Abstract
In the silkworm Bombyx mori, dietary flavonoids are metabolized and accumulate in cocoons, thereby causing green coloration. Classical genetic studies suggest that more than seven independent loci are associated with this trait; however, because of the complex inheritance pattern, none of these loci have been characterized molecularly, and a plausible and comprehensive model for their action has not been proposed. Here, we report the identification of the gene responsible for the Green b (Gb) locus involving the green cocoon trait. In +Gb animals, glucosylation at the 5-O position of dietary quercetin did not occur, and the total amount of flavonoids in tissues and cocoons was dramatically reduced. We performed positional cloning of Gb and found a 38-kb deletion in a UDP-glucosyltransferase (UGT) gene cluster associated with the +Gb allele. RT-PCR and biochemical studies suggested that deletion of Bm-UGT10286 (UGT) is responsible for Gb and Bm-UGT10286 is virtually the sole source of UGT activity toward the 5-O position of quercetin. Our data show that the regiospecific glucosylation of flavonoids by the quercetin 5-O-glucosyltransferase can greatly affect the overall bioavailability of flavonoids in animals. Furthermore, we provide evidence that flavonoids increase the UV-shielding activity of cocoons and thus could confer an increased survival advantage to insects contained in these cocoons. This study will lead to greater understanding of mechanisms for metabolism, uptake, and transport of dietary flavonoids, which have a variety of biological activities in animals and beneficial effects on human health.
Keywords: flavonoid, UGT, Bombyx, mulberry
Cocoon-making behavior is the most highly developed in Lepidoptera (moths and butterflies). The silkworm Bombyx mori is a typical cocoon-making insect. Silkworms have been reared for more than 5,000 y for silk production, and more than 1,000 strains including geographical and mutant strains have been collected (1–3). Among them, many cocoon color mutants, including white, yellow, golden yellow, orange, pinkish, and green, have been maintained (2, 3) (Fig. 1A). Silkworm cocoon colors are determined by two main pigments, carotenoids (4) and flavonoids (5), which are derived from mulberry leaves (6). Yellow and pinkish colors are produced from carotenoids (4, 7), and green colors are the products of flavonoids (8, 9). Inheritance for the carotenoid cocoon has been well established and basically follows a classical Mendelian pattern (2, 3, 5, 7). However, the inheritance pattern for the flavonoid cocoon is much more complicated and largely unknown (5). This is mainly because the metabolic processing of flavonoids absorbed from diets is much more complex than that of carotenoids. In fact, green cocoons of some silkworm varieties contain more than 30 kinds of flavonoids that are not present in their diet (8–12), whereas the main constituents of yellow and pinkish cocoons are lutein and β-carotene (5, 7), respectively, both of which are original components in mulberry leaves. In addition, green cocoon is usually observed as a quantitative trait (i.e., continuous color variations ranging from pale or light green to deep or yellowish green), modified variously by environmental conditions, and governed by many loci (2, 3, 5). Extensive efforts to characterize and map the responsible genes have been carried out since the early 20th century, and at least seven and perhaps more than 10 independent loci are suggested to associate with the green cocoon trait (2, 3, 5). These include Ga (Green a; locus unknown) (13), Gb (Green b; 7–7.0) (13), Gc (Green c; 15-?), Grc (Green cocoon; locus unknown), Gre (Green egg shell; 1–46.4), I-gn-1 (Inhibited green 1; 6–28.6), and Yf (Yellow fluorescent; locus unknown) (3). However, because of the previously mentioned difficulties, none of these genes have been characterized molecularly, and a plausible and comprehensive model for their action has not been proposed.
Fig. 1.
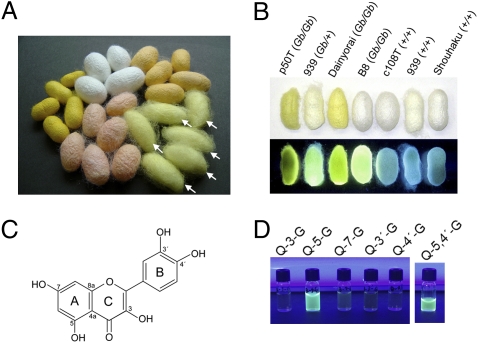
Cocoons from Gb strains contain 5-O-glucosylated quercetins that emit bright yellow fluorescence under UV light. (A) Silkworm cocoons of various colors. Green color is produced from flavonoids (arrows), whereas other colors, yellow, orange and pink, are from carotenoids. (B) Cocoons from several silkworm strains under white light (upper) and UV-B (302 nm) (lower). Strain names and their presumptive genotypes (in parentheses) are indicated above. Cocoons emit yellow fluorescence in the presence of the Gb allele (detailed in Fig. S4). (C) The structural formula of quercetin. (D) Quercetin emits yellow fluorescence when its 5-O position is glucosylated (Q-5-G), but does not do so when other positions, 3-O, 7-O, 3′-O or 4′-O, are glucosylated (Q-3-G, Q-7-G, Q-3′-G, or Q-4′-G, respectively). Fluorescence was also observed in quercetin 5,4′-diglucoside (Q-5,4′-G), the major constituent of the cocoon flavonoids in strain 939 (Gb/+) (Table S1). Concentration of quercetin glucosides in each vial was 100 nmol/mL.
Here, we report the identification of the gene responsible for the Gb locus. We demonstrate that Gb locus encodes a quercetin 5-O-glucosyltransferase (Q5GT) that catalyzes the regioselective formation of quercetin 5-O-glucosides, the major constituent of cocoon flavonoids that emit bright yellow fluorescence under UV light. Our data suggest that this regioselective glucosylation is the first critical step for the biosynthesis of cocoon flavonoids and thereby greatly affects the overall bioavailability of dietary flavonoids in the silkworm. Despite its great importance in sericultural economy and biotechnology (14), the biological significance of the cocoon and its pigments in the silkworm lifecycle has been poorly studied and is largely undetermined. We also provide evidence that flavonoids in the silkworm cocoon can act as a chemical UV shield, protecting the prepupae from harmful effects of sunlight during metamorphosis.
Results
Gb Locus Controls Biosynthesis of the Flavonoid Quercetin 5-O-Glucoside.
The major components of flavonoids in green cocoons are quercetin 5-O-glucosides such as quercetin 5,4′-di-O-glucoside (8). We found that cocoons from silkworm strains carrying the Gb locus emitted a bright yellow fluorescence under UV light (Fig. 1B). This yellow fluorescence seems to be derived from 5-O-glucosylated flavonoids, as only quercetin-5-O-glucoside emitted fluorescence among quercetin monoglucosides (Fig. 1D). To see the effect of the Gb locus on the profile of cocoon flavonoids, we analyzed flavonoids in the cocoon and several tissues of silkworm strain 939 (Table S1). In this strain, the Gb allele is maintained as a heterozygote, and the +Gb allele is linked with the recessive larval body marker gene quail (q) (Fig. S1). (3). Therefore, we could easily distinguish Gb/+ and +/+ larvae by observing their body markings. As shown in Table S1, we observed a dramatic difference in the profile of quercetin metabolites between Gb/+ and +/+ individuals. For example, in silk glands and cocoons, +/+ individuals completely lacked quercetin 5-O-glucosides, which are major constituents of cocoon flavonoids (8), and the total amount of quercetin metabolites was significantly reduced to ≈10–15% of that in Gb/+ individuals. Previous reports have suggested that quercetin 5-O-glucosides are synthesized by UDP-glucosyltransferase (UGT) (8). Thus, we next compared UGT activity between Gb/+ and +/+ larvae (Table 1) and found a clear difference between them. In Gb/+ larvae, the significant UGT activity for the 5-O position of quercetin (quercetin 5-O-glucosyltransferase; Q5GT) was detected in Malpighian tubules, anterior silk glands, midgut, hemocytes, and fat bodies. In contrast, however, +/+ animals completely lost Q5GT activity in Malpighian tubules, anterior silk glands, midgut, and hemocytes, and had almost no activity in fat bodies. Importantly, such dramatic differences in UGT activity were not observed for other positions of quercetin (Table 1), indicating that the loss of the Gb allele specifically reduced Q5GT activity in +/+ larvae of strain 939. Taken together, these results suggest that the Gb locus controls the synthesis of quercetin 5-O-glucosides, probably by regulating the activity of UGT with a preferred 5-O regioselectivity for quercetin.
Table 1.
Glucosyltransferase activity toward quercetin in tissues from Gb/+ and +/+ individuals of strain 939
| UGT activity (nmol/min per mg protein) |
||||||
| Genotype | Tissue | 3-O-glc | 5-O-glc | 7-O-glc | 3′-O-glc | 4´-O-glc |
| Gb/+ | Fat body | 13.31 ± 1.49 | 0.50 ± 0.06* | 0.12 ± 0.04 | 0.12 ± 0.02 | 0.51 ± 0.26 |
| Hemocyte | 0.21 ± 0.04 | 0.35 ± 0.05† | — | — | — | |
| Anterior silk gland | 51.60 ± 14.50 | 1.74 ± 0.51† | — | — | 1.06 ± 0.39 | |
| Middle silk gland | — | — | 9.74 ± 0.98 | — | 0.60 ± 0.09 | |
| Posterior silk gland | — | — | 15.74 ± 2.05* | — | 0.92 ± 0.31 | |
| Midgut | 3.58 ± 1.74 | 1.63 ± 0.30† | 0.04 ± 0.01 | 0.03 ± 0.01 | 0.09 ± 0.03 | |
| Malpighian tubule | 96.56 ± 9.70 | 4.20 ± 0.52† | — | — | 0.71 ± 0.15 | |
| +/+ | Fat body | 12.90 ± 2.58 | 0.08 ± 0.05* | 0.17 ± 0.05 | 0.10 ± 0.04 | 0.78 ± 0.44 |
| Hemocyte | 0.24 ± 0.07 | — | — | — | — | |
| Anterior silk gland | 49.15 ± 11.09 | — | — | — | 0.72 ± 0.22 | |
| Middle silk gland | — | — | 10.44 ± 3.13 | — | 0.47 ± 0.11 | |
| Posterior silk gland | — | — | 7.77 ± 1.01* | — | 0.41 ± 0.02 | |
| Midgut | 2.56 ± 1.17 | — | 0.03 ± 0.01 | 0.01 ± 0.01 | 0.05 ± 0.02 | |
| Malpighian tubule | 86.83 ± 13.55 | — | — | — | 0.51 ± 0.15 | |
Values are mean ± SD (n = 5). —, not detected.
*Activity was significantly different between Gb/+ and +/+ (P < 0.01, t test).
†Activity was not detected in +/+ larvae.
Positional Cloning of the Gb Locus.
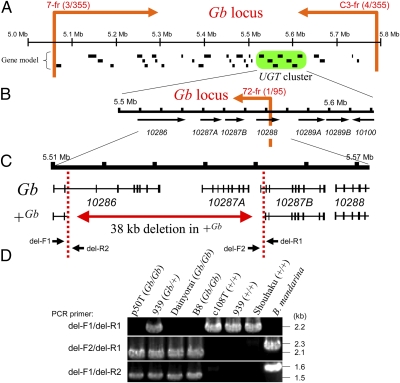
To identify the gene responsible for Gb, we performed positional cloning using F1 progeny obtained from crossing females homozygous for q and +Gb with males heterozygous for both loci from strain 939 (Fig. S1). We first roughly mapped the Gb locus using 355 F1 individuals and were able to narrow the responsible region within ≈750 kb on chromosome 7, scaffold 2986 (Fig. 2A). Notably, we found a UGT gene cluster that harbors seven UGTs (15), termed Bm-UGT10286, 10287A, 10287B, 10288, 10289A, 10289B, and 10100 (Fig. 2B), within this region. We next performed fine mapping using the F1 individuals in which recombination had occurred between Gb and q (Fig. S1). Based on the larval marking, we collected 95 recombinants from ≈1,700 F1 individuals and subjected them to more detailed mapping. These results showed that the Gb locus was located upstream of the sixth intron of Bm-UGT10288 (Fig. 2B and Fig. S1), where we identified a large genomic deletion (38 kb) in the +Gb allele (Fig. 2C). The deletion ranged from the second intron of Bm-UGT10286 to the first intron of the Bm-UGT10287B, thus causing structural defects in Bm-UGT10286, 10287A, and 10287B. This large deletion was conserved in all of the silkworm races carrying the +Gb allele (Fig. 2D), but not in Bombyx mandarina, an ancestral species of B. mori, suggesting a common and single origin of this allele.
Fig. 2.
Positional cloning of Gb. (A) Physical map indicating the results of the initial linkage analysis using 355 F1 progeny obtained from a male informative cross of strain 939. The Gb locus was narrowed to the genomic region flanked by PCR markers 7-fr and C3-fr, as indicated by orange arrows with the number of recombinant animals/total number of scored individuals (Fig. S1). Putative genes predicted by Gene model program (34) are shown below map, and UGT gene cluster harboring seven UGTs (15) is highlighted in green. (B) Physical map resulting from fine mapping using 95 recombinants between Gb and q loci recovered from male informative F1 progeny (~1,700 individuals) of strain 939 (Fig. S1). Gb locus was located upstream of marker 72-fr, as indicated by orange arrow. Black arrows indicate position and orientation of transcription of seven UGTs. (C) Physical map indicating 38-kb deletion found in +Gb allele. Exons of UGTs are indicated by vertical lines. (D) Genotyping of several silkworm strains and B. mandarina for Gb. Genomic PCR was performed using primers (listed in Table S3) that flank the deletion (del-F1 and del-R1) or are located inside the deletion (del-F2 and del-R2), as shown in C. Strain names and their genotypes (in parentheses) are indicated above. Note that these PCR primers can anneal to the genome of B. mandarina and that the deletion was not found in this species.
Characterization of UGT Candidates for the Gb Gene.
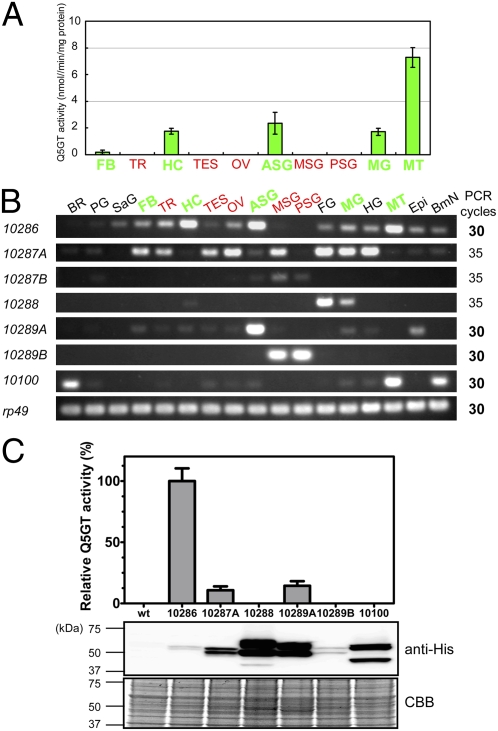
To characterize the UGTs present within the region responsible for Gb (Fig. 2), we first performed RACE experiments and determined their full-length sequences. They shared extensive similarities in genomic structure (eight to nine exons) and coding sequences (64–80% amino acid identity) (Fig. S2). However, despite clustered arrangement and structural similarities of these UGTs, their expression patterns were highly diverse (Fig. 3B). In the p50T strain, we observed expression of Bm-UGT10286 and 10287A in many tissues, whereas those of Bm-UGT10288, 10289A, 10289B, and 10100 were expressed in a strict tissue-specific manner. For example, Bm-UGT10288 was expressed specifically in the foregut and midgut, BmUGT-10289A was highly specific to the anterior silk gland, and BmUGT-10289B was specific to middle and posterior silk glands. To determine which UGT gene is responsible for Gb, we compared the expression pattern of these UGTs with the tissue distribution of UGT activities in p50T. As shown in Fig. 3A and Table S2, we detected significant Q5GT activity in Malpighian tubules, anterior silk gland, midgut, hemocytes, and fat bodies, but not at all in the middle or posterior silk glands. Remarkably, among seven UGTs within the region responsible for Gb, only one UGT, Bm-UGT10286, exhibited an expression pattern closely similar to the tissue distribution of the Q5GT activity. Bm-UGT10286 was strongly expressed in Malpighian tubules, anterior silk glands, and hemocytes, where strong Q5GT activity was detected, but was not expressed in middle or posterior silk glands, where no Q5GT activity was detected, suggesting that BmUGT-10286 is a strong candidate for Gb.
Fig. 3.
Characterization of seven UGT genes. (A) Q5GT activity in tissues of p50T. UGT activity toward other position of quercetin is listed in Table S2. Tissues that possessed Q5GT activity are shown in green letters and those did not are in red letters. See below for abbreviations of tissues. Bars indicate the mean± SD (n = 5). (B) Semiquantitative RT-PCR analysis of seven UGTs. We analyzed total RNA from fifth instar day 3 larvae of the strain p50T or BmN cells. PCR cycles are shown on the right. Note that mRNA expression levels of Bm-UGT10287A, 10287B and 10288 were lower than those of the others. Tissues that possessed Q5GT activity are shown in green; tissues those did not possess Q5GT are shown in red. ASG, anterior silk gland; BmN, BmN-cultured cells; BR, brain; Epi, epidermis; FB, fat body; FG, foregut; HC, hemocyte; HG, hindgut; MG, midgut; MSG, middle silk gland; MT, Malpighian tubules; OV, ovary; PG, prothoracic gland; PSG, posterior silk gland; SaG, salivary gland; TES, testis; TR, trachea. Ribosomal protein 49 gene (rp49) was also analyzed as control. (C) Relative Q5GT activity and expression of recombinant UGTs. Q5GT activity was measured using crude extracts of Sf9 cells infected with WT or recombinant baculoviruses expressing each UGT with a His6-tag at C terminus. Relative expression level of each UGT was determined by immunoblot. Q5GT activity was normalized according to relative expression level of UGTs. Level of Q5GT activity in Bm-UGT10286 was set to 100%. Bm-UGT10287B was not analyzed, as this gene seemed to encode nonfunctional protein due to the truncation at the N terminus (Fig. S2). Bars indicate mean ± SD (n = 5). Results of CBB staining and immunoblot analysis for detection and normalization of recombinant UGTs are also shown below.
To confirm this further, we next examined the enzymatic activity of these UGTs. We expressed recombinant UGTs in Sf9 cells using a baculovirus expression system, and confirmed their expression in extracts of virus-infected cells by immunoblot analysis (Fig. 3C). The observed molecular masses of these UGTs were almost the same as predicted, but each UGT had two bands, suggesting that they were glycosylated. We measured enzymatic activity of these UGTs using extracts from mock- or virus-infected cells (Fig. 3C). Because Sf9 cells had a high background of UGT activity toward C-3, 7 and 4′ positions of quercetin, we could not compare data for these positions. Fortunately, however, we did not detect background activity toward the C-5 position, allowing us to compare the Q5GT activity in extracts of virus-infected cells directly. As shown in Fig. 3C, Bm-UGT10286 showed the highest Q5GT activity. Although we also detected Q5GT activity in Bm-UGT10287A and 10289A, it is unlikely that they are responsible for Gb for the following reasons. First, their Q5GT activities were much lower than the activity of Bm-UGT10286 (less than 20%). Second, the expression pattern of Bm-UGT10287A did not correspond to the tissue distribution of Q5GT activity (Fig. 3A). The expression level of Bm-UGT10287A was extremely low in Malpighian tubules, anterior silk glands, and hemocytes, where strong Q5GT activity was observed (Fig. 3A). Third, Bm-UGT10289A is not deleted in the +Gb allele. The large 38-kb genomic deletion in the +Gb allele might change the expression pattern of UGTs outside of the deletion, but their expression patterns were not affected in +/+ larvae of strain 939 (Fig. S3). Taken together, our data suggest that Bm-UGT10268, which is deleted in the +Gb allele, is the major source of Q5GT activity in the silkworm and thus is responsible for the Gb locus.
Flavonoids in Green Cocoons Act as Chemical Shields Against UV.
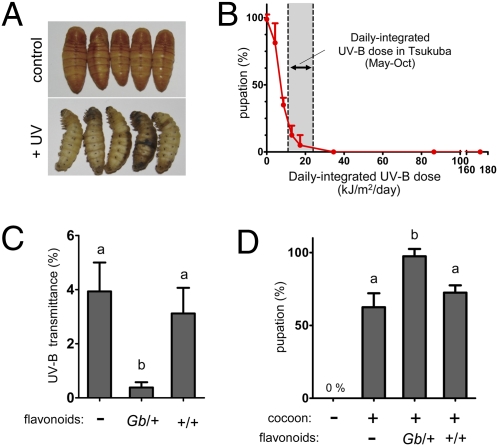
Silkworm larvae begin to spin when they fully develop and stop eating. After ~2 d, they cease spinning silk thread and enter the state called prepupae for ~24 h until pupal ecdysis (5).We found that silkworm prepupae are highly sensitive to UV-B irradiation when they are removed from their protective cocoon covering. When we irradiated naked prepupae with UV-B, their bodies turned black (Fig. 4A), and their pupation rates dramatically decreased with the increase of the UV-B dose (Fig. 4B). No naked prepupae became pupae above a UV-B dose of 34.6 kJ/m2/d. In contrast, ~60% of prepupae placed in cocoons of the strain Annam successfully became pupae even when irradiated with UV-B at 172.8 kJ/m2/d (Fig. 4D). This clear difference in UV-B tolerance indicates that the cocoon serves as a shield that protects the silkworm from UV-B during the larval–pupal transformation. According to records of the Japan Meteorological Agency, the daily-integrated UV-B dose in the field in Tsukuba, Japan, is 11–23 kJ/m2/d during May to October. Therefore, it is noteworthy that only 5.0% of naked prepupae became pupae when irradiated with UV-B at 17.3 kJ/m2/d (Fig. 4B). Our data suggest that the pupation rate of the silkworm in the field is quite low if prepupae are not protected in cocoons from UV-B–containing solar radiation. We also investigated the effect of irradiation of UV-A, which is the predominant component of solar UV radiation and is thought to be less harmful to animals. As expected, we did not see a harmful effect of UV-A irradiation on the pupation rate even at a maximum dose of 518.4 kJ/m2/d. Thus, we focused on the effect of UV-B irradiation in the following experiments.
Fig. 4.
UV-shielding effect of flavonoids in the cocoon. (A) UV-B-irradiation at the prepupal stage impairs the normal larval-pupal transition. Two days after the onset of spinning, we irradiated naked prepupae with UV-B (200 μW/cm2) for 24 h (172.8 kJ/m2/d). (B) Effect of UV-B intensity on the pupation rate. We irradiated naked prepupae with different doses of UV-B (0, 4.3, 8.6, 13.0, 17.3, 34.6, 86.4, and 172.8 kJ/m2/d) as described in A and measured the pupation rates. The values for the daily-integrated UV-B dose in Tsukuba, Japan, were retrieved from the Japan Meteorological Agency. We repeated the experiment eight times with 10 individuals in each treatment. The points indicate the mean± SD (n = 8). (C) Transmittance ratio of UV-B through the cocoon shells of the strain Annam with or without (-) application of flavonoids. Flavonoids were extracted from cocoons of each genotype of strain 939 (Gb/+ and +/+) and applied to cocoons of Annam. Bars indicate mean ± SD (n = 6). Bars with different letters represent significantly different values (P < 0.01, Tukey test). (D) UV-shielding effect of flavonoids applied to the cocoon. Prepupae of Annam were removed from their own cocoons and placed in Annam cocoons, with or without (−) application of flavonoids extracted from cocoons of each genotype of strain 939 (Gb/+ and +/+). Insects were then irradiated with UV-B (172.8 kJ/m2/d). Experiment was repeated four times with 10 individuals in each treatment. Bars indicate mean± SD (n = 4). Bars with different letters represent significantly different values (P < 0.05, Tukey test).
We next investigated whether flavonoids could increase the UV-shielding activity of cocoons. To do this, we extracted flavonoids from cocoons of each genotype of strain 939 (Gb/+ and +Gb/+Gb) and applied them to cocoons of Annam, which originally contain low amounts of flavonoids (0.26 μmol/g cocoon) (9). We first compared the level of transmittance of UV-B radiation through these cocoon shells (Fig. 4C). The average transmittance of UV-B through the cocoon shells of Annam was 3.9% (Fig. 4C). When we applied the extract of Gb/+ cocoons to the cocoons of Annam, this value decreased 10-fold to 0.38%. However, when we applied the extract of +Gb/+Gb cocoons, the transmittance did not decrease significantly (3.1%). There was also a clear difference in pupation rates when naked prepupae were placed in Annam cocoons with or without applied flavonoids. As shown in Fig. 4D, the application of cocoon extracts from Gb/+ conferred almost perfect tolerance to UV-B irradiation, but this was not observed when we used the extract from +/+. These results suggest that 5-O-glucosylated quercetins, synthesized primarily by Bm-UGT10286, can act as chemical potentiators of UV-shielding effects of cocoons.
Discussion
The data presented here have two important implications. First, we clearly uncovered the genetic basis for the regioselective formation of quercetin 5-O-glucoside, which is the first step for the biosynthesis of cocoon flavonoids (see the proposed metabolic pathway of dietary quercetin in the silkworm; Fig. S4) and is a critical determinant of the bioavailability of flavonoids. Second, we presented unique evidence for the functional consequence of flavonoids contained in an insect cocoon.
The green color of the silkworm cocoon is due to flavonoids, one of the two main pigments of the silkworm cocoon (5, 6). In contrast to carotenoids, the other major pigments that control the yellow and pinkish color of cocoons, the genetic basis of green cocoon traits has remained largely obscure. Our data clearly demonstrate that a gene for UDP-glucosyltransferase of the silkworm, Bm-UGT10286, corresponds to the Gb locus, and that its product is essential for the biosynthesis of quercetin 5-O-glucosides, major constituents of cocoon flavonoids. A distinctive character of Bm-UGT10286 is its unusual regioselective activity. The OH group at the C-5 position of flavone is the most inert site for glycosylation, as the carbonyl group at C-4 prevents glycosylation of OH at C-5 by producing steric hindrance and strong intramolecular hydrogen bonding (16). Thus, 5-O-glycosylated flavones and flavonols are rare compounds in nature (17, 18). Despite the high copy number (more than 42) of UGT genes in the silkworm genome (15), Q5GT activity was almost lacking in +Gb/+Gb individuals (Table 1). This result indicates that the Gb locus is virtually the sole source of a UGT that possesses Q5GT activity in the silkworm, reflecting the unusual property of Bm-UGT10286 with a preferred 5-O regioselectivity for quercetin.
The presence of high amounts of 5-O-glucosylated quercetins in silkworm tissues and cocoons (Table S1) (8, 12) raises the question of why the most inert position among hydroxyl groups of quercetin is regioselectively glucosylated. One may predict that quercetin 5-O-glucosides possess unique (or higher) biological activity that is not seen in other quercetin glucosides. However, it is more likely that 5-O-glucosylation of quercetin dramatically increases the bioavailability of quercetin in the silkworm by changing its solubility, permeability, metabolism, excretion, target tissue uptake, or disposition. In mammals, after absorption from the small intestine, flavonoids undergo extensive metabolism, generating different conjugation forms such as glucuronides and sulfates (19, 20). The bioavailability of flavonoids is generally low and can be problematic in pharmacological studies (19, 21, 22), as these conjugation reactions can enhance the elimination of lipophilic xenobiotics (19, 21). It has been shown that glucuronidation of flavonoids can influence the bioavailability of flavonoids in mammals partly by changing their permeability and interactions with their putative transporters such as ATP-binding cassette superfamily transporters (19–21, 23, 24). However, exact mechanisms for absorption, transport, and disposition of flavonoids are not well understood in any animal system (19, 21), and the degree to which the site specificity in the glucuronidation step is important for the bioavailability of flavonoids remains unclear (19, 21, 24). It is notable that the glucosylation reaction at the 5-O position of quercetin caused a marked increase in the amount of flavonoids in silk glands, hemolymph, and cocoon (Table S1). This example clearly shows how glucosylation of flavonoids and its site specificity can affect the overall bioavailability of flavonoids in an animal. Moreover, our data imply that silkworm has a genetically facilitated system for the selective transport of quercetin 5-O-glucosides. According to previous reports (8), dietary quercetin is glucosylated at the 5-O position in the midgut as the first-pass metabolite after oral absorption and is then glucosylated at the 4′-O position in the fat body and silk gland. The significant decrease of flavonoids in +Gb/+Gb tissues and cocoons (Table S1) suggests that glucosylation at the 5-O position of quercetin in the midgut may be a critical determinant of its overall bioavailability, as shown in Fig. S4. In mammals, flavonoid glucuronides are excreted out of enterocytes to both the serosal and luminal sides of intestinal epithelium via efflux transporters (19, 23, 24). Thus, we speculate that glucosylation at the 5-O position might allow or facilitate the efficient uptake and transport of quercetin from the midgut to the silk glands by altering interactions with efflux and/or influx transporters.
To address these questions, we must investigate the detailed mechanisms for the uptake and transport of flavonoids. It is noteworthy that some silkworm mutants may have genetic defects in this process. Ga, which produces green-colored cocoons only when it coexists with Gb (13), is a candidate locus for the transport of flavonoids. Implantation of Ga/Ga silk glands to +Ga/+Ga individuals results in the uptake of hemolymph flavonoids to Ga/Ga silk glands, suggesting that the Ga locus controls the uptake of flavonoids in the silk gland (25) (Fig. S4). The results of our work will facilitate the isolation of Ga and other loci associated with the green cocoon trait and consequently pave the way for a greater understanding of the metabolism and uptake of flavonoids in animals. We also expect that our findings will lead to the molecular breeding and genetic engineering of silkworms for silk fabric containing flavonoids (26) and to the development of a regioselective biocatalyst for the potential production of pharmaceuticals.
Flavonoids play various roles in a wide variety of organisms as antioxidants, antibacterial agents, and UV absorbers (19, 22, 27). We asked whether flavonoids in cocoon shells have functional consequences in silkworm biology. We provide evidence that flavonoids could increase the UV-protection activity of cocoons and thus confer an increased survival advantage (Fig. 4). It is believed that the silkworm was domesticated from the B. mandarina in China (28). Our data suggest that the green cocoon is likely the ancestral form of the silkworm cocoon, as the Gb locus is conserved in B. mandarina (Fig. 2). Thus, we speculate that cocoon flavonoids play a more important role in B. mandarina, whose cocoons are much thinner and more fragile. A number of pigments, such as carotenoids (4), flavonoids (6), bilins (29), and their metabolites are contained in many insect cocoons. In addition to pigments, a pyralid moth, Uresiphita reversalis, accumulates toxic alkaloids derived from host plants in its cocoons (30), and cocoons of a leaf-rolling moth, Dactylioglypha tonica, are densely covered by a blue mold that could protect pupae from bacterial infections (31, 32). Furthermore, the duration of the pupal stage varies in different insects from a few days to several months, suggesting that they may have evolved additional novel protective mechanisms. These observations, along with the present data, provide an expanded view of insect cocoons: cocoons are not only simple physical shields for insects but can be chemically or biologically modified by insects using diverse strategies for more extensive forms of protection. Our findings further imply that pigments and chemicals contained in cocoons might be a useful source for novel bioactive substances for textiles, medicines, cosmetics, and agrochemicals.
Materials and Methods
A detailed description of materials and methods is provided in SI Materials and Methods.
Determination of UGT Activity.
UGT activity was measured as described previously (8). Details are given in SI Materials and Methods.
Positional Cloning.
Positional cloning of Gb was performed as previously described (33). Codominant PCR markers were generated at each position on nscaf2986 (chromosome 7) (Fig. 2) (34), and used for the genetic analysis (Fig. S1). The presence of Gb allele was confirmed by yellow cocoon fluorescence under a UV lamp (360 nm).
Expression and UGT Activity of Recombinant UGTs.
Expression of recombinant UGTs in insect cells and the measurement of UGT activity in virus-infected cells were performed as described previously (8, 35) and in SI Materials and Methods.
Application of Flavonoids to Annam Cocoons.
Flavonoids were extracted from cocoons of strain 939 and applied to cocoons of Annam, which originally contain low amounts of flavonoids (0.26 μmol/g cocoon). Flavonoids were extracted from the cocoon shells by MeOH-H2O (7:3, V/V) at 60 °C for 2 h, concentrated by evaporation, and diluted with distilled water. The aqueous solutions were applied to a solid-phase extraction cartridge (Oasis HLB, 35 mL; Waters Milford). After washing with distilled water, the column was eluted with MeOH. After checking the flavonoid content in the eluate by HPLC, the methanolic solution was evenly applied to the surface of cocoon shells of Annam with a micropipette. After the application, the cocoon shells were kept in the dark and dried naturally at 25 °C. The resulting amount of flavonoids in the treated cocoons was 7.0 (Gb/+) and 1.0 (+/+) μmol/g cocoon shell, respectively. These values were greater than the amount of flavonoids in the cocoon of strain 939 (Table S1), but less than that of the green cocoon strain Pure-Mysore (15.1 μmol/g cocoon shell) (9).
Measurement of UV-B Transmittance.
UV-B irradiation was performed using a FL-20S UV lamp (Kyokko Denki). UV intensity was controlled by the distance from the UV lamp to the target. The irradiance was measured by a UV light meter (UV-340; Lutron). To measure the transmittance ratio of UV-B through the cocoon shells, samples were cut into 1 × 1-cm square pieces, which were used to cover the sensor of the UV light meter, and the irradiance passing through each sample was determined. The transmittance was calculated by the following equation: Transmittance (%) = UV-B irradiance passing through sample/original UV-B intensity × 100.
UV-B Irradiation Experiments.
Larvae of Annam were reared on mulberry leaves throughout development. Two days after the onset of spinning, the larvae were carefully removed from their cocoons using a cutter blade, and each experimental group consisting of 10 naked prepupae was placed on a Petri dish and irradiated with varying doses of UV-B (0–172.8 kJ/m2/d). After 24 h irradiation, each group was kept in the dark for 2 d at 25 °C for pupal ecdysis. In another set of experiments to study the UV-shielding function of cocoon flavonoids, the naked prepupae were placed in cocoons applied with flavonoids extracted from the cocoons of strain 939 (Gb/+ and +/+), and the cocoon shells were sealed with glue before UV irradiation (172.8 kJ/m2/d).
Supplementary Material
Acknowledgments
We thank M. R. Goldsmith for encouragement and critical review of the manuscript; Y. Banno for valuable discussion; M. Kawamoto for technical assistance; and three anonymous reviewers for constructive suggestions. This work was supported by Ministry of Education, Culture, Sports, Science and Technology Grants 17018007 (to T.S.), 21248006 (to T.D. and T.S.), and 20688003 (to T.D.); Ministry of Agriculture, Forestry and Fisheries–National Institute of Agrobiological Sciences (Agrigenome Research Program), and Japan Science and Technology Agency (Professional Program for Agricultural Bioinformatics), Japan.
Footnotes
The authors declare no conflict of interest.
Data deposition: The nucleotide sequences reported in this paper have been deposited in DDBJ/EMBL/GenBank databases (accession nos. AB539963–AB539969).
This article is a PNAS Direct Submission. W.S.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000479107/-/DCSupplemental.
References
- 1.Goldsmith MR, Shimada T, Abe H. The genetics and genomics of the silkworm, Bombyx mori. Annu Rev Entomol. 2004;50:71–100. doi: 10.1146/annurev.ento.50.071803.130456. [DOI] [PubMed] [Google Scholar]
- 2.Fujii H, Banno Y, Doira H, Kihara H, Kawaguchi Y. Genetical Stocks and Mutations of Bombyx mori: Important Genetic Resources. Fukuoka, Japan: Institute of Genetic Resources, Kyushu Univ; 1998. [Google Scholar]
- 3.Banno Y, et al. A Guide to the Silkworm Mutants 2005—Gene Name and Gene. Fukuoka, Japan: Silkworm Genetics Division, Institute of Genetic Resorces, Kyushu Univ; 2005. [Google Scholar]
- 4.Harizuka M. Physiological genetics of the carotenoids in Bombyx mori, with special reference to the pink cocoon. Bull Seric Exp Stn Japan. 1953;14:141–156. [Google Scholar]
- 5.Tazima Y. The Silkworm: An Important Tool. Tokyo: Kodansha; 1978. [Google Scholar]
- 6.Fujimoto N, Hayashiya K, Nakajima K. Studies on the pigments of cocoon. (IV) The formation and translocation of the pigments of green cocoon in the silkworm larvae. J Seric Sci Jpn. 1959;28:30–32. [Google Scholar]
- 7.Sakudoh T, et al. Carotenoid silk coloration is controlled by a carotenoid-binding protein, a product of the Yellow blood gene. Proc Natl Acad Sci USA. 2007;104:8941–8946. doi: 10.1073/pnas.0702860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirayama C, Ono H, Tamura Y, Konno K, Nakamura M. Regioselective formation of quercetin 5-O-glucoside from orally administered quercetin in the silkworm, Bombyx mori. Phytochemistry. 2008;69:1141–1149. doi: 10.1016/j.phytochem.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Hirayama C, Kosegawa E, Tamura Y, Nakamura M. Analysis of flavonoids in the cocoon layer of the silkworm regional races by LC-MS. Sanshi-Konchu Biotec. 2009;78:57–63. [Google Scholar]
- 10.Hirayama C, Ono H, Tamura Y, Nakamura M. C-prolinylquercetins from the yellow cocoon shell of the silkworm, Bombyx mori. Phytochemistry. 2006;67:579–583. doi: 10.1016/j.phytochem.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 11.Kurioka A, Yamazaki M. Purification and identification of flavonoids from the yellow green cocoon shell (Sasamayu) of the silkworm, Bombyx mori. Biosci Biotechnol Biochem. 2002;66:1396–1399. doi: 10.1271/bbb.66.1396. [DOI] [PubMed] [Google Scholar]
- 12.Tamura Y, Nakajima K, Nagayasu K, Takabayashi C. Flavonoid 5-glucosides from the cocoon shell of the silkworm, Bombyx mori. Phytochemistry. 2002;59:275–278. doi: 10.1016/s0031-9422(01)00477-0. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto H. Linkage studies in the silkworm. IV. Inheritance of the green cocoons. Bull Seric Exp Stn Japan. 1941;10:347–358. [Google Scholar]
- 14.Tamura T, et al. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat Biotechnol. 2000;18:81–84. doi: 10.1038/71978. [DOI] [PubMed] [Google Scholar]
- 15.Huang FF, et al. The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genomics. 2008;9 doi: 10.1186/1471-2164-9-563. 10.1186/1471-2164-9-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Lin G, Zuo Z. Position preference on glucuronidation of mono-hydroxylflavones in human intestine. Life Sci. 2006;78:2772–2780. doi: 10.1016/j.lfs.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 17.Iwashina T, Matsumoto S, Nishida M, Nakaike T. New and rare flavonol glycosides from Asplenium trichomanes ramosum as stable chemotaxonomic markers. Biochem Syst Ecol. 1995;23:283–290. [Google Scholar]
- 18.Harborne JB. Comparative biochemistry of flavonoids. V. Luteolin 5-glucoside and its occurrence in the Umbelliferae. Phytochemistry. 1967;6:1569–1573. [Google Scholar]
- 19.Passamonti S, et al. Bioavailability of flavonoids: A review of their membrane transport and the function of bilitranslocase in animal and plant organisms. Curr Drug Metab. 2009;10:369–394. doi: 10.2174/138920009788498950. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Zuo Z, Lin G. Intestinal and hepatic glucuronidation of flavonoids. Mol Pharm. 2007;4:833–845. doi: 10.1021/mp700077z. [DOI] [PubMed] [Google Scholar]
- 21.Hu M. Commentary: Bioavailability of flavonoids and polyphenols: Call to arms. Mol Pharm. 2007;4:803–806. doi: 10.1021/mp7001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srinivas NR. Structurally modified ‘dietary flavonoids’: Are these viable drug candidates for chemoprevention? Curr Clin Pharmacol. 2009;4:67–70. doi: 10.2174/157488409787236065. [DOI] [PubMed] [Google Scholar]
- 23.Nait Chabane M, Al Ahmad A, Peluso J, Muller CD, Ubeaud G. Quercetin and naringenin transport across human intestinal Caco-2 cells. J Pharm Pharmacol. 2009;61:1473–1483. doi: 10.1211/jpp/61.11.0006. [DOI] [PubMed] [Google Scholar]
- 24.Williamson G, et al. Interaction of positional isomers of quercetin glucuronides with the transporter ABCC2 (cMOAT, MRP2) Drug Metab Dispos. 2007;35:1262–1268. doi: 10.1124/dmd.106.014241. [DOI] [PubMed] [Google Scholar]
- 25.Fujimoto N. Studies on the coloration of silk glands of green cocoon strains. J Seric Sci Jpn. 1949;18:215–219. [Google Scholar]
- 26.Mase K, et al. Breeding of the silkworm race “Sericin Flavo” for production of Sericin cocoons containing flavonol. J Insect Biotec Seric. 2008;77:171–174. [Google Scholar]
- 27.Bischoff SC. Quercetin: Potentials in the prevention and therapy of disease. Curr Opin Clin Nutr Metab Care. 2008;11:733–740. doi: 10.1097/MCO.0b013e32831394b8. [DOI] [PubMed] [Google Scholar]
- 28.Xia Q, et al. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx) Science. 2009;326:433–436. doi: 10.1126/science.1176620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada H, Kato Y. Green colouration of cocoons in Antheraea yamamai (Lepidoptera: Saturniidae): Light-induced production of blue bilin in the larval haemolymph. J Insect Physiol. 2004;50:393–401. doi: 10.1016/j.jinsphys.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Nishida R. Sequestration of defensive substances from plants by Lepidoptera. Annu Rev Entomol. 2002;47:57–92. doi: 10.1146/annurev.ento.47.091201.145121. [DOI] [PubMed] [Google Scholar]
- 31.Imamura N, et al. An antibiotic from Penicillium sp. covering the cocoon of the leaf-rolling moth, Dactylioglypha tonica. Biosci Biotechnol Biochem. 2000;64:2216–2217. doi: 10.1271/bbb.64.2216. [DOI] [PubMed] [Google Scholar]
- 32.Imamura N, et al. The relationship between a leaf-rolling moth (Dactylioglypha tonica) and fungi covering the cocoon. Biosci Biotechnol Biochem. 2001;65:1965–1969. doi: 10.1271/bbb.65.1965. [DOI] [PubMed] [Google Scholar]
- 33.Ito K, et al. Deletion of a gene encoding an amino acid transporter in the midgut membrane causes resistance to a Bombyx parvo-like virus. Proc Natl Acad Sci USA. 2008;105:7523–7527. doi: 10.1073/pnas.0711841105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.International Silkworm Genome Consortium The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol. 2008;38:1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Daimon T, Katsuma S, Iwanaga M, Kang W, Shimada T. The BmChi-h gene, a bacterial-type chitinase gene of Bombyx mori, encodes a functional exochitinase that plays a role in the chitin degradation during the molting process. Insect Biochem Mol Biol. 2005;35:1112–1123. doi: 10.1016/j.ibmb.2005.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.