Abstract
AV411 (ibudilast; 3-isobutyryl-2-isopropylpyrazolo-[1,5-a]pyridine) is an antiinflammatory drug that was initially developed for the treatment of bronchial asthma but which also has been used for cerebrovascular and ocular indications. It is a nonselective inhibitor of various phosphodiesterases (PDEs) and has varied antiinflammatory activity. More recently, AV411 has been studied as a possible therapeutic for the treatment of neuropathic pain and opioid withdrawal through its actions on glial cells. As described herein, the PDE inhibitor AV411 and its PDE-inhibition-compromised analog AV1013 inhibit the catalytic and chemotactic functions of the proinflammatory protein, macrophage migration inhibitory factor (MIF). Enzymatic analysis indicates that these compounds are noncompetitive inhibitors of the p-hydroxyphenylpyruvate (HPP) tautomerase activity of MIF and an allosteric binding site of AV411 and AV1013 is detected by NMR. The allosteric inhibition mechanism is further elucidated by X-ray crystallography based on the MIF/AV1013 binary and MIF/AV1013/HPP ternary complexes. In addition, our antibody experiments directed against MIF receptors indicate that CXCR2 is the major receptor for MIF-mediated chemotaxis of peripheral blood mononuclear cells.
Keywords: cross-reactivity, drug repositioning, cytokine, inflammation
AV411 (ibudilast; 3-isobutyryl-2-isopropylpyrazolo-[1,5-a]pyridine) is an antiinflammatory drug used clinically in Japan as a treatment for bronchial asthma at least in part through its effects on the inhibition of phosphodiesterases (PDEs). This action leads to increased levels of cGMP and inhibition of tracheal smooth muscle contractility (1). Because these increased levels of cGMP result in a potentiating effect on the antiplatelet function of endothelial cell nitric oxide, AV411 also has been used for cerebrovascular disorders, such as poststroke complications (2). An increasingly recognized approach for treating various neurological conditions including chronic pain involves attenuation of glial cell activation in the CNS (3). Indeed, experiments involving AV411 on glial cells have led to animal and clinical studies of its effects on neuropathic pain, opioid withdrawal, and potentiation of acute opioid analgesia (4, 5). Animal studies reveal that AV411 is effective in attenuating neuropathic pain in a mechanical allodynia rat model (6) and in reducing glial proinflammatory responses in opioid withdrawal (4). These CNS effects could not all be explained by PDE inhibition because AV1013, a structural analog with 3–50-fold lower PDE inhibition compared to AV411, exhibits similar efficacy in rat models of neuropathic pain and opioid withdrawal (5, 6). Significantly, the macrophage migration inhibitory factor (MIF), originally described as a T-cell lymphokine (7, 8), secreted from microglial cells has been found to contribute to hyperalgesia (9). The cerebral spinal fluid MIF-mediated pain is prevented and reversed in rats by the MIF active site inhibitor (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole acetic acid methyl ester (ISO-1) (9).
Due to the antiinflammatory and glial cell effects of AV411, the inability to explain these effects only by PDE inhibition, and the possibility that antagonism of glial cell expressed MIF may be an alternate or additional mechanism of AV411 action (9, 10), we tested MIF as a potential target for this drug. The proinflammatory protein MIF is stored in the cytosol but released from cells activated by various agents including endotoxin, exotoxins, cytokines, and glucocorticoids (11, 12). Once released, MIF activates the receptor CD74 in a complex with CD44 (13), or the chemokine receptors CXCR2 (14) and CXCR4 (15), initiating a cascade of intracellular signaling (16). One aspect of MIF that remains enigmatic is the presence of a catalytic site, highly conserved among all MIFs and present between subunits of the trimer (16–19). No physiologic substrate for MIF has been identified, but “pseudosubstrates” have been found that allow for screening inhibitors at this site (20, 21). Small molecule inhibitors as well as active site mutants have been used to verify the importance of this site in MIF action in both cellular studies (22, 23) and in vivo mouse models of severe sepsis (24, 25).
In this study, we report that phosphodiesterase inhibitor AV411 and its analog AV1013 also inhibit MIF catalytic and chemotactic activity. MIF chemotactic inhibition occurs at concentrations of AV411 demonstrated to be clinically relevant in its therapeutic use in humans (26). NMR spectroscopic data mapped the AV411 and AV1013 binding sites on the MIF trimer, and crystal structures of the binary rhMIF/AV1013 and ternary rhMIF/(R)-AV1013/p-hydroxyphenylpyruvate (HPP) complexes have been determined. These findings provide a mechanism by which AV411 and AV1013 act as allosteric inhibitors of MIF and may contribute to related therapeutic functions.
Results
AV411 and AV1013 are Noncompetitive Inhibitors of the MIF Tautomerase Activity.
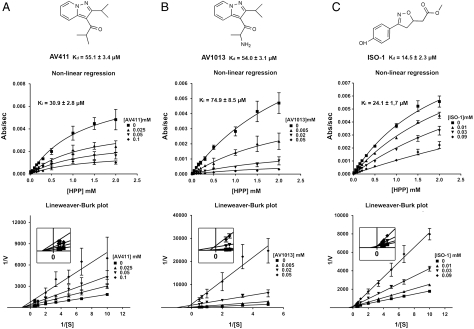
The AV411 and AV1013 structures are depicted in Fig. 1. AV1013 was designed as an analog to AV411 but exhibits 3–50-fold reduced PDE isoform inhibition activity (Table S1). Both compounds were characterized at different concentrations to determine the type of inhibition and the inhibition constant (Ki). Nonlinear regression of dose-response curves from these experiments reveals noncompetitive inhibition as shown in Fig. 1A for AV411. Although the inhibition kinetics of AV1013 were best fit to a noncompetitive inhibition, Lineweaver–Burk analysis does reveal a small variation in apparent KM indicating a possible small effect on substrate binding (Fig. 1B). The finding that these inhibitors were not competitive led us to examine ISO-1 as a positive control for competitive inhibition, which we previously cocrystallized with rhMIF and found to occupy the active site (23). In contrast to the other inhibitors, the nonlinear regression analysis of ISO-1 inhibition data reveals competitive inhibition as expected (Fig. 1C).
Fig. 1.
Chemical structures and kinetic graphs of AV411, AV1013, and ISO-1. Each Lineweaver–Burk plot reveals the noncompetitive inhibition pattern of AV411 (A) and AV1013 (B), and the competitive inhibition pattern of ISO-1 (C). The Lineweaver–Burk of AV1013 demonstrates a small effect on Km (B). The inserts in the Lineweaver–Burk plots show the intersections near the origin. Dissociation constants (Kds) measured by Trp (AV411 and AV1013) and Tyr (ISO-1) fluorescence are presented below the chemical structures.
We also measured the Kd by fluorescence upon the addition of various concentrations of AV411, AV1013, and ISO-1 (Fig. 1). The measurements were performed in the same buffer system as the kinetic assays but results were similar in 20 mM Tris, 20 mM NaCl, pH 7.4. Quenching of Trp fluorescence was observed in the presence of AV411 and AV1013, but not ISO-1. MIF has only one tryptophan, which is distant from the catalytic Pro1 and exposed to the solvent. Trp fluorescence shifted from 340 to 350 nm when the concentration of AV411 and AV1013 increased. For ISO-1, quenching of Tyr fluorescence was observed. This is consistent with a previously reported crystal structure of ISO-1 with MIF (23) where ISO-1 and Tyr95 make a perpendicular phenol–phenol ring interaction.
AV411 and AV1013 Inhibit MIF Chemotactic Activity.
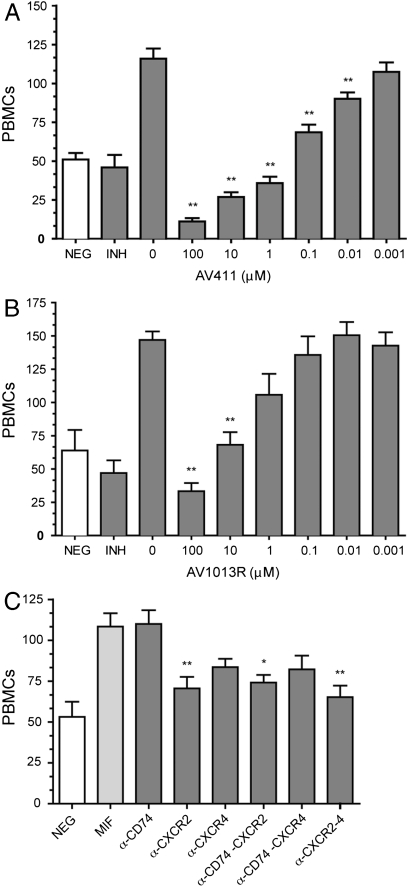
To determine the effect of AV411 and AV1013 on rhMIF-induced peripheral blood mononuclear cell (PBMC) migration, plate-based cell chemotaxis assays were employed. The PBMC migration inhibitory activity of AV411 was determined to be dose dependent, with statistically significant inhibition observed at concentrations as low as 10 nM (Fig. 2A). Consistent with the reduced MIF tautomerase activity, the R and S enantiomers of AV1013 were less potent in terms of their ability to inhibit PBMC migration with significant inhibition only observed at 10 μM and no significant difference for either enantiomer (Fig. 2B and Fig. S1A). The specificity of compounds AV411 and AV1013 for rhMIF-induced migration was determined by testing their ability to inhibit MIP-1α (CCL3)-induced PBMC chemotaxis. Neither AV411 nor AV1013 reduced the ability of MIP-1α stimulated PBMCs to transmigrate relative to controls and also verified that phosphodiesterase mechanisms are not involved in migration due to MIP-1α (Fig. S1B). Similarly, IL-8 stimulated PBMC chemotaxis was unaffected by AV411 (Fig. S1C). As a control on the effect of AV411 on PDE inhibition, we added AV411on the top of the migration chamber in the presence and absence of MIF in the bottom chamber (Fig. S1D). There is no statistically significant difference in MIF-induced migration with or without AV411 in the upper chamber (p > 0.05) indicating that the effect of AV411 on MIF-mediated PBMC migration is not due to PDE inhibition.
Fig. 2.
PBMC cell migration inhibition. PBMC migration dose response for (A) AV411 and (B) (R)-AV1013. For all panels, negative control (NEG) is without rhMIF. Positive control is with rhMIF but no drug added. P values represent significant difference relative to positive control. One, two, or three asterisks represent p < 0.05, 0.01, or 0.001, respectively. Inhibitors (INH) in (A) and (B) are AV411 and (R)-AV1013, respectively. (C) Inhibition of MIF-induced chemotaxis of PBMCs in the presence of individual or paired combinations of monoclonal antibodies (10 μg/mL each) directed against MIF receptors (CD74, CXCR2, and CXCR4). P values represent comparison to the MIF-mediated migration in the absence of the antibodies.
To identify the receptor(s) responsible for MIF-mediated migration, antibodies directed against MIF receptors were tested. Anti-human CD74 did not inhibit MIF-mediated PBMC migration, whereas anti-human CXCR2 inhibited chemotaxis by 69% (Fig. 2C and Table S2). An anti-human CXCR4 monoclonal antibody shows a trend toward inhibition that is not statistically significant (Fig. 2C). Neither a single antibody nor combinations of antibodies completely abrogated migration (Fig. 2C and Fig. S1E). These studies show that CXCR2 is the major receptor that mediates MIF-induced PBMC chemotaxis.
NMR and X-Ray Studies Elucidate an Allosteric Inhibition Mechanism of MIF.
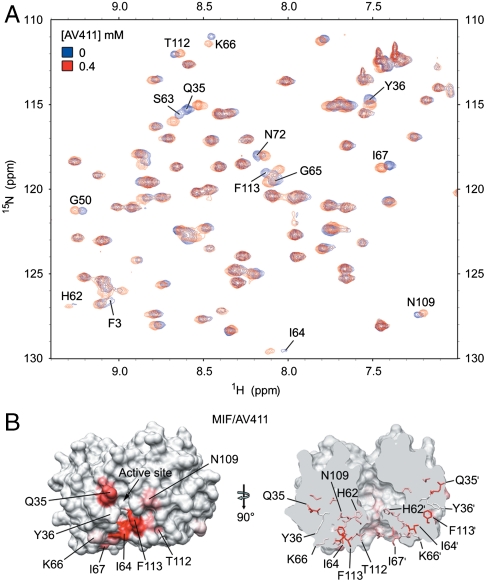
Heteronuclear single quantum coherence (HSQC) (Fig. 3A and Fig. S2A) and transverse relaxation optimized spectroscopy (TROSY) (Fig. S2 B and C) data were collected on 15N-rhMIF in the absence and presence of AV411 or AV1013, respectively. Mapping these changes on the three-dimensional trimeric structure of rhMIF shows regions associated with these changes in chemical shifts. For AV411, one region surrounds the active site with some overlapping active site residues, Ile 64 and Phe113 (Fig. 3B Left). The second region is located behind the active site as shown in Fig. 3B with residue His62 near the pore of the trimer (Fig. 3B Right). By implementing TROSY experiments that allow for higher resolution and sensitivity relative to HSQC, we measured chemical shifts for AV1013 to rhMIF (Fig. S2 B and C). Mapping the chemical shift changes on the structure of rhMIF shows some residues that are identical to those observed with AV411. Among these residues are active site residues Tyr36 and Ile-64 (Fig. S2D and Table S3), although the enzymatic analysis revealed that both AV411 and AV1013 were noncompetitive inhibitors. To more fully characterize the binding site of these noncompetitive inhibitors, crystallographic studies were performed.
Fig. 3.
AV411 and AV1013 interactions with rhMIF in solution. (A) HSQC spectra of rhMIF with (red) or without (blue) AV411 are overlaid. (B) The surface of a rhMIF trimer is shown with the degree of chemical shift change, colored in red, upon binding of AV411 (Left). The highest intensity indicates 0.094 ppm of chemical shift change. On the right, is the surface of the trimer rotated by 90° along the trimer axis and clipped to show relative locations of the mapped residues. The residues are colored in the same manner as with the left. Only the residues revealing large chemical shifts are labeled.
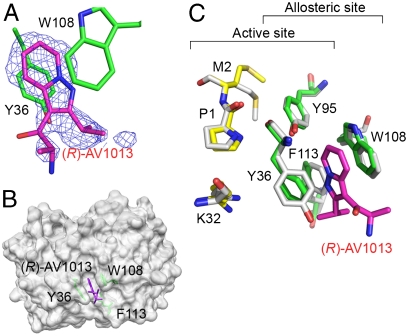
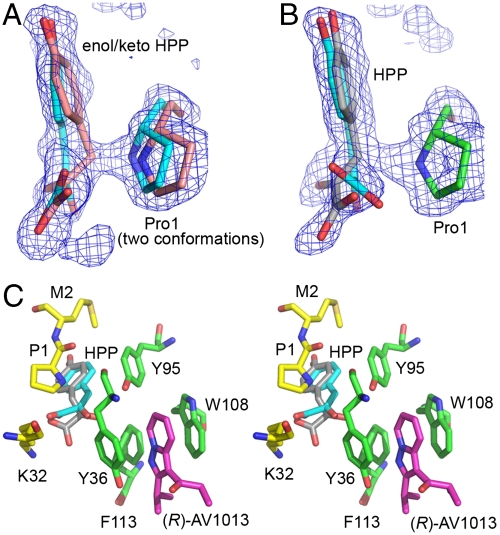
MIF was cocrystallized with the R-isomer of AV1013 alone and with the substrate HPP. The structures of the binary and ternary complex were determined to 1.7 and 1.25 Å resolution, respectively (Table S4). In the binary complex, extra electron density reveals the binding site of AV1013 between the side chains of Trp108 and the active site residue Tyr36 (Fig. 4 A and B) at a location that is distal to the active site. This is consistent with the results of the fluorescence experiment that demonstrates quenching of Trp fluorescence. (In contrast, ISO-1 only quenches tyrosines present in the active site, but has no effect Trp fluorescence.) The chiral center and the amine that distinguishes AV1013 from AV411 are exposed to solvent. Aromatic–aromatic interactions are made by the fused aromatic ring of AV1013 with the phenol ring of Tyr36 and the indole ring of Trp108, and hydrophobic interactions occur with the side chain of Phe113. In the unbound structure, these three amino acids pack together tightly, and there is no cavity in which a molecule can bind (PDB 1MIF) (18). The binding of AV1013 in this new pocket results in a conformational change in the position of residues 33–36 adjacent to the active site. The backbone atoms of Tyr36 are displaced from their original positions and the aromatic side chain is rotated away from Trp108 toward the active site cavity relative to its position in the unbound protein (Fig. 4C). The location of this site agrees with one of the sites indicated by the TROSY study (Fig. S2D).
Fig. 4.
rhMIF/(R)-AV1013 crystal structure. (A) Simulated annealing electron density calculated omitting (R)-AV1013 and contoured at 3.0σ, showing AV1013 (in magenta) between Tyr36 and Trp108. (B) Surface representation of AV1013-bound MIF in the same orientation displayed in Fig. 3 and Fig. S2. The inhibitor binds in a site predicted by the NMR results. (C) Superposition of the complex structure of rhMIF-(R)AV1013 (carbons in green for the allosteric site and yellow for the active site) and apo-rhMIF (carbons in white) showing the conformational shift of Tyr36 that allows AV1013 (all atoms in magenta) to bind. The superposition was performed by overlaying the positions of Cα atoms of residues 2–30 and 37–114 due to larger rms deviations for Pro1 and the loop including residues 31–36.
The ternary complex shows substrate in three active sites but AV1013 only in one allosteric site. The active sites with HPP but no inhibitor are similar and are near symmetry-related proteins that preclude binding or result in ejection of inhibitor during crystal formation. The high resolution of this structure allows us to discern the binding of both the enol and keto forms in these two active sites, with the C3 atom of the keto form very close to the nitrogen atom of the catalytic base, Pro1 (Fig. 5A). The distance of the C3-N interaction is closer than a typical van der Waals contact with continuous electron density between the two atoms, implying that C3 is sharing a proton with the nitrogen. The stereospecificity of the reaction can be resolved as the pro-R proton of the keto form. The enol form on the other hand, which is in the cis conformation, is bound at the center of the active site.
Fig. 5.
rhMIF-HPP-(R)AV1013 ternary complex. (A) An active site with only HPP bound. HPP can be modeled as the enol (carbon atoms in cyan) and keto (carbon atoms in pink) form within the same active site, suggesting that interconversion between these two forms occurs in the active site. The simulated annealing electron density displayed in A and B is 3.0σ Fo-Fc density calculated omitting all of substrate molecules as well as Pro1. (B) The ternary complex with HPP in the active site and (R)-AV1013 in the neighboring allosteric site. (C) Proximity of allosterically bound (R)-AV1013 to the active site bound HPP shown in stereo with carbon atoms of the protein shown in green for the allosteric site and yellow for the active site, those of the inhibitor shown in magenta, those of the enol form of HPP in cyan, and those of the Pro1 proximal HPP in gray. The conformational shift of Tyr36 observed in the binary structure (Fig. 4B) is also seen here in the ternary complex in the (R)-AV1013-bound subunit.
The remaining active site containing the inhibitor in the ternary complex has similar conformational changes of allosteric site residues as in the binary MIF/(R)-AV1013 structure (Figs. 4C and 5C, and Movie S1). The active site also contains two forms of the substrate. The enol form, however, extends quite close to the phenyl ring of Tyr36 (2.5 Å), which has average B factors approximately 50% greater than Tyr36 in the other two subunits. This suggests the possibility that the conformational change trapped by the inhibitor places Tyr36 in a position that makes binding of the enol form of HPP less favorable. The second form of HPP found in the inhibited active site is positioned with the C3 atom very close to Pro1, but attempts to fit either the keto or enol form were unsuccessful. The angle at atom C3 is too wide for the keto form (which would have tetrahedral geometry at C3) and the C2-C3 dihedral angle is neither cis nor trans as required for the enol form. The dihedral and bond angle restraints were relaxed in order to fit the molecule into the electron density (Fig. 5B). This observed form occupies the same position as the keto form in the other two active sites except the distance between C3 and the nitrogen of Pro1 is longer, but still closer than expected (2.5 Å) for van der Waals contacts. The oxygen on C2 is near Phe113, which has two conformations in both the inhibited and an uninhibited active site. The primary conformation of Phe113 in the absence of inhibitor is farther from the substrate. However, it is found closer to the substrate in the presence of the inhibitor (Fig. S3). The dynamic movement of Phe113 into the active site may suppress access of water molecules that could function in proton transfer with the C2 oxygen atom during catalysis. In addition, the movement of the side chain of Phe113 into the active site may slow the movement of HPP into or out of the active site. These changes in active site residues can be explained by the proximity of the allosteric binding site to the active site (Fig. 5C).
Discussion
In this study, we identified the proinflammatory protein MIF as a potential target for a clinically used therapeutic, ibudilast (AV411) in bronchial asthma and poststroke complications. The neuroprotective mechanism of AV411 includes suppression of leukotriene B4, IL-1β, TNF-α, IL-6, and nitric oxide production, and an increase of the expression of the antiinflammatory cytokine IL-10 (27). In neuron and microglia cultures, AV411 also induces the production of NGF, glial-derived neurotrophic factor, and neurotrophin, which adds to its neuroprotective effects (27). We show that AV411 and a close analog, AV1013, which replaces a methyl group with an amine and has reduced activity against phosphodiesterases (approximately 3–57-fold lower inhibition), inhibit the catalytic activity of MIF. These compounds are noncompetitive inhibitors of the MIF tautomerase activity, which allows for binding of inhibitor to substrate- or product-bound enzyme or to free enzyme.
The structures of rhMIF with the stereoisomer (R)-AV1013 in the presence and absence of the substrate HPP were determined at high resolution. In both the binary and ternary forms, there is extra electron density for an aromatic molecule adjacent to the active site residue Tyr36. This binding site requires a conformational change with respect to the apo-structure among tightly packed side chains of Tyr36, Phe113, and the backbone atoms of residues 33–36 in a loop near the entrance of the active site. We speculate that this site forms during normal protein dynamics and AV1013 is able to bind at this site to form aromatic interactions with Tyr36 and Trp108 and induce a conformational change of the active site residue Tyr36. Flexibility of the protein backbone in the vicinity of Tyr36 has been observed in other rhMIF small-molecule complexes. In the case of rhMIF bound to the active site inhibitor (E)-4-hydroxybenzaldehyde-O-cyclohexanecarbonyloxime (PDB 2OOH), Tyr36 of chain B, including main-chain atoms, has two conformations (25). In the structure of an acetaminophen dimer complexed to rhMIF (PDB 3DJI), there is a side-chain torsion change in Tyr36 (28). These examples reflect the inherent flexibility of Tyr36. The dynamic motion of Tyr36 allows various small molecules to bind on either side of the side chain, either in the active site or in a previously unidentified allosteric site as shown in the present work.
In the ternary crystal form (1.25 Å), we are able to observe AV1013 binding to one allosteric site and HPP binding to all three active sites. In the substrate-only active sites, the presence of enol and keto forms of HPP is apparent because both are modeled in the active sites with similar occupancies. Modeling these forms as separate entities illustrates the movement within the active site during catalysis as the enol and keto forms interconvert. The HPP-(R)-AV1013 active site contains the enol form as in the other active sites. However, the other observed ligand binds in a location of the active site lodged between Phe113 and the nitrogen of Pro1, which is close to carbon C3 of the ligand. The binding of AV1013 causes the dynamic movement of Phe113 into the active site resulting in the changes noted above.
Inhibition of PBMC chemotaxis by AV411 and AV1013 is robust with no inhibition occurring when MIP-1α or IL-8 is used as the chemoattractant, thereby confirming the specificity of these inhibitors for the MIF pathway (Fig. 2 and Fig. S1 B and C). AV411 inhibits MIF-mediated chemotaxis in clinically relevant concentrations (6, 26) (Fig. 2A). This chemotactic inhibition at low concentrations is incongruous with the Ki or Kd because only a small percentage of MIF would be inhibited. However, the Ki and Kd may be overestimated in the nonphysiological conditions of these experiments. For example, the substrate HPP used to measure the Ki is a pseudosubstrate because the Km is more than 1,000-fold higher than the in vivo concentration of HPP (20, 29). MIF has been reported to form complexes with over a dozen proteins (15, 30–40). Potential interactions between the enzyme and physiological molecules (substrate or protein) may enhance the affinity of the allosteric inhibitors for MIF and result in their robust chemotactic inhibitory effects.
The current results have a number of implications. The noncompetitive inhibition of MIF is consistent with recent findings that dynamics in proteins form allosteric sites leading to specific binding (41). The MIF allosteric site is absent from the crystal forms of apo-MIF. A minority must be present as a preexisting conformation as it is difficult to grasp how AV1013 or AV411 could induce this site. Once a small molecule occupies this site, a conformational change involving Tyr36 and Phe113 impinges upon the active site. This allosteric site provides a foundation for the discovery and development of MIF inhibitors as therapeutics for disorders including inflammatory, autoimmune, oncogenic, and neurological diseases. Our studies also provide a possible pharmacological mechanism by which clinically relevant AV411 concentrations may contribute therapeutically to MIF-associated bronchial asthma (42, 43), glial attenuation, and related chronic pain (4, 6, 9, 26).
Methods
Materials.
The human MIF was prepared as previously described (17). For NMR experiments, minimal media that contained 15NH4Cl (Cambridge Isotope Laboratories, Inc.) was used as the nitrogen source to express 15N-rhMIF. AV411 was purchased from Sanyo Chemical Laboratory Co., Ltd. Synthesis and resolution of AV1013 is described in Scheme S1 and SI Methods.
Enzymatic and Chemotactic Inhibition Assays.
p-Hydroxyphenylpyruvate assay was performed as described previously with a minor modification (44). The effects of AV411 and AV1013 on 8 nM rhMIF-induced chemotaxis were measured using a plate-based chemotaxis assay as previously described (44). Details are reported in SI Text.
Fluorescence Spectroscopy.
Dose responses were performed by serial addition of inhibitor (14–84 μM) into 14 μM MIF. For these experiments, MIF was titrated with inhibitor and excited at 285 nm for Trp (all inhibitors) and 274 nm for Tyr (for ISO-1). As a reference, the same volume of DMSO was mixed with MIF. Trp fluorescence emission was scanned from 300 to 380 nm. Emission of Tyr fluorescence was scanned from 290 to 310 nm. Kds were obtained by plotting the data according to the modified Stern–Volmer equation (45). More details can be found in the SI Text.
NMR Spectroscopy.
For the HSQC experiment, 356 μM of 15N-labeled rhMIF in 10% D2O, 1.1% DMSO, with or without 534 μM AV411 was prepared. For TROSY experiments, 200 μM of 15N-labeled rhMIF with 10% D2O was mixed with either 88 μL of 50 mM of AV1013 or the same volume of DMSO as a reference. All experiments were performed at 25 ºC. Amide assignments of MIF without ligands were facilitated by previously published data (46). Details are listed in the SI Text.
X-Ray Crystallography.
MIF-(R)AV1013 cocrystals were grown with or without HPP as described previously (23) with a few modifications, most notably that crystals were grown at 37 ºC. The structures were determined by molecular replacement using unliganded MIF (PDB 3DJH) (28). During refinement, dihedral and bond angle parameters in the dictionary files were relaxed for one of the HPP forms in the inhibited site to fit the 1.25 Å electron density. Details are presented in the SI Text.
Supplementary Material
Acknowledgments.
We thank the staff at beamline X29 of the National Synchrotron Light Source at Brookhaven National Laboratory for assistance in data collection. Financial support was provided by Avigen, Inc. (E.J.L.), National Institutes of Health (NIH) R01 AR049610 (to R.B.) and R01 AI-065029 and AI-082295 (to E.J.L.), Alliance for Lupus Research (R.B.), and a fellowship from a NIH Neuropharmacology Training Grant T32 NS007136 (to Y.C.).
Footnotes
Conflict of interest statement: Some financial support was provided by Avigen, Inc. (E.J.L.).
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3IJG and 3IJJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002716107/-/DCSupplemental.
References
- 1.Nakahara T, et al. Relaxation and potentiation of cGMP-mediated response by ibudilast in bovine tracheal smooth muscle. N-S Arch Pharmakol. 2002;366:262–269. doi: 10.1007/s00210-002-0585-3. [DOI] [PubMed] [Google Scholar]
- 2.Kishi Y, et al. Ibudilast modulates platelet-endothelium interaction mainly through cyclic gmp-dependent mechanism. J Cardiovasc Pharma. 2000;36:65–70. doi: 10.1097/00005344-200007000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Milligan ED, Watkins LR. Pathological and protective roles of glia in chronic pain. Nat Rev Neurosci. 2009;10:23–36. doi: 10.1038/nrn2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchinson MR, et al. Reduction of opioid withdrawal and potentiation of acute opioid analgesia by systemic AV411 (ibudilast) Brain Behav Immun. 2009;23:240–250. doi: 10.1016/j.bbi.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ledeboer A, Hutchinson MR, Watkins LR, Johnson KW. Ibudilast (AV-411). A new class therapeutic candidate for neuropathic pain and opioid withdrawal syndromes. Expert Opin Inv Drug. 2007;16:935–950. doi: 10.1517/13543784.16.7.935. [DOI] [PubMed] [Google Scholar]
- 6.Ledeboer A, et al. The glial modulatory drug AV411 attenuates mechanical allodynia in rat models of neuropathic pain. Neuron Glia Biol. 2006;2:279–291. doi: 10.1017/S1740925X0700035X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153:80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- 8.David JR. Delayed hypersensitivity in vitro: Its mediation by cell-free substances formed by lymphoid cell-antigen interaction. Proc Natl Acad Sci USA. 1966;56:72–77. doi: 10.1073/pnas.56.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F, et al. Spinal macrophage migration inhibitory factor contributes to the pathogenesis of inflammatory hyperalgesia in rats. Pain. 2010;148:275–283. doi: 10.1016/j.pain.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 10.Nishibori M, et al. Presence of macrophage migration inhibitory factor (MIF) in ependyma, astrocytes and neurons in the bovine brain. Neurosci Lett. 1996;213:193–196. doi: 10.1016/0304-3940(96)12864-0. [DOI] [PubMed] [Google Scholar]
- 11.Lolis E. Glucocorticoid counter regulation: Macrophage migration inhibitory factor as a target for drug discovery. Curr Opin Pharmacol. 2001;1:662–668. doi: 10.1016/s1471-4892(01)00112-6. [DOI] [PubMed] [Google Scholar]
- 12.Calandra T, Roger T. Macrophage migration inhibitory factor: A regulator of innate immunity. Nat Rev Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi X, et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber C, et al. Structural determinants of MIF functions in CXCR2-mediated inflammatory and atherogenic leukocyte recruitment. Proc Natl Acad Sci USA. 2008;105:16278–16283. doi: 10.1073/pnas.0804017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernhagen J, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13:587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 16.Lolis E, Bucala R. Macrophage migration inhibitory factor. Expert Opin Ther Tar. 2003;7:153–164. doi: 10.1517/14728222.7.2.153. [DOI] [PubMed] [Google Scholar]
- 17.Sun HW, et al. The subunit structure of human macrophage migration inhibitory factor: Evidence for a trimer. Protein Eng. 1996;9:631–635. doi: 10.1093/protein/9.8.631. [DOI] [PubMed] [Google Scholar]
- 18.Sun HW, Bernhagen J, Bucala R, Lolis E. Crystal structure at 2.6-A resolution of human macrophage migration inhibitory factor. Proc Natl Acad Sci USA. 1996;93:5191–5196. doi: 10.1073/pnas.93.11.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swope MD, Lolis E. Macrophage migration inhibitory factor: Cytokine, hormone, or enzyme. Rev Physiol Biochem Pharmacol. 1999;139:1–32. doi: 10.1007/BFb0033647. [DOI] [PubMed] [Google Scholar]
- 20.Rosengren E, et al. The macrophage migration inhibitory factor MIF is a phenylpyruvate tautomerase. FEBS Lett. 1997;417:85–88. doi: 10.1016/s0014-5793(97)01261-1. [DOI] [PubMed] [Google Scholar]
- 21.Rosengren E, et al. The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol Med. 1996;2:143–149. [PMC free article] [PubMed] [Google Scholar]
- 22.Swope M, Sun HW, Blake PR, Lolis E. Direct link between cytokine activity and a catalytic site for macrophage migration inhibitory factor. EMBO J. 1998;17:3534–3541. doi: 10.1093/emboj/17.13.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lubetsky JB, et al. The tautomerase active site of macrophage migration inhibitory factor is a potential target for discovery of novel anti-inflammatory agents. J Biol Chem. 2002;277:24976–24982. doi: 10.1074/jbc.M203220200. [DOI] [PubMed] [Google Scholar]
- 24.Al-Abed Y, et al. ISO-1 binding to the tautomerase active site of mif inhibits its pro-inflammatory activity and increases survival in severe sepsis. J Biol Chem. 2005;280:36541–36544. doi: 10.1074/jbc.C500243200. [DOI] [PubMed] [Google Scholar]
- 25.Crichlow GV, et al. Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor. J Biol Chem. 2007;282:23089–23095. doi: 10.1074/jbc.M701825200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rolan P, et al. Ibudilast in healthy volunteers: Safety, tolerability and pharmacokinetics with single and multiple doses. Brit J Clin Pharmaco. 2008;66:792–801. doi: 10.1111/j.1365-2125.2008.03270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuno T, et al. Neuroprotective role of phosphodiesterase inhibitor ibudilast on neuronal cell death induced by activated microglia. Neuropharmacology. 2004;46:404–411. doi: 10.1016/j.neuropharm.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 28.Crichlow GV, Lubetsky JB, Leng L, Bucala R, Lolis EJ. Structural and kinetic analyses of macrophage migration inhibitory factor active site interactions. Biochemistry. 2009;48:132–139. doi: 10.1021/bi8014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deutsch JC. Determination of p-hydroxyphenylpyruvate, p-hydroxyphenyllactate and tyrosine in normal human plasma by gas chromatography-mass spectrometry isotope-dilution assay. J Chromatogr B. 1997;690:1–6. doi: 10.1016/s0378-4347(96)00411-2. [DOI] [PubMed] [Google Scholar]
- 30.Kleemann R, et al. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature. 2000;408:211–216. doi: 10.1038/35041591. [DOI] [PubMed] [Google Scholar]
- 31.Jung H, Kim T, Chae HZ, Kim KT, Ha H. Regulation of macrophage migration inhibitory factor and thiol-specific antioxidant protein PAG by direct interaction. J Biol Chem. 2001;276:15504–15510. doi: 10.1074/jbc.M009620200. [DOI] [PubMed] [Google Scholar]
- 32.Wadgaonkar R, et al. Intracellular interaction of myosin light chain kinase with macrophage migration inhibition factor (MIF) in endothelium. J Cell Biochem. 2005;95:849–858. doi: 10.1002/jcb.20472. [DOI] [PubMed] [Google Scholar]
- 33.Leng L, et al. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Lu C, Xing G, Zhu Y, He F. Macrophage migration inhibitory factor directly interacts with hepatopoietin and regulates the proliferation of hepatoma cell. Exp Cell Res. 2004;300:379–387. doi: 10.1016/j.yexcr.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 35.Jung H, Seong H-A, Ha H. Critical role of cysteine residue 81 of macrophage migration inhibitory factor (MIF) in MIF-induced inhibition of p53 activity. J Biol Chem. 2008;283:20383–20396. doi: 10.1074/jbc.M800050200. [DOI] [PubMed] [Google Scholar]
- 36.Jung H, Seong H-A, Ha H. Direct interaction between Nm23-H1 and macrophage amigration inhibitory factor (MIF) ss critical for alleviation of MIF-mediated suppression of p53 activity. J Biol Chem. 2008;283:32669–32679. doi: 10.1074/jbc.M806225200. [DOI] [PubMed] [Google Scholar]
- 37.Liu L, et al. Macrophage migration inhibitory factor (MIF) interacts with Bim and inhibits Bim-mediated apoptosis. Mol Cells. 2008;26:193–199. [PubMed] [Google Scholar]
- 38.Son A, et al. Direct association of thioredoxin-1 (TRX) with macrophage migration inhibitory factor (MIF): Regulatory role of TRX on MIF internalization and signaling. Antioxid Redox Sign. 2009;11:2595–2605. doi: 10.1089/ars.2009.2522. [DOI] [PubMed] [Google Scholar]
- 39.Merk M, et al. The golgi-associated protein p115 mediates the secretion of macrophage migration inhibitory factor. J Immunol. 2009;182:6896–6906. doi: 10.4049/jimmunol.0803710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer-Siegler KL, Cox J, Leng L, Bucala R, Vera PL. Macrophage migration inhibitory factor anti-thrombin III complexes are decreased in bladder cancer patient serum: Complex formation as a mechanism of inactivation. Cancer Lett. 2010;290:49–57. doi: 10.1016/j.canlet.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee GM, Craik CS. Trapping moving targets with small molecules. Science. 2009;324:213–215. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizue Y, et al. Role for macrophage migration inhibitory factor in asthma. Proc Natl Acad Sci USA. 2005;102:14410–14415. doi: 10.1073/pnas.0507189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi E, et al. Macrophage migration inhibitory factor (MIF) in bronchial asthma. Clin Exp Allergy. 2000;30:1244–1249. doi: 10.1046/j.1365-2222.2000.00888.x. [DOI] [PubMed] [Google Scholar]
- 44.Cho Y, et al. Structural and functional characterization of a secreted hookworm Macrophage Migration Inhibitory Factor (MIF) that interacts with the human MIF receptor CD74. J Biol Chem. 2007;282:23447–23456. doi: 10.1074/jbc.M702950200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samworth CM, Degli Esposti M, Lenaz G. Quenching of the intrinsic tryptophan fluorescence of mitochondrial ubiquinol—cytochrome-c reductase by the binding of ubiquinone. Eur J Biochem. 1988;171:81–86. doi: 10.1111/j.1432-1033.1988.tb13761.x. [DOI] [PubMed] [Google Scholar]
- 46.Muhlhahn P, et al. NMR characterization of structure, backbone dynamics, and glutathione binding of the human macrophage migration inhibitory factor (MIF) Protein Sci. 1996;5:2095–2103. doi: 10.1002/pro.5560051016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.