Abstract
Avermectin and its analogues are produced by the actinomycete Streptomyces avermitilis and are widely used in the field of animal health, agriculture, and human health. Here we have adopted a practical approach to successfully improve avermectin production in an industrial overproducer. Transcriptional levels of the wild-type strain and industrial overproducer in production cultures were monitored using microarray analysis. The avermectin biosynthetic genes, especially the pathway-specific regulatory gene, aveR, were up-regulated in the high-producing strain. The upstream promoter region of aveR was predicted and proved to be directly recognized by σhrdB in vitro. A mutant library of hrdB gene was constructed by error-prone PCR and selected by high-throughput screening. As a result of evolved hrdB expressed in the modified avermectin high-producing strain, 6.38 g/L of avermectin B1a was produced with over 50% yield improvement, in which the transcription level of aveR was significantly increased. The relevant residues were identified to center in the conserved regions. Engineering of the hrdB gene can not only elicit the overexpression of aveR but also allows for simultaneous transcription of many other genes. The results indicate that manipulating the key genes revealed by reverse engineering can effectively improve the yield of the target metabolites, providing a route to optimize production in these complex regulatory systems.
Keywords: precision engineering, RNA polymerase, overproduction
Most drugs and drug precursors found in natural organisms need to be produced on a large scale by microbial fermentation in the pharmaceutical industry (1, 2). Because these compounds are produced in very small quantities under natural conditions and their total synthesis is difficult and impractical because of their complexity and diversity of chemical structure, strain improvement has become a critical part of the drug development process (3, 4). Random mutagenesis and screening has traditionally been used for strain improvement as it is technically simple to perform and little knowledge is needed about the strains. With these methods, the productivity of a penicillin strain used in industry was improved by 100,000-fold compared with the original Fleming strain (5). Even though a tremendous increase in production has been achieved, there are certain drawbacks. The traditional mutate-and-screen methods suffer from being time- and labor-intensive; iterative rounds of mutagenesis and selection are normally needed for yield improvement, which risk accumulation of unwanted mutations resulting in crippled strains; and the molecular genetic basis underlying such yield enhancement is largely unknown. Thus the overproduction mechanisms cannot be applied for the rational engineering of a better strain (6).
Since recombinant DNA technology has become increasingly sophisticated, the ability to modify specific genes and pathways for optimizing the production of commercial- significant microbial metabolites has advanced rapidly (7). The “omics”-guided technologies have also seen rapid development in recent years and are likely to become valuable tools for strain development (3, 6, 8–10). Reverse engineering an organism with a desirable property is carried out by identifying the genetic basis of the property and then directly engineering the key genes, the aim of which is to rapidly gain insight into the complex mechanisms controlling microbial metabolism. Reverse engineering has successfully improved the yield of lovastatin in Aspergillus terreus by association analysis of transcriptional and metabolite profiling data to elucidate the key genetic components and physiological traits that impact the production of lovastatin (11). This approach can be utilized to elucidate the interrelationships between physiological traits and more efficiently direct the engineering of target compound-producing strains to synthesize high yields of these natural products.
Avermectin and its analogues, produced by the actinomycete Streptomyces avermitilis, are a series of 16-membered pentacyclic macrolactone type I polyketides that exhibit an excellent anthelminthic activity against a variety of nematode and arthropod parasites. With few side effects against the host organism, they are widely used in the field of animal health, agriculture, and human infections (12–14). With the published genomic information of S. avermitilis, the avermectin biosynthetic pathway was investigated, and it was deduced whether the strain had the ability to produce novel secondary metabolites (15, 16). The avermectin biosynthetic gene cluster (SAV935–SAV953) encodes four multifunctional modular polyketide synthase components, AVES 1, 2, 3, and 4, which each catalyze a specific round of polyketide chain elongation, and includes 15 additional proteins that are responsible for the biosynthesis of oleandrose, glycosylation, postpolyketide modification, and the regulation of avermectin production (17). Seven acetate units and five propionate units are condensed to the starting units derived from 2-methylbutyrate or isobutyrate, catalyzed by the polyketide synthases, in a stepwise process. After cyclization, the aglycone undergoes further modification including dehydration at C-22 and C-23, O-methylation at C-5, and glycosylation at C-13 (18). The biosynthesis of avermectins is closely related to the production of metabolites in primary metabolic pathways, and the expression levels of genes involved in those precursor metabolic-related pathways will influence the metabolic flux for avermectin biosynthesis.
The goal of this study was to identify the genetic basis of the high yield of avermectin B1a in an industrial avermectin overproducer generated by a successful mutate-and-screen program for strain improvement. By analyzing thousands of genes in parallel, reverse engineering should greatly facilitate the characterization of existing overproducing strains and generate information for guiding of more efficient engineering to synthesize higher yields of avermectin B1a. In this paper, an industrial strain, which was obtained by traditional strain improvement with avermectin B1a production of 4,167 mg/L, was chosen as the parent strain for genetic manipulation. It is difficult to successfully or continuously improve its yield through the traditional mutate-and-select method. It was hoped that the genetic information obtained would lead to better understanding of the metabolic basis of the industrial avermectin overproducer and ultimately trigger further improvement.
Results
Transcriptional Comparative Analysis Reveals Altered Gene Expression in the Overproducer.
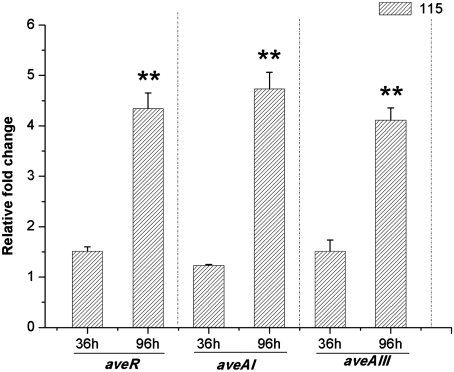
The avermectin B1a overproducing strain of S. avermitilis 3-115 (19, 20) and the wild-type strain ATCC31267 were grown in a fermentation medium. Comparative analysis of the microarray data revealed that 125 genes were differentially expressed and 31 genes were up-regulated by at least twofold at the stationary phase of 3-115 growth (Table S1). The twofold differentially expressed genes were divided into seven groups according to their biological functions defined by the Kyoto Encyclopedia of Genes and Genomes Pathway Database. Most of them were found to be involved in sig-regulons (e.g., sig37, sig3, and rsbW), gene transcription and regulation, signal transport (e.g., pitH1), and avermectin biosynthesis (e.g., aveR, aveAI, and aveAIII); the remainder function in bioenergetics pathways (e.g., sucC1, ackA, and ppc), amino acid metabolism (e.g., thrA), and morphological differentiation (ssgB). Avermectin biosynthetic genes and their relative expression levels are listed in Table S2. It was noticed that the high expression of aveR, aveAI, and aveAIII was statistically significant in the overproducer, which was further confirmed by RT-PCR analysis (Fig. 1). When compared with the expression level of genes in the low producer, that of aveR, aveAI, and aveAIII in strain 3-115 significantly increased by about fourfold in the stationary phase of growth, whereas there was no remarkable change in the vegetative phase of growth. Sequence analysis revealed that the pathway-specific regulatory gene, aveR, and its promoter region in the 3-115 strain was the same as that in the strain ATCC31267, which encouraged us to explore the upstream regulatory factors. Several studies revealed that the major sigma factor, σhrdB, in Streptomyces coelicolor (21–23) recognizes and binds to the consensus sequence: 5′-TGACA-N17,20-AN3T-3′. In S. avermitilis 3-115, five of these up-regulated genes were predicted to be transcribed by the major holoenzyme containing σhrdB (Table S1), which was consistent with the pleiotropic regulating effect of σhrdB. The sequence of the predicted aveRp promoter was quite similar to the -10 and -35 hexamer consensus sequences of rrnDp2 and dagAp4 (Fig. 2A), which were proved to be transcribed by holoenzyme containing σ66 (σhrdB) in S. coelicolor A3(2) (24). This result indicated that the transcription of aveR may be initiated by the principal and essential σhrdB.
Fig. 1.
Expression levels of aveAI, aveAIII, and aveR in S. avermitilis 3-115 relative to those in ATCC 31267. Samples were collected from 3-115 and ATCC 31267 after 36 h (vegetative growth) and 96 h (stationary phase and avermectin producing). We used 16 s rDNA as an RNA integrity control. Standard deviations are marked with error bars (n = 3). All the genes were examined by relative quantification real-time RT-PCR with gene-specific primers. ∗∗P < 0.01 vs. control strain.
Fig. 2.
Recognition of the aveRp promoter by σhrdB-containing RNA polymerase. (A) Comparison of the putative aveRp promoter of S. avermitilis with rrnD-p2 and dagA-p4 of S. coelicolor. The conserved nucleotides within the -10 and -35 regions as well as transcription initiation sites are marked in bold. (B) In vitro transcription assay of the aveRp promoter by RNA polymerase containing σhrdB (EσhrdB). The promoter of the rrnD-homologous gene in S. avermitilis was used as a positive control. In all lanes, the EσhrdB holoenzyme was reconstituted by 0.25 pmol E. coli RNA polymerase core enzyme (EPICENTRE) with the indicated amounts of σhrdB. The ratio of sigma factor to the core RNA polymerase is indicated.
Transcription of aveR Was Activated by RNA Polymerase (RNAP) Containing σhrdB in Vitro.
To provide a biochemical basis for the assertion (based on sequence alignments) that the transcription of aveR was specifically recognized and activated by σhrdB, it was necessary to purify σhrdB RNA polymerase holoenzyme. Therefore, hrdB of S. avermitilis 3-115 was overexpressed in Escherichia coli using the pET28b vector. A prominent band of Mr 57,000 corresponding closely to the Mr predicted for σhrdB was purified (Fig. S1). To determine whether the purified protein possessed σ factor activity, and to evaluate whether in vivo the aveR promoter is directly dependent on σhrdB, titrated amounts of σhrdB were added to the purified core RNA polymerase from E. coli to reconstitute EσhrdB holoenzyme and were used for in vitro runoff transcription experiments with linear DNA templates harboring the predicted promoters. As shown in Fig. 2B, a template containing the S. avermitilis rrnDp was used as a positive control. Transcription assays with a template containing the aveRp resulted in transcripts whose levels increased as the σhrdB concentration increased. The identity of the transcript seen with core RNA polymerase alone is not clear, but it may indicate initiation at aveR promoters, perhaps by a contaminating holoenzyme form. The in vitro transcription assays demonstrated biochemically that aveRp is recognized by EσhrdB in the absence of other transcriptional activators.
High-Throughput Screening of a hrdB Mutant Library.
To identify the effect of σhrdB on aveR and avermectin biosynthesis, especially its function on the regulation of avermectin biosynthesis, a library was constructed by random mutagenesis of the hrdB gene at a moderate mutation frequency (4.5– 9 mutations/kb), which contained approximately 106 white colonies. The first round high-throughput screening was performed in 96 deep-well microplates, and 403 viable single colonies on MS plates were screened, from which 89 mutants were selected for the second fermentation in Erlenmeyer flasks to confirm the productivity. Reference strains C1 and C2 were simultaneously cultured to investigate the influence of the plasmid and the additional copy of hrdB. The selection results are shown in Fig. 3, in which the A56 strain (pSET152-hrdBA56) and the A393 strain (pSET152-hrdBA393) showed increased production of avermectin B1a compared with the control (100% line). Production of avermectin B1a in strain C1 was comparable to that in strain C2, with slight reduction when compared with the parental strain 3-115. Because the aim of this work was to obtain a yield-improved strain, we chose to use the avermectin B1a production of parent strain containing only the chromosomal hrdB gene as the control for further studies.
Fig. 3.
Screening of avermectin B1a overproduction mutants transformed with optimal mutant hrdB gene. S. avermitilis 3-115 was used as the control (dashed line -100%), and the error bars represent standard deviations. All samples were cultured in Erlenmeyer flasks and measured in triplicate. C1: strain containing a blank pSET152; C2: strain containing an additional copy of wild-type hrdB gene.
Characterization of the Mutant hrdB Genes.
To identify whether the yield improvement of A56 or A393 was elicited by the introduction of mutant hrdB, the mutant fragments were rescued and reconjugationally transferred into 3-115. Strains 3-115, A56, and A393 were used as reference strains (Table S3). Strains 3-115::hrdBA56 and 3-115::hrdBA393 have comparable levels of avermectin production to that of A56 and A393, respectively. The result revealed that the yield-improved character of A56, and A393, was directly related to the addition of the mutant hrdB fragment. The mutant HrdBA56 was overexpressed and purified from E. coli. The transcriptional activity of holoenzyme reconstituted by the purified His6-HrdBA56 and core enzyme was analyzed. The reconstituted HrdBA56-holoenzyme showed greater than threefold transcriptional activity compared with HrdB115-holoenzyme at the same concentration (Fig. 4). These results revealed that the enhanced transcriptional activity was responsible for the yield improvement.
Fig. 4.
Comparative analysis of in vitro transcriptional assay of reconstituted holoenzyme with HrdB115 protein or its mutant HrdBA56 protein on aveRp promoter. In all lanes, the holoenzyme was reconstituted by 0.25 pmol E. coli RNA polymerase core enzyme (EPICENTRE) with the indicated amounts of σhrdB. The ratio of sigma factor to the core RNA polymerase is indicated.
Sequence Analysis of the Mutant hrdB Genes.
For the mutants A56 and A393 that showed the remarkable yield increase and stability, the fragments hrdBA56 and hrdBA393 were sequenced. As shown in Fig. 5, the mutant sites were concentrated in region 1.1 and region 2.4. In the 512 AA of hrdBA56, there occurred six site mutations of A137S, K139E, E163G, M356V, V357A, and M389I. For the hrdBA393, there were seven site mutations of M70I, V83L, D120V, K139E, E163G, D345H, and M356V. Among these mutation sites, three site mutations located in region 1.1 (K139E and E163G) and region 2.4 (M356V) were common in the two strains. The results suggested that the mutant hrdB genes from A56 and A393 might be responsible for the high avermectin production.
Fig. 5.
Mutations for the best clone A56 and A393 isolated from the hrdB mutant library are shown mapped onto a schematic of critical functional components of σhrdB. Conserved regions and their proposed functions are shown. The hrdB from parent strain 3-115 is also shown.
Increased Production of A56 in a Large-Scale Fermentor.
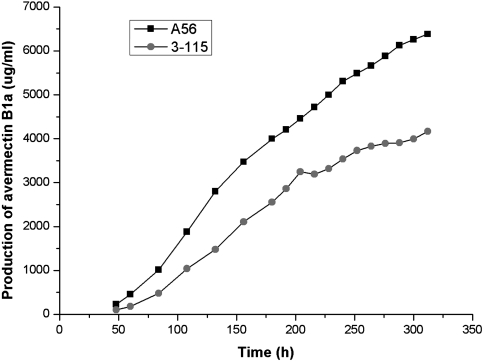
To investigate the reproducibility of yield-improved mutants in large scale, A56 was cultivated in a 180-m3 fermentor, in which A56 exhibited an excellent productivity. As shown in Fig. 6, A56 had a higher avermectin-producing rate, and the yield of avermectin B1a in A56 has an increase of 53% more than that in the parent strain (3-115). This indicated that A56 maintained the industrial character of the parent strain and could be well adapted to industrial-scale production.
Fig. 6.
Time course of avermectin B1a production of parent strain 3-115 and mutant A56 in a 180-m3 fermentor. (One of the representative datasets is shown.)
Discussion
The complexity of cellular networks makes it difficult to build a complete understanding of multiple processes but also offers innumerable possibilities for modification to obtain yield-improved strains. Reverse engineering facilitates elucidation of overproduction mechanisms and the identification of genetic targets for modification. Because of the wide use of avermectins, many other efforts have been made to optimize production systems and maximize productivity, as it is not easy to improve the titer in an overproducing strain. In this study, we attempted to precisely engineer an industrial actinomycete strain and successfully obtained yield-improved mutants.
Our analysis of the differentially expressed level between the wild-type strain and the high producer identified 31 different genes that had diverse functions, including gene transcription and regulation, avermectin biosynthesis, sig-regulons, energy metabolism, and others. It is reasonable to assume that many of the observed alternations are somewhat related to avermectin high producing in stain 3-115. It has been reported that sigma factors play a central role in restructuring the transcriptome responses to environmental signals (25). Among these differentially expressed genes, although sig-regulons (such as sig37, sig3, and rsbW) have not been previously studied in detail, those genes may be involved in counteracting, e.g., the nutrient limitation, or may be potential mediators through which σhrdB exerts its control on downstream targets. Most of the up-regulated genes are involved in gene transcription and regulator, several of which are believed to be involved in carbohydrate metabolism (26), such as the LacI/GalR family transcriptional regulator (SAV1728), and in regulation of a wide range of cellular activities including antibiotic production and amino acid metabolism (27), such as TetR-family transcriptional regulator. Some other genes involved in energy metabolism, amino acid metabolism, phosphotransferase system, and ATP-binding cassette (ABC) transporter were also up-regulated in 3-115. The up-regulation of those genes is likely to be necessary to rapidly supply the cell with the essential elements (28, 29) (amino acids, carbohydrates, and nucleosides) for rapid growth and antibiotic biosynthesis. The high expression level of avermectin polyketide biosynthetic genes means more of the polyketide moiety produced in the overproducer. Enhanced expression of the pathway-specific regulator AveR in 3-115 indicated increased transcription of avermectin polyketide synthases genes, and therefore high avermectin production. This observation is consistent with its activity as a positive pathway-specific regulator (30).
The principal sigma factor, σhrdB, is essential for morphogenesis and antibiotic production in S. coelicolor A3(2) (31). It is suspected that a cascade of other sigma factors, pleiotropic regulators and pathway-specific regulators, might be triggered to express by σhrdB (32). Engineering the binding factors of RNA polymerase has been successfully applied in the improvement of ethanol tolerance and productivity in Saccharomyces cerevisiae (33), and in the yield improvement of products expressed in the heterogeneous host E. coli (34, 35). By binding to and affecting the promoter specificity of the RNA polymerase core enzyme, σhrdB directs the selective transcription of gene sets and acts cooperatively with one or more Streptomyces extracytoplasmic function sigma factors to exert its control on downstream targets. So the expression level and the structural character of σhrdB can affect the expression of its functional protein. As none of the close homologues of σhrdB (σhrdD, σhrdA, and σhrdC) was required in vivo for differentiation and antibiotic production in S. coelicolor, it is likely that the promoter of the pathway-specific regulator genes, actIIORF4p and redDp, is recognized by RNAP containing σhrdB (36). It has been reported that HrdB can recognize the proposed consensus sequences (TTGACN-16 to 18 bp-AN3T) of promoters (37). Based on the consensus promoter-binding sequence, aveRp was the candidate σhrdB-specific promoter. Since we detected a σhrdB-dose dependent level of in vitro transcription from aveRp with reconstituted EσhrdB holoenzyme, we surmise that σhrdB perhaps recognizes aveRp in vivo; the potential -10 and -35 sequence of aveRp was similar to the proposed consensus sequence for the major holoenzyme. While the aveRp may be recognized in vivo by at least one other minor σ factor, it is possible that the ability of σhrdB to recognize aveRp in vitro is an artifact of the assay conditions used. Our data indicated that engineering of hrdB can affect the transcription activity of aveR, which induces positive regulation and high expression of the avermectin biosynthetic genes. Engineering of σhrdB may also make higher-level modifications, which tune the multiple, simultaneous alterations of diverse pathways and gene expression to obtain avermectin improvement.
While introduction of the wild-type hrdB ORF into the avermectin-high-producing strain 3-115 had no significant influence on avermectin production, the mutant hrdB ORF rescued from A56 and A393 introduced back into 3-115 endowed the host with a comparable level of production improvement as A56 and A393, suggesting that the mutant hrdB ORF was responsible for the yield improvement and played a dominant role when competing with the wild-type hrdB in vivo. It has been reported that the hrdB gene is essential for strain survival (31), so in our system, we enhanced the copy number of the hrdB gene in the parent strain 3-115 to examine its influence on avermectin production. The gene structure of hrdB was also optimized for avermectin production. The strain identified in this paper was cultured in a fermentor, and the yield indicated that the improved strain with mutant σhrdB could be successfully applied and well-adapted to industrial fermentation processes. This mutant A56 was genetically stable and has the potential to greatly improve the industrial productivity of avermectin B1a in industry.
σhrdB with mutations on the conserve region 1.2 was reported to result in antibiotic deficiency through reduced transcription of pathway-specific regulatory genes (38). In the high avermectin producing strain 3-115, we found three site mutations mainly located in the conserved region 1.1 and 1.2 in the predicted hrdB protein sequence. Region 1.1 can function as an autoinhibitory domain that masks DNA binding determinants in free σ70. The high negative charge of region 1.1 allows it to act as a mimic of downstream DNA in its interactions with the active site channel, which might bind DNA recognition determinants in free σ, thereby outcompeting promoter DNA (39). Depending on the promoter, the site mutation on the region 1.1 either promoted or inhibited transcription initiation by influencing open complex formation (38, 40, 41). Region 2.4 has been reported to be highly conserved and suggested to be involved in recognizing the promoters at -10 regions. We found two mutations occurred at the terminus of region 2.4 in hrdBA56 and hrdBA393. Amino acid substitution in this region might change the recognition of some promoters, therefore affecting the transcription of the functional proteins, which would be expected to cause avermectin overproduction in the mutant A56. As the components of global cellular transcription machinery, the site mutations in the conserved region of hrdB may directly affect the transcriptional level of aveR and thus the avermectin biosynthetic genes. The mechanism of the mutant hrdB effect on avermectin biosynthesis relative pathway genes needs further study by global transcriptional analysis.
In conclusion, this study represents preliminary attempts at reverse engineering to rapidly and effectively improve avermectin production in an industrial overproducer. The approach of reverse engineering facilitates whole-cell engineering through the selection and identification of key genes responsible for a variety of improved cellular phenotypes. This study has revealed that engineering on the target genes can effectively enhance the yield of desired natural products. The methods described in this paper have broad application to other actinomycetes in elucidating the correlation between the production and limitation steps in the biosynthetic pathway.
Materials and Methods
S. avermitilis ATCC31267 was used as the reference wild-type strain in microarray analysis. S. avermitilis 3-115 (CGMCC No. 3229), a high-level industrial producer of avermectin B1a (4,167 mg/L) obtained by traditional improvement programs with 35 × higher avermectin titers compared with ATCC31267, was collected in our laboratory. Strain 3-115 was used as the parent strains for genetic manipulation. Recombinant proteins for in vitro transcriptional assay were expressed and purified from E. coli. Detailed information on reagents, transcriptional analysis, expression and purification of recombinant proteins, data collection, high-throughput screening, and library construction is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
The authors are very grateful for the technical support provided by Huimin Yu from Tsinghua University. They also thank Elizabeth Ashforth, Huarong Tan, Zixin Deng, Linquan Bai, Zhiheng Liu, Jibin Sun, Keqian Yang, Xiaopeng Jia, and Yaqiao Li for critical reading and helpful discussions. This work was supported in part by grants from National 863 Project (2006AA09Z402, 2007AA09Z443), Key Project for International Cooperation (2007DFB31620), National Natural Science Foundation of China (30560001, 30600001, 30700015), National Key Technology R&D Program (2007BAI26B02), the CAS Pillar Program (KSCX2-YW-R-164), and Important National Science & Technology Specific Projects (2008ZX09401-05, 2009ZX09302-004). L.-X.Z. received funding from the Hundred Talents Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006085107/-/DCSupplemental.
References
- 1.Demain AL. Pharmaceutically active secondary metabolites of microorganisms. Appl Microbiol Biotechnol. 1999;52:455–463. doi: 10.1007/s002530051546. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L. In: Natural Products: Drug Discovery and Therapeutic Medicine. Demain AL, Zhang L, editors. Totowa, NJ: Humana Press; 2005. pp. 33–55. [Google Scholar]
- 3.Baltz RH. Genetic methods and strategies for secondary metabolite yield improvement in actinomycetes. Antonie van Leeuwenhoek. 2001;79:251–259. doi: 10.1023/a:1012020918624. [DOI] [PubMed] [Google Scholar]
- 4.Parekh S, Vinci VA, Strobel RJ. Improvement of microbial strains and fermentation processes. Appl Microbiol Biotechnol. 2000;54:287–301. doi: 10.1007/s002530000403. [DOI] [PubMed] [Google Scholar]
- 5.Rokem JS, Lantz AE, Nielsen J. Systems biology of antibiotic production by microorganisms. Nat Prod Rep. 2007;24:1262–1287. doi: 10.1039/b617765b. [DOI] [PubMed] [Google Scholar]
- 6.Lum AM, Huang J, Hutchinson CR, Kao CM. Reverse engineering of industrial pharmaceutical-producing actinomycete strains using DNA microarrays. Metab Eng. 2004;6:186–196. doi: 10.1016/j.ymben.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, et al. Exploring novel bioactive compounds from marine microbes. Curr Opin Microbiol. 2005;8:276–281. doi: 10.1016/j.mib.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Kang SH, et al. Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. J Bacteriol. 2007;189:4315–4319. doi: 10.1128/JB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin P, et al. Isolation of soluble proteins from an industrial strain Streptomyces avermitilis in complex culture medium for two-dimensional gel electrophoresis. J Microbiol Meth. 2008;73:105–110. doi: 10.1016/j.mimet.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Wang C, et al. SigN is responsible for differentiation and stress responses based on comparative proteomic analyses of Streptomyces coelicolor wild-type and sigN deletion strains. Microbiol Res. 2010;165:221–231. doi: 10.1016/j.micres.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Askenazi M, et al. Integrating transcriptional and metabolite profiles to direct the engineering of lovastatin-producing fungal strains. Nat Biotechnol. 2003;21:150–156. doi: 10.1038/nbt781. [DOI] [PubMed] [Google Scholar]
- 12.Burg RW, et al. Avermectins, new family of potent anthelmintic agents: Producing organism and fermentation. Antimicrob Agents Ch. 1979;15:361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egerton JR, et al. Avermectins, new family of potent anthelmintic agents: Efficacy of the B1a component. Antimicrob Agents Ch. 1979;15:372–378. doi: 10.1128/aac.15.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korystov YN, et al. Avermectins inhibit multidrug resistance of tumor cells. Eur J Pharmacol. 2004;493:57–64. doi: 10.1016/j.ejphar.2004.03.067. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda H, et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 16.Omura S, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: Deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon YJ, Kim ES, Hwang YS, Choi CY. Avermectin: Biochemical and molecular basis of its biosynthesis and regulation. Appl Microbiol Biot. 2004;63:626–634. doi: 10.1007/s00253-003-1491-4. [DOI] [PubMed] [Google Scholar]
- 18.MacNeil T, Gewain KM, MacNeil DJ. Deletion analysis of the avermectin biosynthetic genes of Streptomyces avermitilis by gene cluster displacement. J Bacteriol. 1993;175:2552–2563. doi: 10.1128/jb.175.9.2552-2563.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao H, et al. Medium optimization for the production of avermectin B1a by Streptomyces avermitilis 14-12A using response surface methodology. Bioresource Technol. 2009;100:4012–4016. doi: 10.1016/j.biortech.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Gao H, et al. Identification of avermectin-high-producing strains by high-throughput screening methods. Appl Microbiol Biotechnol. 2009;85:1219–1225. doi: 10.1007/s00253-009-2345-5. [DOI] [PubMed] [Google Scholar]
- 21.Touzain F, et al. SIGffRid: A tool to search for sigma factor binding sites in bacterial genomes using comparative approach and biologically driven statistics. BMC Bioinformatics. 2008;9:73. doi: 10.1186/1471-2105-9-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown KL, Wood S, Buttner MJ. Isolation and characterization of the major vegetative RNA polymerase of Streptomyces coelicolor A3(2); renaturation of a sigma subunit using GroEL. Mol Microbiol. 1992;6:1133–1139. doi: 10.1111/j.1365-2958.1992.tb01551.x. [DOI] [PubMed] [Google Scholar]
- 23.Delic I, Robbins P, Westpheling J. Direct repeat sequences are implicated in the regulation of two Streptomyces chitinase promoters that are subject to carbon catabolite control. Proc Natl Acad Sci USA. 1992;89:1885–1889. doi: 10.1073/pnas.89.5.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang JG, Hahn MY, Ishihama A, Roe JH. Identification of sigma factors for growth phase-related promoter selectivity of RNA polymerases from Streptomyces coelicolor A3(2) Nucleic Acids Res. 1997;25:2566–2573. doi: 10.1093/nar/25.13.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weickert MJ, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 27.Ramos JL, et al. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlosser A, Kampers T, Schrempf H. The Streptomyces ATP-binding component MsiK assists in cellobiose and maltose transport. J Bacteriol. 1997;179:2092–2095. doi: 10.1128/jb.179.6.2092-2095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao X, et al. The novel Streptomyces olivaceoviridis ABC transporter Ngc mediates uptake of N-acetylglucosamine and N,N'-diacetylchitobiose. Mol Genet Genomics. 2002;267:429–439. doi: 10.1007/s00438-002-0640-2. [DOI] [PubMed] [Google Scholar]
- 30.Kitani S, Ikeda H, Sakamoto T, Noguchi S, Nihira T. Characterization of a regulatory gene, aveR, for the biosynthesis of avermectin in Streptomyces avermitilis. Appl Microbiol Biotechnol. 2009;82:1089–1096. doi: 10.1007/s00253-008-1850-2. [DOI] [PubMed] [Google Scholar]
- 31.Buttner MJ, Chater KF, Bibb MJ. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2) J Bacteriol. 1990;172:3367–3378. doi: 10.1128/jb.172.6.3367-3378.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aigle B, Wietzorrek A, Takano E, Bibb MJ. A single amino acid substitution in region 1.2 of the principal sigma factor of Streptomyces coelicolor A3(2) results in pleiotropic loss of antibiotic production. Mol Microbiol. 2000;37:995–1004. doi: 10.1046/j.1365-2958.2000.02022.x. [DOI] [PubMed] [Google Scholar]
- 33.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 34.Alper H, Stephanopoulos G. Global transcription machinery engineering: A new approach for improving cellular phenotype. Metab Eng. 2007;9:258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Yu H, Tyo K, Alper H, Klein-Marcuschamer D, Stephanopoulos G. A high-throughput screen for hyaluronic acid accumulation in recombinant Escherichia coli transformed by libraries of engineered sigma factors. Biotechnol Bioeng. 2008;101:788–796. doi: 10.1002/bit.21947. [DOI] [PubMed] [Google Scholar]
- 36.Buttner MJ, Lewis CG. Construction and characterization of Streptomyces coelicolor A3(2) mutants that are multiply deficient in the nonessential hrd-encoded RNA polymerase sigma factors. J Bacteriol. 1992;174:5165–5167. doi: 10.1128/jb.174.15.5165-5167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strohl WR. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vuthoori S, Bowers CW, McCracken A, Dombroski AJ, Hinton DM. Domain 1.1 of the sigma(70) subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes. J Mol Biol. 2001;309:561–572. doi: 10.1006/jmbi.2001.4690. [DOI] [PubMed] [Google Scholar]
- 39.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 40.Bowers CW, Dombroski AJ. A mutation in region 1.1 of sigma70 affects promoter DNA binding by Escherichia coli RNA polymerase holoenzyme. EMBO J. 1999;18:709–716. doi: 10.1093/emboj/18.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson C, Dombroski AJ. Region 1 of sigma70 is required for efficient isomerization and initiation of transcription by Escherichia coli RNA polymerase. J Mol Biol. 1997;267:60–74. doi: 10.1006/jmbi.1997.0875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.