Abstract
Escherichia coli is a normal inhabitant of the human gut. However, E. coli strains of phylogenetic group B2 harbor a genomic island called “pks” that codes for the production of a polyketide-peptide genotoxin, Colibactin. Here we report that in vivo infection with E. coli harboring the pks island, but not with a pks isogenic mutant, induced the formation of phosphorylated H2AX foci in mouse enterocytes. We show that a single, short exposure of cultured mammalian epithelial cells to live pks+ E. coli at low infectious doses induced a transient DNA damage response followed by cell division with signs of incomplete DNA repair, leading to anaphase bridges and chromosome aberrations. Micronuclei, aneuploidy, ring chromosomes, and anaphase bridges persisted in dividing cells up to 21 d after infection, indicating occurrence of breakage–fusion–bridge cycles and chromosomal instability. Exposed cells exhibited a significant increase in gene mutation frequency and anchorage-independent colony formation, demonstrating the infection mutagenic and transforming potential. Therefore, colon colonization with these E. coli strains harboring the pks island could contribute to the development of sporadic colorectal cancer.
Keywords: bacteria, genotoxin, aneuploidy, chromosomal instability, cancer
The dense bacterial consortium, called “microbiota,” that inhabits the intestinal tract is recognized increasingly as playing a major role in human health and disease. The microbiota generally influences the host in a beneficial fashion by shaping gastrointestinal and immune functions, exerting protection against pathogens, and contributing to metabolic pathways (1). Escherichia coli is a consistent member of the human intestinal microbiota, colonizing the intestine within a few days after birth and persisting throughout the life of the host. The E. coli strain population can be categorized in at least four major phylogenetic groups (A, B1, B2, and D), each group being more specifically associated with certain ecological niches. E. coli strains belonging to group B2 are recovered from the environment less frequently but can persist longer in the colon than other groups and represent 30–50% of strains isolated from the feces of healthy humans living in high-income countries (2, 3). We recently discovered that up to 34% of commensal E. coli strains of the phylogenetic group B2 carry a conserved genomic island named “pks island” (4 –6). This gene cluster codes for nonribosomal peptide synthetases (NRPS) and polyketide synthetases (PKS) that allow production of a putative hybrid peptide-polyketide genotoxin, Colibactin. In vitro infection with these strains induces DNA double-strand breaks (DSBs) in cultivated human cells, but the pks island was not proved to cause DNA damage in vivo (4).
In this study, we wished to explore whether those bacteria were able to induce genetic damage in vivo on the colonic mucosa and to characterize the consequences of this damage on mammalian cells in relation with the number of infecting bacteria. We report that pks+ E. coli induced DSBs in vivo. In addition, infection of various mammalian cells with pks+ E. coli induced, at very low multiplicity of infection (MOI), reversible DNA damage response that did not repair all DSBs, leading to chronic mitotic and chromosomal aberrations together with increased frequency of gene mutation and anchorage-independent growth. Taken together, these findings strongly suggest that these pks+ strains are genotoxic in vivo and provide insights into mechanisms by which common E. coli strains may contribute to cellular transformation and possibly sporadic colorectal cancer tumorigenesis.
Results
pks+ E. coli Induces γH2AX Foci in Vivo.
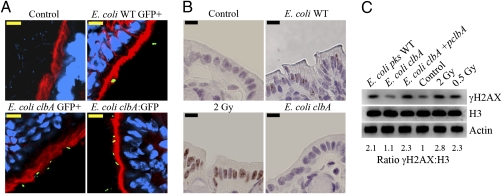
To test whether pks+ E. coli expressed Colibactin genes and induced DNA damage in vivo, we first used a mouse intestinal loop model. Colon loops were infected with E. coli harboring the pks island and a plasmid-encoded GFP under control of either a constitutive promoter or the native clbA promoter. The clbA gene is localized on the pks island and encodes a phosphopantetheinyl transferase required for Colibactin biosynthesis by the NRPS and PKS enzymes (4). We observed that GFP-expressing bacteria were localized close to or in contact with the intestinal brush border (Fig. 1A). Next, we assessed by immunohistology and Western blotting the S139-phosphorylation of histone H2AX (γH2AX), a sensitive marker of DSBs (7). Gamma-irradiated mice (0.5 or 2 Gy) were used as positive controls. Colon loops were infected with wild-type pks+ E. coli, the isogenic clbA mutant, or the mutant complemented with a plasmid bearing the clbA allele. After 6 h of incubation, tissue samples were collected. Significant numbers of γH2AX foci were found in the nuclei of enterocytes exposed to pks+ E. coli as compared with the isogenic clbA mutant and negative control (Fig. 1B). γH2AX foci were found in 22.7% of enterocytes infected with wild-type pks+ E. coli, three times more than in enterocytes infected with the isogenic clbA mutant (P < 0.001, ANOVA). Western blot analyses of colonocytes also indicated increased γH2AX in the intestinal loops infected with the wild-type pks+ E. coli or with the complemented clbA mutant, similar to levels seen with 0.5-Gy gamma irradiation (Fig. 1C).
Fig. 1.
pks+ E. coli induces DSBs in vivo. Ligated colon loops were prepared in BALB/c mice, and then the loops were injected with sterile culture medium or with 3 × 109 wild-type pks+ E. coli (WT), the isogenic clbA mutant impaired for biosynthesis by the NRPS-PKS enzymes, or the clbA mutant complemented with a plasmid-encoded clbA allele. After 6-h incubation, the loops were removed and processed for immunohistology or Western blot analysis. (A) Frozen tissue sections were stained for DNA (blue) and F-actin (red) and then were examined by confocal microcopy. Bacteria expressing GFP (constitutive promoter) or GFP under control of the clbA promoter (clbA:GFP) were detected in the green channel. (Scale bars, 10 μm.) (B) Paraffin tissue sections were stained for γH2AX (brown) and counterstained with hematoxylin. Gamma-irradiated mice (whole-body, 2 Gy) were used as positive controls. (Scale bars, 10 μm.) (C) Western blot analysis of γH2AX in colonocytes 6 h after inoculation of colon loops with wild-type pks+ E. coli, the clbA isogenic mutant, or the mutant complemented with a plasmid coding for clbA. Gamma-irradiated mice (whole-body, 2 or 0.5 Gy) were use as positive controls. Histone H3 and actin were probed as protein-loading controls. γH2AX levels relative to histone H3 content were estimated by densitometry.
To substantiate these results, we used a second in vivo model in which antibiotic-treated mice were given the pks+ bacteria per os. Similar clbA:gfp expression and γH2AX foci were observed in the colons of mice treated for 5 d with streptomycin-bacitracin-neomycin and then inoculated by gastric gavage with pks+ E. coli (Fig. S1). Together these results indicated that pks+ E. coli induced γH2AX in vivo, suggesting DNA damage to colonic epithelial cells.
Transient DNA Damage Response, Incomplete DNA Repair, and Cell Division After Low-Dose Infection with pks+ E. coli.
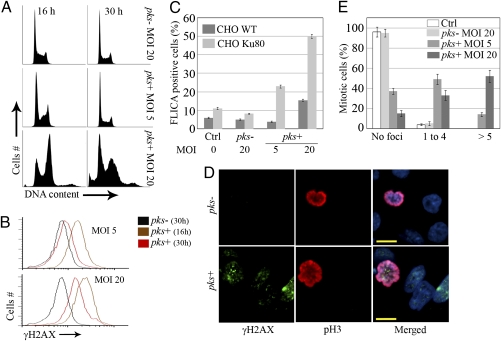
We next examined in vitro the consequences of the DNA damage inflicted on mammalian cells by infection with low numbers of pks+ E. coli as observed in vivo. Chromosomally stable CHO cells were infected with live pks− or pks+ E. coli. Because the estimated in vivo infectious dose was about four bacteria per enterocyte (SI Materials and Methods), we used MOIs of 1–20 bacteria per cell. After a 4-h infection, cells were washed and incubated with antibiotics to kill remaining bacteria. Cell-cycle distribution, γH2AX response, and cell death were monitored 16–30 h later. Cells exposed to pks− E. coli exhibited a normal cell cycle and background levels of γH2AX, whereas cells infected with pks+ E. coli shown a transient increase in G2/M population and γH2AX response that was stronger at MOI 20 than at MOI 5 (Fig. 2 A and B). Sub-G1 peak and active caspases levels indicated that the cell death remained at background levels for MOI 5 and below 15% for MOI 20 (Fig. 2 A and C). In contrast to wild-type CHO cells, Ku80 mutant cells, which are deficient for the nonhomologous end-joining (NHEJ) repair pathway and thus are hypersensitive to DSBs (8), died massively (Fig. 2C). These results indicate that cells exposed to low numbers of pks+ bacteria suffered DNA damage but were able to repair, primarily by the NHEJ pathway, and resumed cell cycle and division. However, 24 h after infection 50% of mitotic cells previously infected with pks+ E. coli still harbored one to four γH2AX foci, in contrast to control cells or cells infected with pks− bacteria showing no foci (Fig. 2 D and E). These γH2AX foci on mitotic chromosomes represent scars of repaired lesions or even unrepaired DNA breaks (9, 10). Thus, low-dose infection with pks+ E. coli induces reversible activation of the DNA damage response followed by cell division with signs of damaged DNA.
Fig. 2.
DNA damage repair, cell death, and division after low-dose infection with pks+ E. coli. CHO cells were infected for 4 h with live pks+ or pks− E. coli with an MOI of 5–20 bacteria per cell or were left uninfected (Ctrl). At the end of the infection, the cells were washed and grown with gentamicin. (A) Cell-cycle analysis 16 and 30 h after infection. (B) γH2AX levels were quantified by flow cytometry 16 or 30 h after infection. (C) CHO or xrs-6 Ku80-defective cells were infected; 24 h later, apoptotic cells were labeled with a carboxyfluorescein fluoromethyl ketone peptide inhibitor of caspases (FLICA) for 1 h and quantified by flow cytometry. Error bars represent SE from three experiments. (D) The cells were examined by confocal microscopy for DNA (blue), Ser10-phosphorylated histone H3 (pH3, red), and γH2AX (green) 24 h after infection. (Scale bars, 10 μm.) (E) Quantification of γH2AX foci in mitotic cells. Error bars represent SEs from three experiments.
Infection with pks+ E. coli Induces Anaphase Bridging and Chromosome Aberrations.
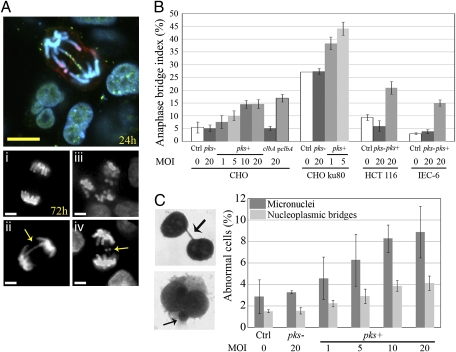
Misrepaired DSBs can induce chromosome fusions resulting in chromatin bridges that often break during anaphase (11). Such anaphase bridges harboring γH2AX foci could be detected 24 h after infection with pks+ E. coli (Fig. 3A). Moreover, anaphase bridges, lagging chromosomes, and multipolar mitosis also were observed in the cell population 72 h (three to four cell divisions) after infection with pks+ bacteria (Fig. 3A). The anaphase bridge index increased with the MOI of pks+ E. coli, whereas it remained at background level in cells exposed to pks− or to the clbA mutant and was restored to pks+ level upon mutant complementation (Fig. 3B). NHEJ-deficient Ku80 mutant cells with a constitutively enhanced rate of bridge formation (12) also showed an increase in bridging after infection with pks+ E. coli. Anaphase bridges were found not only in CHO cells but also in human colon cancer HCT-116 cells and nontransformed rat intestinal epithelial IEC-6 cells 3 d after infection with pks+ E. coli (Fig. 3B). To corroborate these results, we performed a cytokinesis block assay that allows scoring of lagging or acentric chromosomes, which give rise to micronuclei, and anaphase bridges that result in nucleoplasmic bridges (13). Three days after infection with pks+ but not with pks− E. coli, binucleate cells harbored substantial numbers of micronuclei and nucleoplasmic bridges that increased with the MOI (Fig. 3C).
Fig. 3.
Infection with pks+ E. coli induces anaphase bridges and micronuclei. (A) (Upper) Anaphase bridge 24 h after infection with pks+ E. coli (DNA shown in blue, γH2AX in green, and pH3 in red). (Lower) Seventy-two hours after infection (i) normal anaphase, (ii) anaphase bridge (arrow), (iii) multipolar mitosis, (iv) lagging chromosomes (arrow). (Scale bars, 10 μm.) (B) Anaphase bridge index in CHO, ku80-defective CHO, HCT-116, and IEC-6 cells 3 d after infection. (C) Cytochalasin-B–induced cytokinesis block assay. (Left) Images and arrows show a nucleoplasmic bridge (formed by anaphase bridging) and a micronucleus (formed by lagging or acentric chromosomes) in CHO cells 3 d after infection. (Right) Micronuclei and nucleoplasmic bridges were counted in 1,000 binucleated cells. Error bars in B and C represent the SE from three experiments.
We next assessed metaphasic chromosomes 24 h and 3 d after infection. A variety of chromosome aberrations were observed, including translocations, chromatid breaks, and dicentric and ring chromosomes (Fig. S2A). The number of aberrant chromosomes diminished at 3 d compared with 24 h after infection (Fig. S2B). Heavily damaged cells likely died after the first division, consistent with the cell death that was measured 24 h after infection with pks+ E. coli (Fig. 2C). Three days after pks+ E. coli infection, we found 2% metaphases with centric rings and 5% with translocations; 21 d later, 1% of metaphases still harbored ring chromosomes. Together these results indicate that cells infected with pks+ E. coli seem to propagate lasting chromosome aberrations and cycles of breakage–fusion–bridges.
Infection with pks+ E. coli Induces Aneuploidy and Tetraploidy.
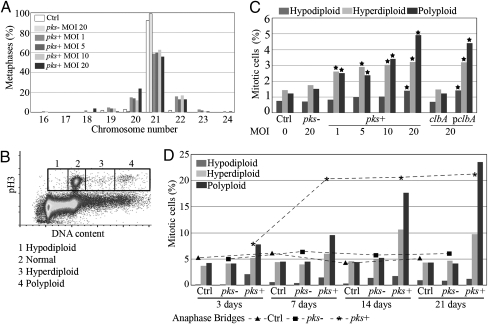
Anaphase bridging can induce aneuploidy (gain or loss of chromosomes) and tetraploidy (8n DNA content for a cell in G2) (14, 15). Chromosomes were counted in metaphase spreads 3 d after infection. About 40% of CHO cells infected with pks+ E. coli harbored abnormal chromosome numbers, even at the lowest infection dose (one bacterium per cell) (Fig. 4A). To evaluate aneuploidy and tetraploidy further in CHO, HCT-116, and IEC-6 cells, mitotic cells were stained for phosphorylated histone H3, and their DNA content was determined by flow cytometry (16) 3 d after infection (Fig. 4B). We found significant increases in aneuploid (hypodiploid and hyperdiploid) and tetraploid cells after infection with pks+ or complemented clbA mutant E. coli compared with the control population or cells that were infected with the pks− or clbA mutant E. coli (Fig. 4C and Table S1). This increment of cells with abnormal DNA content persisted up to 21 d after pks + E. coli infection, and the number of polyploid cells even increased. In parallel, the anaphase bridge index increased to 20% at 7 d and persisted (Fig. 4D). It thus appears that cells exposed to low infectious doses of pks+ E. coli propagated breakage–fusion–bridge cycles, generating chromosome numerical instability in the long term.
Fig. 4.
Infection with pks+ E. coli induces aneuploidy and tetraploidy. (A) Chromosome counts in CHO cells 3 d after infection. At least 100 metaphase spreads were examined in each group. (B) Aneuploidy assay by flow cytometry (16). At least 2 × 105 mitotic (Ser10-phosphorylated histone H3-positive) cells were analyzed for DNA content. Normal (4n), hypodiploid (<4n), hyperdiploid (4 < n < 8) and polyploid (8n) cells were gated and counted. (C) Three days after infection (MOI of 1–20), CHO cells were analyzed for aneuploidy as in B. *, P < 0.001, exact Fisher's test. Similar results were found with IEC-6 and HCT-116 cells (Table S1). (D) Cells were infected (MOI = 20) and then were grown for 21 d (with six passages). Aneuploidy (histograms) was assessed as in B, and the anaphase bridge index (curves) was scored as in Fig 2B.
Increased Gene Mutation Frequency and Anchorage-Independent Growth After Infection with pks+ E. coli.
Chromosome rearrangements generate gene mutations, and, conversely, gene mutations are required to allow maintenance of chromosomal instability and aneuploidy (17). Therefore, we tested gene mutation frequencies at the hypoxanthine-guanine phosphoribosyltransferase (hprt) and thymidine kinase (tk) loci after infection of CHO or HCT-116 cells (Table 1). We found a significant increase in 6-thioguanine–resistant (hprt mutant) colonies after infection with the pks+ or complemented clbA mutant E. coli compared with uninfected cells or cells that were infected with pks− or clbA mutant E. coli. Infection with pks+ E. coli also resulted in a significant increase of tk mutants selected with trifluorothymidine.
Table 1.
Increased mutation frequencies at the hprt and tk loci after infection with pks+ E. coli
| Locus | Cells | Infection | MF ± SE × 10−5 |
| hprt | CHO | Control | 1.68 ± 1.17 |
| E. coli pks− | 2.89 ± 2.02 | ||
| E. coli pks+ | 11.40 ± 1.16* | ||
| E. coli clbA | 1.54 ± 1.11 | ||
| E. coli clbA + pclbA | 11.80 ± 1.14* | ||
| tk | CHO | Control | 31.7 ± 2.44 |
| E. coli pks− | 29.1 ± 3.18 | ||
| E. coli pks+ | 48.3 ± 2.02* | ||
| hprt | HCT-116 | Control | 1.52 ± 0.18 |
| E. coli pks− | 1.52 ± 0.27 | ||
| E. coli pks+ | 3.58 ± 0.20* |
CHO and HCT-116 cells were infected (MOI = 20) or left uninfected, then grown for 7 or 3 d before plating with 6-thioguanine or trifluorothymidine to select for hprt or tk mutants, respectively.
*P < 0.05, x 2- Mc Nemar test compared with control, pks−, and clbA. MF, mutation frequency.
To examine further whether this increased frequency of gene mutation correlated with a transformed phenotype (18), we tested the cell anchorage-independent proliferation in soft agar after infection of CHO, HCT-116, or IEC-6 cells (Table 2). The cells infected with the pks+ or complemented mutant E. coli formed a significantly higher number of colonies in soft agar compared with pks−- or clbA-infected and control cells. Together these results indicate the mutagenic and transforming potential of the infection with pks+ E. coli.
Table 2.
Anchorage-independent growth in soft agar after infection with pks+ E. coli
| Infection | CFU ± SE per 10,000 cells |
| CHO | |
| Control | 0.00 |
| E. coli pks− | 0.00 |
| E. coli pks+ | 3.33 ± 0.41** |
| E. coli clbA | 0.00 |
| E. coli clbA + pclbA | 4.33 ± 0.58** |
| IEC-6 | |
| Control | 0.33 ± 0.25 |
| E. coli pks− | 0.00 |
| E. coli pks+ | 10.33 ± 0.62** |
| HCT-116 | |
| Control | 4.67 ± 0.36 |
| E. coli pks− | 2.67 ± 0.41 |
| E. coli pks+ | 12.33 ± 0.41*** |
CHO, IEC-6, and HCT-116 cells were infected (MOI = 20) or left uninfected, then grown for 3 d before plating in 0.3% soft agar. Colonies (>50 cells) were counted after 7 d.
**P < 0.01 and ***P < 0.001 by ANOVA with Bonferroni posttest.
Discussion
Colorectal cancer is a disease primarily occuring in high-income countries, where it represents the second most frequently diagnosed malignancy and accounts for 500,000 (3.3%) human deaths annually (19). Sporadic colorectal cancer essentially is a genetic disease in which chromosomal instability, found in 85% of cases, is central to the tumorigenesis process (20, 21). Age together with dietary practices, alcohol and tobacco consumption, physical activity, and body weight are major risk factors (19), but several studies also have implicated the colonic microbiota in the development of colorectal cancer (22). However, epidemiological approaches attempting to identify links between colonic bacteria and colorectal cancer are hampered by the long (20–40 yr) lag time between initiation and disease, by the enormous complexity of the microbiota, and by bacterial strain-to-strain variations (23, 24). Nonetheless, bacterial strains producing metabolites or toxins that insult host DNA (25 –27) represent an important factor for chronic DNA damage in the colon and thus constitute a potential etiologic component of sporadic colorectal cancer.
Our previous findings (4) and the present study (showing the higher sensitivity of NHEJ-deficient cells) demonstrate that infection of eukaryotic cells with pks+ E. coli strains induces host-cell DNA DSBs and activation of the DNA damage signaling cascade, including the ATM–CHK–CDC25–CDK1 pathway and Ser139 phosphorylation of histone H2AX. Infection with high numbers of toxigenic bacteria induces an irreversible cell cycle arrest and eventually apoptotic cell death (cells with sub-G1 DNA content and activated caspases). In this study, we also observed that when eukaryotic cells were infected with infectious doses more relevant to those occuring in vivo with commensal bacteria, most cells exhibited only a transient DNA damage response and rapidly resumed division cycles. However, these cycling cells frequently displayed γH2AX foci during the subsequent mitosis. The small numbers of gamma foci were below the threshold level of 10–20 foci activating the G2/M checkpoint maintenance, thus allowing the generation of chromosome breaks during mitosis (28 –30). These observations raise the question of the impaired reparability of Colibactin-induced DSBs. Consistent with this model, we observed mitotic aberrations such as lagging chromosomes and anaphase bridges. Bridging is known to result from the fusion of broken chromosome ends, generating dicentric chromosomes that are pulled to both poles of the mitotic spindle (11). After breakage of the bridge and rejoining in the next interphase, breakage–fusion–bridge cycles propagated and further generated genetic aberrations including aneuploidy/polyploidy and gene mutation (31). Propagation of this chromosomal instability phenotype continued, because a significant fraction of the cells harbored chromosome rings, aneuploid/polyploid DNA content, and anaphase bridging 21 d after transient infection with pks+ E. coli. The clonogenic survival assays confirmed that cells infected with pks+ E. coli continued to proliferate in the presence of DNA damage and exhibited increased mutation frequency and transformed phenotype.
E. coli is primarily a commensal inhabitant of the mammalian colon, colonizing the gut early after birth and remaining resident throughout the life of the host, but commensal strains differ in their colonization efficiency. E. coli strains belonging to phylogenetic group B2 seem to have a superior capacity to colonize and persist in the colonic microbiota (3), and during the last decade the incidence of B2 strains has risen quickly in colonic microflora of human beings living in high-income countries (2). Surveys have shown that the pks island is present in up to 34% of commensal isolates of phylogenetic group B2 from healthy individuals (4 –6). A recent comparative metagenomic analysis of fecal samples from 13 healthy individuals of various ages, including unweaned infants, showed that pks island genes are found in 40% of samples, especially in infants (32). Thus, the pks island is hosted by commensal E. coli strains commonly found in the human microbiota at birth, and these strains could remain in the colon for years or even decades. A key finding in the present study is that the Colibactin genes are expressed in vivo and induce γH2AX foci in enterocytes. The effect was estimated to be similar to that of a 0.5-Gy gamma ray whole-body irradiation and was enough to trigger in vitro breakage–fusion–bridge cycles, chromosomal instability, aneuploidy, and gene mutations in mammalian cells. In conclusion, our results indicate that common pks+ E. coli strains could play a role in colon carcinogenesis by repeatedly provoking low-grade DNA damage at the enterocyte level. The role of the human intestinal microflora in colon cancer has been overlooked (33) and deserves further scrutiny.
Materials and Methods
Bacterial Strains and Cell Lines.
The wild-type E. coli B2 pks+ strain SP15 (4) was used for in vivo experiments. An isogenic mutant was constructed by allelic replacement of the clbA gene on the pks island with a kanamycin resistance cassette, and the mutant was complemented with the wild-type clbA allele cloned on a plasmid (pclbA), as previously described (4). To visualize the bacteria in vivo, the strains were transformed with plasmids that encode a gfp gene under control of a constitutive promoter (pFPV25.1) (34) or gfp under control of the clbA promoter/regulatory region (pJN871; clbA:gfp). For in vitro infections we used the E. coli strain DH10B hosting a BAC bearing the pks island (pBACpks; pks+ E. coli), or hosting the pBeloBAC11 vector (pks− E. coli), the isogenic DH10B pBACpks clbA mutant, or the complemented mutant (4). The cell lines used were CHO AA8, CHO xrs-6 (Ku80-deficient), nontransformed rat intestinal epithelial IEC-6 cells (ATCC CRL-1592), and human colon cancer cells HCT-116 (ATCC CCL-247).
Murine Colon Loop and Antibiotic Treatment Models.
Animal experimentations were carried out in accordance with European Guidelines for the Care and Use of Animals for Research Purposes. For the colon loop assay, BALB/cJ mice were anesthetized, a midline abdominal incision was made, a ligature was placed at the proximal and distal ends of the colon, and 3 × 109 bacteria (300 μL) were injected into the colonic lumen. The incision was sutured, and animals were allowed to recover. The mice were killed 6 h later, and the colons were collected immediately. For the antibiotic treatment assay, BALB/cJ mice were treated for 5 d with streptomycin (2 g/L), bacitracin (2 g/L), and neomycin (1 g/L) in the drinking water. Antibiotic treatment was stopped 24 h before gastric gavage with 109 bacteria twice at 24-h intervals. Mice were killed 12 h later, and the colons were collected immediately. Gamma-irradiated mice (0.5- or 2-Gy 137Cs whole-body irradiation) were used as positive controls for DNA damage. For Western blot analyses, colonocytes were prepared in a 1.5-mM EDTA, 0.5-mM DTT buffer. Western blot and immunohistological analyses were done following standard procedures.
In Vitro Infection Assay.
Cells (~75% confluent) were washed four times and incubated in infection medium based on DMEM for IEC-6 and HCT-116 cells or MEMα for CHO cells, supplemented with 25 mM Hepes and 5% FCS (Invitrogen). Bacteria were pregrown in infection medium to the midlogarithmic phase; then the infection dose was calculated according to an MOI of 1–20 (number of bacteria per cell at the onset of infection). After a 4-h infection at 37 °C and 5% CO2, cells were washed four to six times and incubated until analysis in cell culture medium supplemented with gentamicin.
Genomic instability Analyses.
Cell cycle, γH2AX flow cytometry, and microscopic analyses were done as before (4). Apoptotic cell death was quantified by staining with carboxyfluorescein-labeled fluoromethyl ketone caspase inhibitor (Chemicon) and flow cytometric measurement of sub-G1 peak. Anaphase bridges (35), micronuclei, and nucleoplasmic bridges (13) were scored as described. Metaphase spreads were prepared and analyzed using standard cytogenetic procedures. DNA quantification in mitotic (phospho-histone H3-positive) cells was done as previously described (16). For tk and hprt gene mutation assay, established methods were used (36). Anchorage-independent growth was determined using a soft agar colony formation assay (37).
Statistical Analyses.
All microscopic quantifications (γH2AX foci, chromosome aberrations, micronuclei, and nucleoplasmic bridges) were done blindly. Experiments were repeated at least three times. For statistical analyses a P value was considered significant if P < 0.05, P < 0.01, or P < 0.001. Gene mutation frequencies were calculated as described (36).
Materials and methods are detailed thoroughly in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Pinton for help in the cytogenetic studies, M. Defais (Centre National de la Recherche Scientifique, Toulouse, France) for the donation of the CHO cell lines and for advice, F. Pierre for help in the clonogenic assay, and J.-S. Hoffmann and B. Ducommun for helpful discussions and suggestions. This work was supported by grants from French Association pour la Recherche sur le Cancer and La Ligue Contre le Cancer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001261107/-/DCSupplemental.
References
- 1.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escobar-Páramo P, et al. Large-scale population structure of human commensal Escherichia coli isolates. Appl Environ Microbiol. 2004;70:5698–5700. doi: 10.1128/AEM.70.9.5698-5700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nowrouzian FL, Wold AE, Adlerberth I. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J Infect Dis. 2005;191:1078–1083. doi: 10.1086/427996. [DOI] [PubMed] [Google Scholar]
- 4.Nougayrède JP, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 5.Putze J, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun. 2009;77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson JR, Johnston B, Kuskowski MA, Nougayrede JP, Oswald E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J Clin Microbiol. 2008;46:3906–3911. doi: 10.1128/JCM.00949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 8.Taccioli GE, et al. Ku80: Product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki M, Suzuki K, Kodama S, Watanabe M. Phosphorylated histone H2AX foci persist on rejoined mitotic chromosomes in normal human diploid cells exposed to ionizing radiation. Radiat Res. 2006;165:269–276. doi: 10.1667/rr3508.1. [DOI] [PubMed] [Google Scholar]
- 10.Syljuåsen RG, Jensen S, Bartek J, Lukas J. Adaptation to the ionizing radiation-induced G2 checkpoint occurs in human cells and depends on checkpoint kinase 1 and Polo-like kinase 1 kinases. Cancer Res. 2006;66:10253–10257. doi: 10.1158/0008-5472.CAN-06-2144. [DOI] [PubMed] [Google Scholar]
- 11.McClintock B. The association of mutants with homozygous deficiencies in Zea mays . Genetics. 1941;26:542–571. doi: 10.1093/genetics/26.5.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acilan C, Potter DM, Saunders WS. DNA repair pathways involved in anaphase bridge formation. Genes Chromosomes Cancer. 2007;46:522–531. doi: 10.1002/gcc.20425. [DOI] [PubMed] [Google Scholar]
- 13.Fenech M. Cytokinesis-block micronucleus cytome assay. Nat Protoc. 2007;2:1084–1104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- 14.Stewénius Y, et al. Structural and numerical chromosome changes in colon cancer develop through telomere-mediated anaphase bridges, not through mitotic multipolarity. Proc Natl Acad Sci USA. 2005;102:5541–5546. doi: 10.1073/pnas.0408454102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- 16.Muehlbauer PA, Schuler MJ. Detection of numerical chromosomal aberrations by flow cytometry: A novel process for identifying aneugenic agents. Mutat Res. 2005;585:156–169. doi: 10.1016/j.mrgentox.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin SI, Freedman VH, Risser R, Pollack R. Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc Natl Acad Sci USA. 1975;72:4435–4439. doi: 10.1073/pnas.72.11.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vainio H, Miller AB. Primary and secondary prevention in colorectal cancer. Acta Oncol. 2003;42:809–815. doi: 10.1080/02841860310010673. [DOI] [PubMed] [Google Scholar]
- 20.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 21.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hope ME, Hold GL, Kain R, El-Omar EM. Sporadic colorectal cancer—role of the commensal microbiota. FEMS Microbiol Lett. 2005;244:1–7. doi: 10.1016/j.femsle.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 23.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lax AJ. Opinion: Bacterial toxins and cancer—a case to answer? Nat Rev Microbiol. 2005;3:343–349. doi: 10.1038/nrmicro1130. [DOI] [PubMed] [Google Scholar]
- 26.Lara-Tejero M, Galán JE. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science. 2000;290:354–357. doi: 10.1126/science.290.5490.354. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Huycke MM. Extracellular superoxide production by Enterococcus faecalis promotes chromosomal instability in mammalian cells. Gastroenterology. 2007;132:551–561. doi: 10.1053/j.gastro.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 28.Löbrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 29.Deckbar D, et al. Chromosome breakage after G2 checkpoint release. J Cell Biol. 2007;176:749–755. doi: 10.1083/jcb.200612047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krempler A, Deckbar D, Jeggo PA, Löbrich M. An imperfect G2M checkpoint contributes to chromosome instability following irradiation of S and G2 phase cells. Cell Cycle. 2007;6:1682–1686. doi: 10.4161/cc.6.14.4480. [DOI] [PubMed] [Google Scholar]
- 31.Gisselsson D, et al. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci USA. 2000;97:5357–5362. doi: 10.1073/pnas.090013497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinicrope FA. Sporadic colorectal cancer: An infectious disease? Gastroenterology. 2007;132:797–801. doi: 10.1053/j.gastro.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Valdivia RH, Falkow S. Bacterial genetics by flow cytometry: Rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol. 1996;22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 35.Luo LZ, Werner KM, Gollin SM, Saunders WS. Cigarette smoke induces anaphase bridges and genomic imbalances in normal cells. Mutat Res. 2004;554:375–385. doi: 10.1016/j.mrfmmm.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 36.Arlett CF, et al. In: Statistical Evaluation of Mutagenenicity Test Data. Kirkland DJ, editor. Cambridge, UK: Cambridge Univ. Press; 2008. pp. 66–101. [Google Scholar]
- 37.Andreassen A, et al. 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) induces genetic changes in murine intestinal tumours and cells with ApcMin mutation. Mutat Res. 2006;604:60–70. doi: 10.1016/j.mrgentox.2006.01.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.