Abstract
Most biological processes are performed by multiprotein complexes. Traditionally described as static entities, evidence is now emerging that their components can be highly dynamic, exchanging constantly with cellular pools. The bacterial flagellar motor contains ∼13 different proteins and provides an ideal system to study functional molecular complexes. It is powered by transmembrane ion flux through a ring of stator complexes that push on a central rotor. The Escherichia coli motor switches direction stochastically in response to binding of the response regulator CheY to the rotor switch component FliM. Much is known of the static motor structure, but we are just beginning to understand the dynamics of its individual components. Here we measure the stoichiometry and turnover of FliM in functioning flagellar motors, by using high-resolution fluorescence microscopy of E. coli expressing genomically encoded YPet derivatives of FliM at physiological levels. We show that the ∼30 FliM molecules per motor exist in two discrete populations, one tightly associated with the motor and the other undergoing stochastic turnover. This turnover of FliM molecules depends on the presence of active CheY, suggesting a potential role in the process of motor switching. In many ways the bacterial flagellar motor is as an archetype macromolecular assembly, and our results may have further implications for the functional relevance of protein turnover in other large molecular complexes.
Keywords: chemotaxis, single molecule, total internal reflection fluorescence, in vivo microscopy, molecular motor
The bacterial flagellar motor is one of the most complex biological nanomachines (1), ideal for investigating turnover within a multimeric complex. It is the result of the coordinated, sequential expression of about 50 genes (2) producing a structure that spans the cell membrane and rotates an extracellular filament extending several micrometers from the cell surface, at speed of hundreds of hertz. The motor is about is about 45 nm in diameter and is composed of at least 13 different proteins, all in different copy numbers. It is powered by a transmembrane ion flux (1, 3, 4) and consists of a core rotating against a ring of stator proteins (1, 5–7). The C ring, also called the “switch complex,” is part of the rotor and localized to the cytoplasmic motor region. The response regulator CheY-P binds one of the C ring components, FliM, causing the rotor to switch rotational direction, thus making FliM the interface with the chemosensory pathway (8–11).
Much is known of the static motor structure (5–7), but the dynamics and interactions of its constituents under natural conditions in living cells are poorly understood. Recent results showed that molecules of the stator protein MotB in the flagellar motor, fused to GFP, exchange with a membrane pool of freely diffusing MotB on a time scale of minutes (12–14). This observation raised the question of whether protein turnover is a general feature of molecular complexes or is a peculiarity of MotB.
To address this question, we investigated FliM-YPet stoichiometry and turnover in Escherichia coli. We found that two FliM populations coexist within the same motor, one undergoing stochastic turnover and one remaining “fixed.” Surprisingly, exchange within the dynamic population relies on the presence of an active form of the response regulator protein CheY. These results illustrate the highly dynamic and adaptive nature of the bacterial flagellar motor and possibly extend to many other large molecular complexes. Stoichiometry experiments revealed the presence of ∼25 FliM-YPet spots per cell in total, 2–8 times more than the typical number (4–8) of complete flagella. Analysis of the FliM stoichiometry of all these complexes gave a heterogeneous distribution, with a peak at ∼32 molecules, close to that observed for tethered spinning motors, and another peak at ∼18 molecules that might reflect preassembly stages of the C ring.
Results and Discussion
Stoichiometry of FliM in Functioning Flagellar Motors.
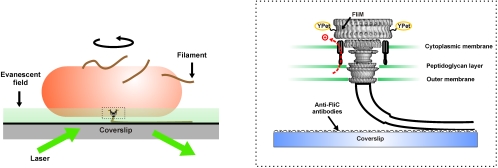
We replaced genomic fliM with fliM-yPet in E. coli. The fliM-yPet strain showed wild-type levels of protein expression. A bead assay showed that the fliM-yPet cells have similar average motor speeds to wild-type cells (82 ± 11 Hz compared to 77 ± 5 Hz for wild-type cells) and no distinguishable differences were seen in the motor switching frequency (run-tumble behavior). However, a small reduction in chemotaxis was observed on swarm plates, with the swarm ring diameters of the fliM-yPet strain being about 75% of those of wild-type E. coli. We observed cells attached to a microscope coverslip via antifilament antibody using bright-field and total internal reflection fluorescence (TIRF) microscopy as described previously (12) (Fig. 1). Cells either rotated freely around a single motor or were immobile, presumably because of the cell body or multiple filaments attaching to the surface (12).
Fig. 1.
TIRF microscopy of live FliM-YPet cells. Tethered cell assay. The cell is attached to the coverslip via one of its sheared filaments using anti-FliC antibody. (Inset) Schematic of the flagellar motor based on EM images (7).
Bright-field images of rotating cells allowed the center of rotation to be found, and TIRF images always indicated a fluorescent spot at this position, confirming the presence of a functional motor (Fig. 2A). We divided the fluorescence intensity in circular regions of interest centered on each spot (radius 400 nm) into two components: a symmetrical 2D Gaussian function representing the spot and a uniform local background. Custom-written imaging software automatically separated and quantified these components and was capable of detecting spots with a total intensity of 2,000 counts or more. The full width at half maximum of spots was typically 300–350 nm, consistent with a FliM ring of diameter ∼50 nm convolved with the point spread function of a single YPet molecule in our microscope of width 250–300 nm (12, 15, 16).
Fig. 2.
Using stepwise photobleaching to estimate FliM-YPet stoichiometry. (A) Bright-field and sequential TIRF (false-color) images of a tethered FliM-YPet cell rotating around a flagellar motor (red cross on bright-field image and red arrow on TIRF image), in the direction indicated by the white arrow. A second motor is visible at the bottom of the TIRF images, as is a fluorescent spot of lower brightness (Green Arrow). (B) Photobleach trace for the motor indicated by the red arrow in A, showing raw (Blue) and filtered (Red) intensities, with expansion of part of the trace (Inset). (C) Power spectrum following Fourier spectral analysis, indicating a peak in periodicity at ∼1.3 kcounts. Stoichiometry distributions using Gaussian kernel density estimation (width = 3) for (D) tethered, cells (Gray) with Gaussian fit (Black) and (E) immobilized cells (Gray) with combined (Black) and individual (Red) two Gaussian fit. The mean ± SD is shown, with 35 spots analyzed for D and E.
Fig. 2B shows decay in the fluorescence intensity of the tethered spinning motor of Fig. 2A, under continuous TIRF illumination. Decay was stepwise with roughly integer multiples of a unitary spacing IYpet (Fig. 2B, Inset), consistent with irreversible photobleaching of single YPet molecules. Fourier spectral analysis (12) indicated IYpet = 1,300 ± 200 counts (Fig. 2C). We calculated the initial intensity I0 by using an exponential fit to extrapolate the trace back to the start of the bleach and estimated the stoichiometry as I0 divided by IYpet. Unbiased kernel density estimation indicated a unimodal distribution over a range ∼20–70 FliM molecules per motor (Fig. 2D). The peak was fitted by a single Gaussian centered on 30 ± 6 (± SD) molecules. This value is similar, within experimental error, to previously reported copy numbers of 33–35 FliM molecules from cryoEM images (5–7).
Stoichiometry of FliM Complexes in Immobilized Cells.
The same analysis on immobilized cells gave a more heterogeneous distribution, with a peak at 32 ± 5 molecules close to that observed for tethered spinning motors and another peak at 18 ± 6 molecules (Fig. 2E). We counted 4 ± 1 spots per cell from the TIRF images. TIRF microscopy takes advantage of the creation of an evanescent wave propagating into the sample with the intensity of the field decreasing exponentially with perpendicular distance from the coverslip surface such that the wavelength-dependent characteristic 1/e depth of penetration is typically ∼100–200 nm (here we measured this as 110 ± 10 nm) . For this reason, only a narrow part of the sample is illuminated. On our microscope, the average step size corresponding to the photobleaching of a single YPet molecule on the surface of the coverslip was 4,000 ± 1,000 counts. Therefore, IYpet within a motor is consistent with the FliM component of the motor being ∼120 nm from the coverslip. If we approximate the TIRF evanescent field as illuminating an optical slice whose height is given by the 1/e depth of penetration and assume that the cell membrane surface closest to the coverslip is ∼50 nm from the coverslip as estimated previously (12), this indicates that ∼17% of the membrane area is encompassed by the optical slice, and therefore the 4 ± 1 spots per cell observed from our TIRF images indicate 24 ± 6 spots per cell in total, 2–8 times more than the typical number (4–8) of complete flagella (17). The peak at 32 molecules in Fig. 2E contains ∼40% of all spots, indicating that there are ∼10 such spots per cell. The peak at 18 molecules may represent C rings in a preassembly state that have not fully integrated into a functional motor. Sourjik and Berg (18) made similar observations by using overexpressed FliM-YFP from an inducible plasmid. We attribute the ∼15% of spots in each distribution falling outside the fitted peaks to multiple C rings not independently resolved by our microscope and spot-detecting algorithm. This value is consistent with the fraction of randomly distributed C rings expected to fall closer to their nearest neighbor than the width of the microscope point spread function (15); these closely neighboring motors are thus detected as a single spot.
Total Number of FliM Molecules per Cell.
We estimated the contribution of instrumental background and cellular autofluorescence to the measured background intensity by using the non-yPet parental strain under the same imaging conditions. We then subtracted this component from background intensity estimates for the fliM-yPet strain to yield the contribution from FliM-YPet molecules not associated with spots. By modeling these molecules as freely diffusing in the cytoplasm, using a convolution model that utilized the measured 3D point spread function of our microscope and the full functional form of the TIRF evanescent field, we estimated a total of 630 ± 290 FliM-YPet molecules per cell (SI Text). Adding this number to the estimate for the total number of molecules associated with all spots gave a total of 1,450 ± 360 molecules per cell, in very good agreement with the 1,400 ± 200 FliM molecules per cell measured in earlier biochemical studies (19).
FliM Turnover and Evidence for Two FliM Populations in the Flagellar Motor.
We investigated FliM turnover by photobleaching FliM-YPet in tethered, spinning motors with a 300-ms focused laser pulse (12) (Movies S1–S3 and Fig. 3A). Fig. 3A (lower) shows fluorescence recovery after photobleaching (FRAP) in a spinning motor (red arrow), indicating the exchange of photobleached molecules in the motor with unbleached molecules from elsewhere in the cell. The reciprocal technique of fluorescence loss in photobleaching (FLIP), in which unbleached motors lose fluorescence because of exchange with bleached molecules from elsewhere in the cell, proved technically challenging in tethered cells, because of blurring of spot images and fluctuations in the cell body height from the coverslip surface. However, we could perform both FRAP and FLIP on immobilized cells. As in tethered cells, FRAP was clearly visible, both in spots and in the local bleached background. Similarly, fluorescence loss (FLIP) was visible in spots distant from the original laser focus (Fig. 3A, upper). We selected spots with a stoichiometry within 1 SD of the mean of the peak at 32 molecules (probable motors) and combined these traces with those from the tethered cell FRAP experiments (definite motors) to construct mean FRAP and FLIP curves (Fig. 3C). These indicated turnover of FliM-YPet in the motor on a time scale of ∼10 min.
Fig. 3.
Turnover in the FliM-YPet complex. (A, B) Pre- and post-focused-laser bleach images, showing immobilized (Upper) and tethered (Lower) cell images for: (A) FliM-YPet and (B) FliM-YPet/ΔCheY. Typical examples of spots analyzed for FRAP (Red Arrow) and FLIP (Blue Arrow) are shown. (C–F) Mean FRAP (Red) and FLIP (Blue) traces (SEM error bounds shown as dotted lines) for: (C) FliM-YPet, (D) FliM-YPet/ΔCheY, (E) FliM-YPet/ΔCheY/CheYD13K/Y106W, and (F) FliM-YPet/ΔCheY/CheYD57A strains. Each trace is based on 7–11 spots.
The mean FRAP and FLIP curves were asymptotic, consistent with approaching a steady state where the fractions of mobile, photobleached molecules in the spot and in the rest of the cell are the same. We modeled FRAP and FLIP experiments by using stochastic Monte Carlo simulations that included diffusion of cytoplasmic FliM-YPet in the cell, reversible binding to fixed motors in the membrane, and convolution of the FliM-YPet distribution with the microscope point spread function to give the predicted TIRF image (SI Text). Exploring a range of different parameter values in the simulations indicates that the recovery traces could be best fitted by using two different populations of FliM-YPet in each motor: 10 molecules fixed in the motor over the experimental time scale and 20 mobile molecules with a mean dwell time in the motor of 40 s. This scheme is consistent with recent structural models of the C ring that suggest two different FliM conformations (20, 21). Performing a similar analysis on spots from immobilized cells with a stoichiometry within 1 SD of the mean of the peak at 18 molecules indicated no significant turnover within experimental error over the time scale of our observations of ∼1,000 s (SI Text).
FliM Turnover Depends on the Presence of CheY.
We repeated FRAP and FLIP experiments in a nonswitching ΔcheY/fliM-yPet strain. In this strain, we saw spots of comparable brightness to those observed previously in both tethered and immobilized cells, but turnover, as measured by FRAP and FLIP, was abolished (Fig. 3 B and D). To confirm the dependence of turnover on CheY, we complemented the ΔcheY/fliM-yPet strain with the CheY mutant CheYD13K/Y106W that mimics the active form of CheY (22). As before, we saw spots of comparable brightness to those observed previously, but FRAP and FLIP traces showed turnover over similar time scales to the wild-type strain (Fig. 3E). We also complemented the ΔcheY/fliM-yPet strain with the CheY mutant CheYD57A that cannot be phosphorylated and performed the same FRAP and FLIP experiments with this strain. In this case, we observed turnover with final recovery intensity levels ∼2/3 that of the wild-type (Fig. 3F). The lower rate seen when overexpressing CheYD57A may be attributable to weaker binding of the nonphosphorylated form of CheY to FliM in these cells.
Our estimate of ∼30 FliM-YPet proteins per functional motor agrees well with earlier structural determinations (5–7). The observed standard deviation of the fitted peak in the stoichiometry distribution was ∼6 molecules, compared to ∼3 predicted by stochastic photobleach simulations with no assumed variation in stoichiometry, analyzed by the same algorithms (12). The discrepancy could be explained by a natural variation of ∼5 molecules in the number of molecules per motor, similar to that observed by cryoelectron microscopy (6). In immobilized cells, we observed ∼4 times as many FliM-YPet complexes per cell as there are functional motors. The majority of the “extra” complexes had a lower stoichiometry of ∼20 molecules per complex and may reflect switch units in a state of preassembly. Our observation that these putative assembly intermediates do not undergo measurable turnover suggests that they have a greater underlying stability compared to fully assembled, functional motor complexes.
Our estimate for the dynamic fraction of FliM in fully assembled motors is close to structural predictions for the stoichiometry of the rotor protein FliG. Recent structural models of the C ring postulate two populations of FliM (20, 21), with ∼26 out of ∼34 FliM molecules bound to the C-terminal domain of FliG, in a different conformation from the other ∼10, which would be tilted inwards towards the rotor axis and bind the middle region of FliG. Thus, if this model is correct, we can speculate that the dynamic FliM motor-associated molecules may be bound to the C-terminal domain of FliG and the nondynamic FliM molecules to the middle region. In this case, FliG molecules are expected to be nondynamic, whereas FliN molecules bound to the dynamic population of FliM should also exhibit turnover. Fukuoka et al. observed such behavior for both FliG and FliN, but with a much larger dwell time for FliN than observed for FliM in our present study (23). Such an observation might be explained by the differences in the technique used. Sarkar et al. reported a rearrangement between FliM and FliN at the bottom of the C ring upon repellent stimulus (24). Such conformational changes, which might be transmitted to the rest of the C ring, may account for FliM and FliN turnover. This also raises the question of whether FliM and FliN molecules also associate outside the motor.
Our FRAP and FLIP experiments on putative assembly intermediate complexes indicate that FliM turnover occurs significantly only when FliM is incorporated into fully assembled, functional motors. In addition, our earlier observations obtained from strains expressing different CheY backgrounds indicate that the active, phosphorylated form of CheY, CheY-P, the response regulator in the chemotaxis signal transduction pathway, is also required for measurable FliM turnover to occur over the time scale of our experiments. One possible explanation is that when CheY-P molecules bind to FliM in the functional motor they cause a conformational change to the C ring that destabilizes the structure, manifest in our experiments as observable turnover of FliM molecules. Whether CheY-P binds exclusively to FliM in functional motors, or also to putative preassembly complexes, remains to be confirmed. It is as yet unclear whether this speculative conformational change of the C ring is a necessary component of the signaling mechanism, namely, that it triggers switching of the motor, or is a by-product of the C ring conformational changes that occurs as a result of motor switching. However, our observation that only ∼20 out of ∼30 FliM molecules in the motor are dynamic raises the possibility that CheY-P binding to the C ring disrupts the interaction between FliM molecules and the C-terminal domain of the rotor protein FliG.
This work represents direct evidence for signal-dependent dynamic exchange of switch complex components in functioning flagellar motors, raising the possibility that turnover is involved in the signaling mechanism. The estimated dwell time for motor-bound FliM-YPet was ∼40 s, slightly longer than that measured previously for GFP-MotB in stator complexes (12). Turnover of a component of the rotor is even more surprising than stator turnover, given that it was previously known that the number of stator complexes can change while the motor is running (25, 26). Simulations also provide an estimate of ∼7 μm2 s-1 for the diffusion coefficient of cytoplasmic FliM-YPet (SI Text), similar to other cytoplasmic bacterial fusion proteins of comparable molecular weight (16). Our data indicate that FliM turnover requires the presence of an active form of the chemotaxis response regulator CheY. This finding, together with previously reported data on MotB turnover in response to prevailing ionic conditions (13, 14), indicates that protein turnover and exchange may be important factors in the function of the bacterial flagellar motor and not only a by-product of motor maintenance. This biological nanomachine can in many ways be viewed as an archetype for large macromolecular assemblies, and our results here may have more general implications to such molecular complexes.
Materials and Methods
Cell Strains and Preparation.
A construct containing the last 500 bp of fliM, followed by yPet, a repeat of the last 9 codons of FliM and 500 bp downstream of fliM, was inserted into the chromosome of E. coli (wild-type and ΔcheY) by allelic exchange as described in SI Text. CheYD13K/Y106W and CheYD57A were overexpressed from pIND4 (27) in the ΔcheY strain.
Preparation of Cells for Microscopy.
Cells were grown in 20 mL tryptone broth at 30 °C to midlog phase (OD600 nm = 0.4–0.5), centrifuged at 4 °C for 10 min at 1,500 × g, and resuspended in 5 mL of motility buffer (10 mM potassium phosphate, 0.1 mM EDTA, pH 7.0). Filaments were sheared by forcing 1 mL of the cell suspension ∼50 times between two syringes with 26-gauge needles connected by a piece of polyethylene tubing (12 cm long, 0.58 mm inner diameter) (12). The cell suspension was centrifuged 3 times at 4,000 × g and resuspended in an equal volume of motility buffer. A 200-μL aliquot of cell suspension was then incubated with 10–20 μL of a 1/50 dilution of stock anti-FliC antibody in PBS for 5 min on ice. The mixture was injected through the tunnel slide and left to incubate for 20 min. Excess motility buffer was then flushed through the tunnel slide to remove unbound cells. Tethered cells typically rotated with maximum speeds of ∼1 Hz.
Microscopy.
A home-built inverted TIRF microscope with 532 nm excitation wavelength was used, as described previously (12, 15). Fluorescence emission was imaged at 50 nm/pixel in frame-transfer mode at 25 Hz by a 128 × 128-pixel, cooled, back-thinned electron-multiplying charge-coupled device camera (iXon DV860-BI; Andor Technology).
Image Acquisition and Analysis.
For stoichiometry, images were sampled for ∼10 s resulting in complete photobleaching of FliM-YPet complexes within the TIRF field. For FRAP and FLIP experiments, single 40-ms TIRF exposures were taken at intervals up to 1,024 s after bleaching with a focused laser spot for 300 ms, centered either over (FRAP) or ∼1–3 μm from (FLIP) a fluorescent spot. Spot and background components were separated and corrected for cumulative photobleaching (12) and normalized by the mean unitary step size to estimate stoichiometry. Mean curves were generated for FRAP and FLIP traces from spots whose stoichiometry was within 1 SD of either the mean observed for tethered cells (∼32 molecules per spot) or for putative intermediate assembly state complexes (∼18 molecules per spot).
In immobilized cells, custom-written software was used to determine boundaries of cell bodies, a method that permitted estimation of the difference  because of membrane curvature in the distance (z) between motor and slide, with tethered motors defined as z = 0 (15). Spot intensities were corrected by a factor exp(Δz/d), d = 110 ± 10 nm (12) to account for differences in the evanescent field intensity.
because of membrane curvature in the distance (z) between motor and slide, with tethered motors defined as z = 0 (15). Spot intensities were corrected by a factor exp(Δz/d), d = 110 ± 10 nm (12) to account for differences in the evanescent field intensity.
Continuous TIRF intensity data were filtered by a Chung–Kennedy edge-preserving algorithm (28). Fourier spectral analysis of the pairwise intensity-difference histograms was used to determine the unitary step size (12, 29). Cytoplasmic diffusion and turnover at the switch complex were modeled by using Monte Carlo simulations (SI Text).
Supplementary Material
Acknowledgments.
The research was supported by the Biotechnology and Biological Sciences Research Council (N.J.D., G.H.W., Q.X., M.T.B., I.M.D., R.M.B., M.C.L., J.P.A), the Engineering, Physics and Science Research Council (G.R., M.C.L.), the Royal Society (M.C.L.), and Hertford College Oxford (M.C.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 11151.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000284107/-/DCSupplemental.
References
- 1.Sowa Y, Berry RM. Bacterial flagellar motor. Q Rev Biophys. 2008;41:103–132. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- 2.Macnab RM. How bacteria assemble flagella. Ann Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 3.Braun TF, Blair DF. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: Evidence for two H+ channels in the stator complex. Biochemistry. 2001;40:13051–13059. doi: 10.1021/bi011264g. [DOI] [PubMed] [Google Scholar]
- 4.Gabel CV, Berg HC. The speed of the flagellar rotary motor of Escherichia colivaries linearly with protonmotive force. Proc Natl Acad Sci USA. 2003;100:8748–8751. doi: 10.1073/pnas.1533395100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki H, Yonekura K, Namba K. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J Mol Biol. 2004;337:105–113. doi: 10.1016/j.jmb.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 6.Thomas DR, Morgan DG, DeRosier DJ. Rotational symmetry of the C-ring and a mechanism for the flagellar rotary motor. Proc Natl Acad Sci USA. 1999;96:10134–10139. doi: 10.1073/pnas.96.18.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas DR, Francis NR, Xu C, DeRosier DJ. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella entericaserovar Typhimurium. J Bacteriol. 2006;188:7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bren A, Eisenbach M. The N-terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol. 1998;278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 9.McEvoy MM, Bren A, Eisenbach M, Dahlquist FW. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein FliM. J Mol Biol. 1999;289:1423–1433. doi: 10.1006/jmbi.1999.2830. [DOI] [PubMed] [Google Scholar]
- 10.Welch M, Oosawa K, Aizawa S, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 12.Leake MC, et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 13.Fukuoka H, Wada T, Kojima S, Ishijima A, Homma M. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol Microbiol. 2009;71:825–835. doi: 10.1111/j.1365-2958.2008.06569.x. [DOI] [PubMed] [Google Scholar]
- 14.Paulick A, et al. Two different stator systems drive a single polar flagellum in Shewanella oneidensisMR-1. Mol Microbiol. 2009;71:836–850. doi: 10.1111/j.1365-2958.2008.06570.x. [DOI] [PubMed] [Google Scholar]
- 15.Leake MC, et al. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc Natl Acad Sci USA. 2008;105:15376–15381. doi: 10.1073/pnas.0806338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenn T, Leake MC, Mullineaux CW. Clustering and dynamics of cytochrome bd-I complexes in the Escherichia coliplasma membrane in vivo. Mol Microbiol. 2008;70:1397–1407. doi: 10.1111/j.1365-2958.2008.06486.x. [DOI] [PubMed] [Google Scholar]
- 17.Turner L, Ryu WS, Berg HC. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coliusing fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 19.Tang H, Blair DF. Regulated underexpression of the FliM protein of Escherichia coliand evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown PN, Terrazas M, Paul K, Blair DF. Mutational analysis of the flagellar protein FliG: Sites of interaction with FliM and implications for organization of the switch complex. J Bacteriol. 2007;189:305–312. doi: 10.1128/JB.01281-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manson MD. How 34 pegs fit into 26 + 8 holes in the flagellar motor. J Bacteriol. 2007;189:291–293. doi: 10.1128/JB.01556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scharf BE, Fahrner KA, Turner L, Berg HC. Control of direction of flagellar rotation in bacterial chemotaxis. Proc Natl Acad Sci USA. 1998;95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukuoka H, et al. Exchange of rotor components in functioning bacterial flagellar motor. Biochem Biophys Res Commun. 2010;394:130–135. doi: 10.1016/j.bbrc.2010.02.129. [DOI] [PubMed] [Google Scholar]
- 24.Sarkar MK, Paul K, Blair DF. Subunit organization and reversal-associated movements in the flagellar switch of Escherichia coli. J Biol Chem. 2010;285:675–684. doi: 10.1074/jbc.M109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, Berg HC. Torque-speed relationship of the flagellar rotary motor of Escherichia coli. Biophys J. 2000;78:1036–1041. doi: 10.1016/S0006-3495(00)76662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sowa Y, Hotta H, Homma M, Ishijima A. Torque-speed relationship of the Na+-driven flagellar motor of Vibrio alginolyticus. J Mol Biol. 2003;327:1043–1051. doi: 10.1016/s0022-2836(03)00176-1. [DOI] [PubMed] [Google Scholar]
- 27.Ind AC, et al. An inducible expression plasmid for Rhodobacter sphaeroidesand Paracoccus denitrificans. Appl Environ Microbiol. 2009;75:6613–6615. doi: 10.1128/AEM.01587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leake MC, Wilson D, Gautel M, Simmons RM. The elasticity of single titin molecules using a two-bead optical tweezers assay. Biophys J. 2004;87:1112–1135. doi: 10.1529/biophysj.103.033571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.