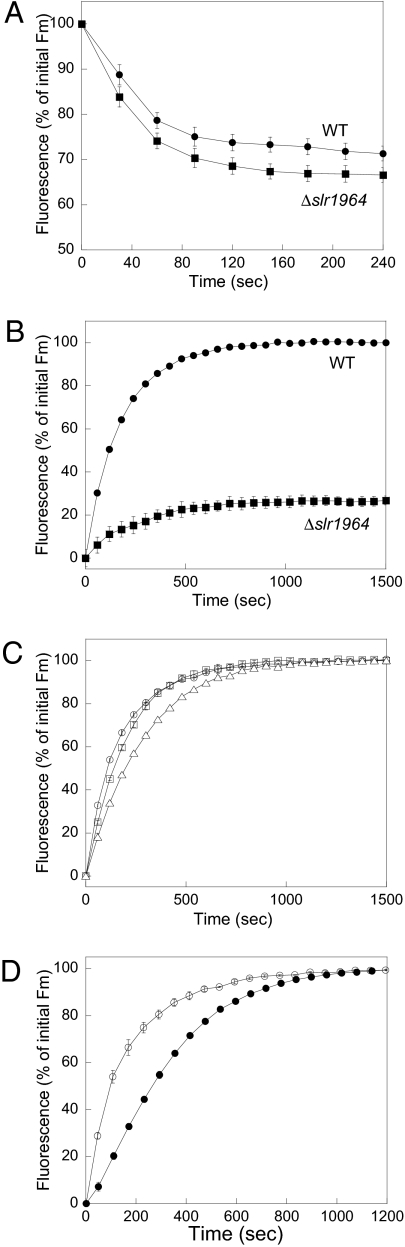
To study the role of Slr1964, various Synechocystis mutants were constructed (SI Materials and Methods and Fig. S2). In a Synechocystis mutant, in which the slr1964 gene was interrupted by an antibiotic cassette, the decrease of fluorescence induced by blue-green light was only slightly increased, indicating that Slr1964 is not involved in the formation of the quenching (Fig. 2A). In contrast, only 20% of fluorescence recovery was observed in this mutant when cells were transferred to low light intensities or darkness (Fig. 2B). When ΔSlr1964 mutant cells were grown under low light conditions, they were not “quenched” (Fig. S3A). The lack of recovery explains the slightly larger fluorescence quenching observed in the ΔSlr1964 mutant (Fig. 2A). This mutant was transformed by plasmids in which the WT slr1964 gene or a 5′ terminal His-tagged (after the GTG coding for the first Met) slr1964 gene or a 3′ terminal His-tagged slr1964 gene was under the control of the strong psbA2 promoter (Fig. 1C and Fig. S2A). In these mutants the psbA2 gene is absent. The absence of this gene did not affect the kinetics of fluorescence quenching or fluorescence recovery in a ΔpsbA2 mutant in which the psbA2 gene was replaced by an antibiotic resistance cassette (Fig. S3B). When quenched cells of the double mutants were transferred to low light intensities, 100% of fluorescence recovery was observed indicating that Slr1964 has an important role in fluorescence recovery (Fig. 2C). Therefore, hereafter we will call Slr1964 the fluorescence recovery protein (FRP).
Fig. 2.
The role of the FRP encoded by the slr1964 gene. (A) Decrease of maximal fluorescence (Fm) during exposure of WT (circles) and Δslr1964 mutant (squares) cells to 740 μmol photons m−2 s−1 of blue-green light (400–550 nm). (B–D). Increase of Fm during exposure to low blue-green light (80 μmol photons m−2 s−1) after high irradiance of cells of WT (B, circles), Δslr1964 (B, squares) overexpressing nontagged FRP (C, squares), overexpressing C-terminal His-tagged FRP (C, triangles), overexpressing N-terminal His-tagged FRP (C and D, open circles) and overexpressing OCP-FRP (D, closed circles) mutants. All curves are the averages of three independent experiments. The cells were diluted to 3 μg Chl/mL
The presence of a His tag in the N-terminal did not affect the recovery kinetics but a His tag in the C-terminal slows down the recovery (Fig. 2C), suggesting that the C-terminal part of the protein is involved in FRP activity. The ΔFRP mutant was also transformed by a plasmid containing the putative operon formed by the slr1963 (ocp) and slr1964 (frp) genes. The operon was under the control of the psbA2 promoter (Fig. S2B). In the mutant obtained, strong blue-green light induced a very large fluorescence quenching as a result of the increased OCP concentration (14). When the quenched cells were transferred to low light, 100% of the fluorescence was recovered but more slowly (t1/2 of 260 s vs. 100 s) than in the mutants containing the slr1964 gene just downstream the psbA2 promoter (Fig. 2D). This result suggests that the ratio of FRP to OCP is lower in the mutant in which the putative operon ocp-frp is under the control of the psbA2 promoter than in the mutant in which the frp gene is directly under the control of this promoter. Thus, the transcript containing both genes, ocp and frp, is very unstable and/or frp is transcribed independently from its own promoter.
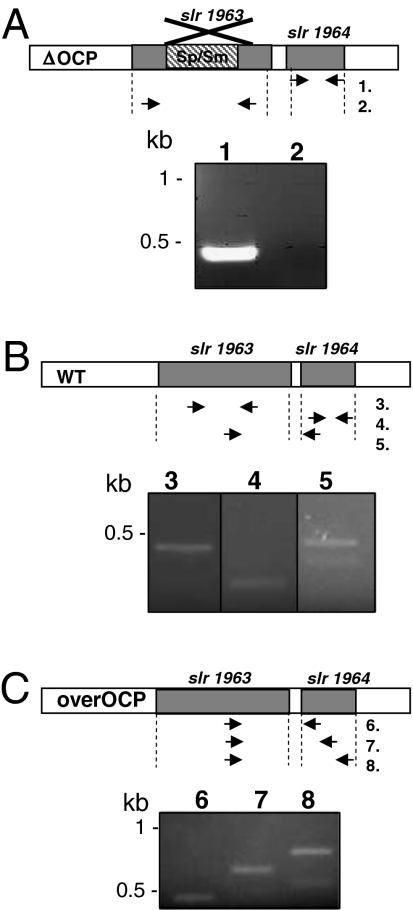
To test the presence of a frp-specific promoter occurring between the ocp and frp genes, the presence of an mRNA containing frp was tested in a mutant in which ocp was interrupted with a spectinomycin/streptinomycin resistance cassette (containing a transcription terminator). In this mutant, no traces of ocp mRNA were detected by reverse transcription followed by PCR amplification. In contrast, frp mRNA was clearly present (Fig. 3A), indicating that frp can be transcribed independently of the ocp and that there exists a frp-specific promoter. Efforts were made to detect transcripts containing both genes in WT cells. Although, ocp and frp mRNAs were present (Fig. 3B), no traces of a mRNA containing both complete genes were detected. The longer mRNA detected contained the ocp gene and only the first 30 nucleotides of the frp gene (Fig. 3B, lane 5). In contrast, long mRNAs containing the whole ocp and frp genes were present in Synechocystis cells in which ocp and frp were downstream the psbA promoter (Fig. 3C), indicating that a cotranscription of both genes may exist, at least in these conditions.
Fig. 3.
The ocp and frp genes can be independently transcribed or cotranscribed. (A) In the ΔOCP mutant, the frp mRNA (lane 1) was present whereas the ocp mRNA was absent (lane 2); (B) In WT cells, ocp (lane 3) and frp (lane 4) mRNAs were present. An mRNA including the ocp gene and the first 30 nucleotides of the frp gene were also detectable (lane 5). (C) In the overexpressing OCP/N-terminal His-tagged FRP strain, mRNAs containing the ocp gene and at least the 30 first nucleotides of frp (lane 6) or half of the frp gene (lane 7) or the whole frp gene (lane 8) were detected. See Table S2 for primer sequences.
The frp gene with an N-terminal His tag (after the first Met) was overexpressed in Escherichia coli to obtain large amounts of purified FRP for in vitro studies. The overexpressed protein precipitated as inclusion bodies. It was purified after solubilization with lithium dodecyl sulfate (LiDS) and renaturation using affinity Ni-chromatography (Fig. S4). This FRP was used to obtain antibodies. Unfortunately, the antibodies recognized only the N-terminal His-tagged FRP protein (like the anti–His tag antibody) but not the native nor the C-terminal His-tagged protein (Fig. S5). Nevertheless, the reaction was slightly better than with the anti–His tag antibody. Thus, it was used in the experiments described later.
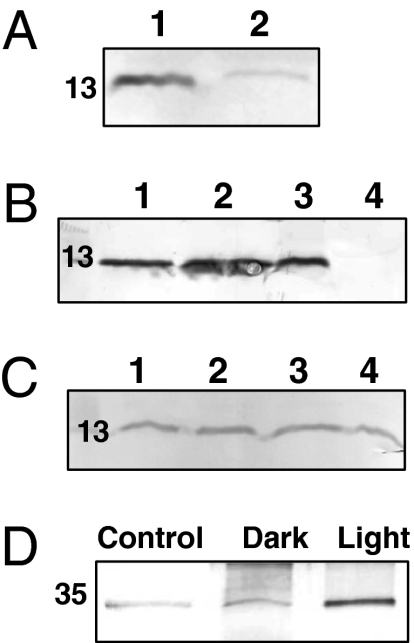
The anti-His tag and anti-FRP antibodies recognized a 13-kDa polypeptide in the strain containing an N-terminal His-tagged FRP in which the frp gene (or the putative ocp-frp operon) is under the psbA2 promoter (Fig. 4A). As predicted by the kinetic experiments, a much larger quantity of FRP was detected in the mutant overexpressing the frp gene compared with that overexpressing the ocp-frp operon (eight to 10 times more), whereas in this mutant there was approximately eight to 10 times more OCP (14) than in the frp-overexpressing mutant. However, the differences in the ratio of FRP to OCP were larger than the differences in the fluorescence recovery kinetics, suggesting that, in the ocp-frp overexpressing mutant, short FRPs, which are not detectable by our antibodies (beginning in the fourth Met and without His tag), could also be present. The possibility of the existence of short FRPs in several strains including the WT is currently being studied in our laboratory.
Fig. 4.
The location of FRP and its interaction with OCP. Immunoblot detection of FRP (A) in total proteins from the overexpressing N-terminal His-tagged FRP strain (1) and from the overexpressing OCP-FRP (N-terminal His-tagged) strain (2); (B) in total proteins (1), in the membrane-PB fraction (2), in the membrane fraction (3), and in the soluble fraction (4) isolated from the overexpressing N-terminal His-tagged FRP. (C) In isolated membranes from the overexpressing N-terminal His-tagged FRP mutant washed with Tris-HCl (1), 0.1 M Na2CO3 (2), 1 M NaCl (3), and 0.75 M NaSCN. Each lane contained 4 μg Chl (or 40 μg protein). (D) Immunoblot detection of OCP in coimmunoprecipitation experiments. The proteins attached to the Sepharose beads via the anti-FRP antibodies were eluted and separated by gel electrophoresis. OCP was then detected. The figure shows the experiment realized in darkness (dark) and under high light conditions (light, 3,000 μmol photons m−2 s−1). A control (i.e., no anti-FRP antibodies) performed under high light conditions is also shown.
To identify the cellular location of the FRP, the cells were broken in a phosphate/citrate buffer or Tris/HCl buffer to obtain PB-associated (MP) or PB-free (M) fractions by centrifugation. Protein separation by SDS/PAGE and detection of the FRP with the anti-FRP antibody showed that the FRP was present in only the membrane fractions (Fig. 4B), suggesting a strong interaction between the FRP and the thylakoids. In contrast, the OCP is present only in the soluble protein fraction when cells are broken in a Tris-HCl buffer (14). Moreover, washing out the thylakoids with 1 M NaCl or 0.75 M NaSCN did not remove the protein from the membrane, indicating a very strong attachment to the membrane and even suggesting that the FRP could be a membrane protein (Fig. 4C). FRP has no predicted hydrophobic domains. Thus, the possibility that FRP is a membrane protein was completely unexpected. Nevertheless, as the PBs and the OCP are stromal extramembrane proteins, FRP is expected to be exposed to the stromal side of the thylakoids.
To elucidate a possible interaction between the OCP and the FRP, coimmunoprecipitation experiments were performed. Total proteins of the mutant containing an N-terminal His-tagged FRP were incubated with FRP antibodies fixed to Sepharose–protein A beads in darkness or under strong white light illumination. After washing off the free proteins, the proteins attached to the anti-FRP were eluted and loaded into an SDS electrophoresis gel and OCP was detected by Western blot. Fig. 4D shows that OCP coprecipitated with the FRP. Much more OCP was coprecipitated with FRP in high light conditions than in darkness, suggesting a stronger interaction between the FRP and the OCPr than with the OCPo.
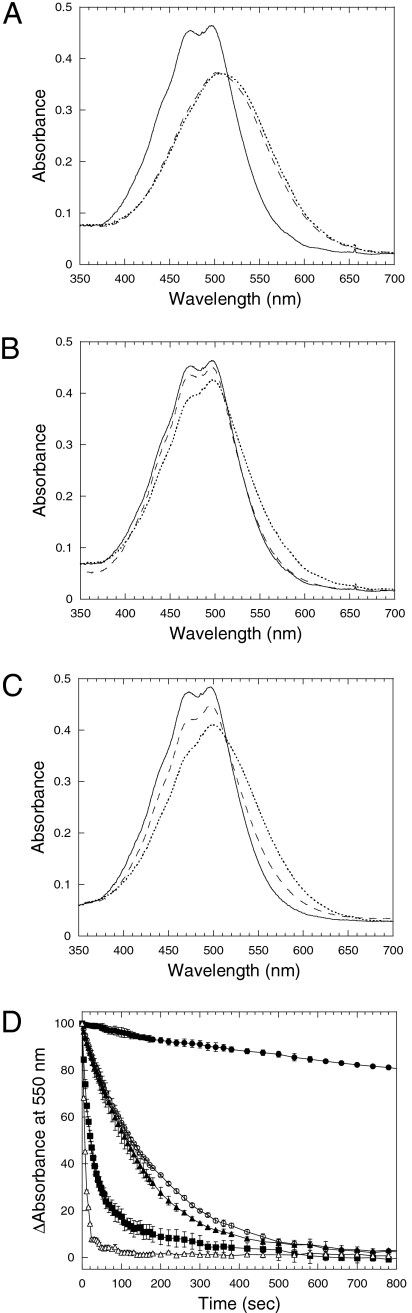
The influence of the FRP on the conversions of OCPo to OCPr and of OCPr to OCPo was also studied. The “long” FRP (His-tagged after the first Met) overexpressed in E. coli could not be used in these experiments because we were unable to obtain this protein completely purified (SI Materials and Methods), and the fraction of solubilized E. coli membrane proteins, which contaminated the FRP, rendered the OCPr completely stable (Fig. S6C). This effect was related to the procedure of solubilization as the presence of E. coli or Synechocystis membranes did not affect the stability of the OCPr (Fig. S6 D–F). Thus, we decided to overexpress a shorter FRP beginning in the Met26, as in almost all other cyanobacteria, FRP begins around this Met (Fig. 1B and Fig. S1). The “short” His-tagged FRP was mostly in the soluble fraction. It was isolated and obtained completely purified in one step using affinity Ni-chromatography (Fig. S4). The isolated protein presented no absorbance in the visible region, strongly suggesting the absence of a bound chromophore. Gel filtration experiments showed that the purified FRP (the short and the long) formed trimeric complexes in solution (Fig. S4). The photoconversion of the OCP was monitored with a spectrophotometer Spercord S600 (AnalyticJena) during illumination with high intensities of white light (5,000 μmol photons m−2 s−1) in the presence of different concentrations of the short FRP at 18 °C and 8 °C. At 18 °C, despite this high irradiance, when at least 1 mol of FRP per mol of OCP was present, almost no accumulation of the OCPr was observed, whereas half FRP concentration allowed the accumulation of some OCPr (Fig. 5). When the illumination was performed at 8 °C using the same FRP concentrations, a larger accumulation of OCPr was observed (Fig. 5). These results suggested that FRP influences the kinetics of OCPr-to-OCPo conversion. Indeed, Fig. 5D shows that the presence of FRP greatly accelerated this conversion whereas the initial rates of the light-induced OCPo-to-OCPr conversion were not affected (Fig. S7). The rates of the dark OCPr-to-OCPo conversion are temperature-dependent (14), with the t1/2 at 8 °C more than 1 h and at 18 °C approximately 120 s in the absence of FRP. The t1/2 was much lower in the presence of FRP: at 8 °C, approximately 110 s (i.e., 1 FRP per 2 OCP) and 20 s (i.e., 1 FRP per 1 OCP); at 18 °C, approximately 5 s (i.e., 1 FRP per 2 OCP).
Fig. 5.
FRP affects OCPr accumulation and accelerates OCPr-to-OCPo conversion. (A–C) Absorbance spectra of the dark (solid line) and light forms (dashed and dotted lines) of the isolated OCP. The OCP (2.3 μM) was illuminated with white light (5,000 μmol photons m−2 s−1) in the absence (A) or presence of 2.3 μM FRP (B) or 1.15 μM FRP (C) at 18 °C (dashed line) or at 8 °C (dotted line). (D) Darkness OCPr to OCPo (2.3 μM) conversion (decrease of the absorbance at 550 nm) in the absence (circle) or in the presence of 2.3 μM FRP (square) or 1.15 μM FRP (triangle) at 8 °C (closed symbols) or at 18 °C (open symbols). Average of three independent experiments is shown.
Discussion
Previously (12, 14), we demonstrated that the OCP is specifically involved in a photoprotective PB-associated mechanism and that the OCP is a photoactive protein sensing light intensity. Strong light induces the conversion between a dark stable orange form, OCPo, and a metastable active red form, OCPr. The accumulation of the OCPr is essential for the induction of the photoprotective mechanism. Here we showed that, in Synechocystis cells, the presence of the FRP, encoded by the slr1964 ORF, is essential for the recovery of a full functional antenna and high fluorescence levels when the cells are no longer under the high light regime. Homologues of the slr1964 gene are present in almost all of the OCP-containing cyanobacteria, indicating that the presence of FRP-like proteins is essential in all cyanobacteria presenting the photoprotective mechanism. The frp gene could be expressed independently of the ocp gene indicating the existence of its own specific promoter. However, the presence of long mRNA containing both genes in the overexpressing strain suggested that the two genes could be cotranscribed under some conditions.
The FRP is a 15-kDa protein (detected as a 13-kDa band in gel electrophoresis) that is strongly associated to the membrane even though its sequence was predicted to have no hydrophobic domains. The fact that the FRP, which mediates fluorescence recovery, is strongly attached to the membrane (or is a membrane protein) could explain the observation that fluorescence recovery is inhibited in membranes in a crystalline rigid state (8) and greatly decreased at low temperatures. It is possible that FRP acts as a multimer. Gel filtration analysis of the isolated E. coli overexpressed short and long FRPs showed that they form complexes of 40 to 42 and 56 to 58 kDa, respectively, suggesting the presence of stable trimeric FRP complexes in Synechocystis cells.
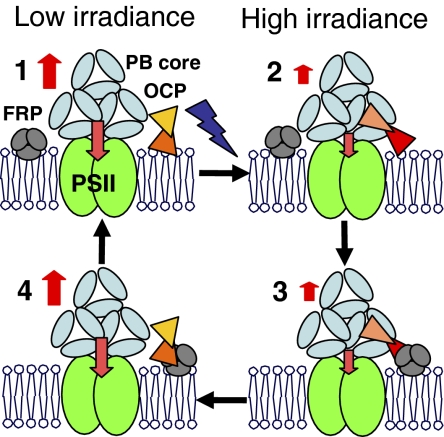
We clearly demonstrated that FRP destabilizes the OCPr. The FRP–OCP interaction accelerates the OCPr-to-OCPo conversion. Our working model is described in Fig. 6. We propose that, in darkness, the OCPo is not attached (or weakly attached) to the PB; however, once it is converted into the red form it becomes strongly attached. We have already discussed the fact that the fluorescence recovery in vivo is much slower than the red to orange reconversion of the isolated OCP (12). As the presence of Synechocystis membranes did not stabilize the OCPr, the stabilizing factor could be the PB. In this case, to be reconverted into the orange form, the OCP must be detached from the PB or interact with a destabilizing factor. By binding to the OCPr, the FRP could help the OCP to detach from the PB and then accelerate the conversion to OCPo. Our working hypothesis proposes that OCP interacts with the trimers of allophycocyanin (forming the core of the PB) via its C-terminal part to allow the interaction between the core chromophores and the OCP carotenoid (14). This proposition was suggested by the structural similarity between the C-terminal part of the OCP and the core linker protein, Lc8.9. We now propose that FRP interacts with the “free” N-terminal part of the OCP. As a higher affinity of the FRP to OCPr than to OCPo was observed, light-induced conformational changes of the OCP apoprotein are essential not only for OCP binding to the PB and fluorescence quenching but also for interaction with the FRP. The small fluorescence recovery observed in the absence of FRP could be related to a population of OCPr more loosely attached to the PB and/or to state transitions.
Fig. 6.
Working model. (1) In darkness and under low irradiance the OCP is mostly orange and not (or weakly) attached to the PB. (2) Absorption of blue-green light induces changes in the carotenoid and the protein converting OCPo to OCPr. The OCPr will bind to the APC trimers of the core of the PBs via its C-terminal region and fluorescence quenching will be induced. The PB protein and chromophore interacting with the OCPr reminds to be elucidated. (3) The FRP, as a trimer, binds to the N-terminal OCPr, (4) the FRP helps the detachment of the OCPr from the PB and accelerates the OCPr-to-OCPo conversion inducing the recovery of fluorescence. Only the core of the PB is presented in the figure.
As the FRP has no chromophore, it cannot be activated or deactivated by light, remaining permanently active, whereas OCP is activated by light absorption and deactivated in darkness. Thus, to maintain maximal photoprotection under high light conditions, a low FRP concentration compared with OCP concentration must be present. A high FRP concentration will prevent OCPr accumulation. This is suggested by the fact that the rate of fluorescence recovery depends on the ratio FRP to OCP, being faster when the ratio is bigger. In addition, a larger fluorescence quenching was observed in the absence of FRP (Fig. 2A), whereas in the frp-overexpressing strain a smaller fluorescence quenching than in the WT was detected.
The results presented in this article provide a more complete vision of the molecular mechanism of the OCP mediated-photoprotection mechanism.
Materials and Methods
Construction of His-Tagged Strains and Purification of His-Tagged FRP.
To obtain a Synechocystis strain overexpressing FRP, the slr1964 gene was amplified and cloned in the pPSBA2 ampicillin-resistant vector (21) to put the slr1964 gene under the control of the psbA2 gene promoter (Fig. 1 and Fig. S2). Nucleotides encoding 6 His were added in the 3′ or the 5′ side of the slr1964 gene and a 2-kb spectinomycin and streptomycin resistance cassette was inserted. The plasmids obtained were used to transform the kanamycin-resistant Δslr1964 mutant of Synechocystis (14). To detect the presence of FRP in the cells, total protein, membrane, and membrane-PB fractions were isolated (13) and analyzed by SDS/PAGE on 17% 2 M urea in a Tris/Mes system (22). The FRP was detected by a rabbit polyclonal antibody against the N-terminal His-tagged FRP (Covalab).
To overexpress long (from M1) and short (from M26) FRP in E. coli, the slr1964 gene was cloned in the pET-15b plasmid (Novagene), and the plasmids obtained, in which the slr1964 was under the control of the lactose promoter, were used to transform the BL21 E. coli strain adapted for protein overexpression (Stratagene). The overexpressed long FRP was obtained as inclusion bodies that were purified and dissolved with 2% LiDS. Then the LiDS was precipitated by adding 150 mM KCl in the presence of 6 M urea and 0.03% β- dodecyl-maltoside (DM). The FRP was renaturated by dialysis against 40 mM Tris-HCl pH 8 and 0.03% β-DM, and purified on a Ni-ProBond resin. The short overexpressed FRP was isolated from the soluble fraction using a Ni-column. More details are given in SI Materials and Methods and Fig. S3.
Coimmunoprecipitation.
Anti-FRP antibodies (Covalab) were fixed to 6B Sepharose–protein A beads (Sigma). Total proteins [resuspended in a buffer containing Triton X-100 (1%) and β-DM (1%) to solubilize membrane proteins] of the mutant overexpressing the FRP with an N-terminal His tag were incubated with the anti-FRP antibodies bound to the beads. The beads were washed to eliminate the nonbound proteins and the proteins attached to the anti-FRP antibodies were eluted with the loading buffer of SDS/PAGE and loaded on the SDS gel. A Western blot using anti-OCP antibodies was performed. The control fraction did not contained anti-FRP antibodies. The experiments were performed in darkness or under a strong white light (3,000 μmol photons m−2 s−1) during the incubation and washing steps.
Total RNA Isolation and RT-PCR.
Total RNA was isolated from 10 mL of Synechocystis cells at a DO800nm of 0.8 using the bead-phenol-chloroform method described by Kim et al. (23) and treated with RNase-free DNase I (Sigma). RT-PCR was carried out using Omniscript reverse transcriptase (Qiagen). The primers 64–33-R, 64–232-R, 64–394-R, 63–744R, and car-R (Table S2) were used to construct the cDNAs. Then, the cDNAs were amplified by PCR using the oligonucleotides described in Table S2.
OCP Purification and Fluorescence Quenching.
The purification of Synechocystis OCP was performed as previously described (14). The kinetics of fluorescence changes were monitored in a modulated PAM fluorometer (Walz) as previously described (8).
Supplementary Material
Acknowledgments
We thank G. Ajlani (CEA Saclay, France) and W. Vermaas (Arizona State University, AZ) for the gift of the plasmid pPSBA2 and A. W. Rutherford, C. Kerfeld, and G. Ajlani for critical reading of the article. This work was supported by grants from Agence Nationale de la Recherche (Caroprotect program) and Marie Curie Research Training Network Grant INTRO2 EU FP6.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002912107/-/DCSupplemental.
References
- 1.Horton P, Ruban AV, Walters RG. Regulation of light harvesting in green plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:655–684. doi: 10.1146/annurev.arplant.47.1.655. [DOI] [PubMed] [Google Scholar]
- 2.Niyogi KK. Photoprotection revisited: Genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- 3.Pascal AA, et al. Molecular basis of photoprotection and control of photosynthetic light-harvesting. Nature. 2005;436:134–137. doi: 10.1038/nature03795. [DOI] [PubMed] [Google Scholar]
- 4.Ruban AV, et al. Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature. 2007;450:575–578. doi: 10.1038/nature06262. [DOI] [PubMed] [Google Scholar]
- 5.Adir N. Elucidation of the molecular structures of components of the phycobilisome: Reconstructing a giant. Photosynth Res. 2005;85:15–32. doi: 10.1007/s11120-004-2143-y. [DOI] [PubMed] [Google Scholar]
- 6.Glazer AN. Light harvesting by phycobilisomes. Annu Rev Biophys Biophys Chem. 1985;14:47–77. doi: 10.1146/annurev.bb.14.060185.000403. [DOI] [PubMed] [Google Scholar]
- 7.MacColl R. Cyanobacterial phycobilisomes. J Struct Biol. 1998;124:311–334. doi: 10.1006/jsbi.1998.4062. [DOI] [PubMed] [Google Scholar]
- 8.El Bissati K, Delphin E, Murata N, Etienne A, Kirilovsky D. Photosystem II fluorescence quenching in the cyanobacterium Synechocystis PCC 6803: Involvement of two different mechanisms. Biochim Biophys Acta. 2000;1457:229–242. doi: 10.1016/s0005-2728(00)00104-3. [DOI] [PubMed] [Google Scholar]
- 9.Rakhimberdieva MG, Elanskaya IV, Vermaas WFJ, Karapetyan NV. Carotenoid-triggered energy dissipation in phycobilisomes of Synechocystis sp. PCC 6803 diverts excitation away from reaction centers of both photosystems. Biochim Biophys Acta. 2010;1797:241–249. doi: 10.1016/j.bbabio.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Rakhimberdieva MG, Stadnichuk IN, Elanskaya IV, Karapetyan NV. Carotenoid-induced quenching of the phycobilisome fluorescence in photosystem II-deficient mutant of Synechocystis sp. FEBS Lett. 2004;574:85–88. doi: 10.1016/j.febslet.2004.07.087. [DOI] [PubMed] [Google Scholar]
- 11.Scott M, et al. Mechanism of the down regulation of photosynthesis by blue light in the Cyanobacterium synechocystis sp. PCC 6803. Biochemistry. 2006;45:8952–8958. doi: 10.1021/bi060767p. [DOI] [PubMed] [Google Scholar]
- 12.Wilson A, et al. A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell. 2006;18:992–1007. doi: 10.1105/tpc.105.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson A, Boulay C, Wilde A, Kerfeld CA, Kirilovsky D. Light-induced energy dissipation in iron-starved cyanobacteria: Roles of OCP and IsiA proteins. Plant Cell. 2007;19:656–672. doi: 10.1105/tpc.106.045351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson A, et al. A photoactive carotenoid protein acting as light intensity sensor. Proc Natl Acad Sci USA. 2008;105:12075–12080. doi: 10.1073/pnas.0804636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Punginelli C, Wilson A, Routaboul JM, Kirilovsky D. Influence of zeaxanthin and echinenone binding on the activity of the orange carotenoid protein. Biochim Biophys Acta. 2009;1787:280–288. doi: 10.1016/j.bbabio.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Boulay C, Wilson A, Kirilovsky D. 2007 Energy from the Sun Proceedings of the 14th International Congress on Photosynthesis. Heidelberg: Springer; 2008. Orange carotenoid protein (OCP) related NPQ in synechocystis PCC 6803 OCP-phycobilisomes interactions in Photosynthesis; pp. 254–254. [Google Scholar]
- 17.Wu YP, Krogmann DW. The orange carotenoid protein of Synechocystis PCC 6803. Biochim Biophys Acta. 1997;1322:1–7. doi: 10.1016/s0005-2728(97)00067-4. [DOI] [PubMed] [Google Scholar]
- 18.Fulda S, et al. Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics. 2006;6:2733–2745. doi: 10.1002/pmic.200500538. [DOI] [PubMed] [Google Scholar]
- 19.Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M. DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell. 2001;13:793–806. doi: 10.1105/tpc.13.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boulay C, Abasova L, Six C, Vass I, Kirilovsky D. Occurrence and function of the orange carotenoid protein in photoprotective mechanisms in various cyanobacteria. Biochim Biophys Acta. 2008;1777:1344–1354. doi: 10.1016/j.bbabio.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Lagarde D, Beuf L, Vermaas W. Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl Environ Microbiol. 2000;66:64–72. doi: 10.1128/aem.66.1.64-72.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kashino Y, Koike H, Satoh K. An improved sodium dodecyl sulfate-polyacrylamide gel electrophoresis system for the analysis of membrane protein complexes. Electrophoresis. 2001;22:1004–1007. doi: 10.1002/1522-2683()22:6<1004::AID-ELPS1004>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 23.Kim B-H, et al. Simple method for RNA preparation from cyanobacteria. J Phycol. 2006;42:1137–1141. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.