Abstract
MicroRNAs (miRNAs) are short noncoding RNAs that exert posttranscriptional gene silencing and regulate gene expression. In addition to the hundreds of conserved cellular miRNAs that have been identified, miRNAs of viral origin have been isolated and found to modulate both the viral life cycle and the cellular transcriptome. Thus far, detection of virus-derived miRNAs has been largely limited to DNA viruses, suggesting that RNA viruses may be unable to exploit this aspect of transcriptional regulation. Lack of RNA virus-produced miRNAs has been attributed to the replicative constraints that would incur following RNase III processing of a genomic hairpin. To ascertain whether the generation of viral miRNAs is limited to DNA viruses, we investigated whether influenza virus could be designed to deliver functional miRNAs without affecting replication. Here, we describe a modified influenza A virus that expresses cellular microRNA-124 (miR-124). Insertion of the miR-124 hairpin into an intron of the nuclear export protein transcript resulted in endogenous processing and functional miR-124. We demonstrate that a viral RNA genome incorporating a hairpin does not result in segment instability or miRNA-mediated genomic targeting, thereby permitting the virus to produce a miRNA without having a negative impact on viral replication. This work demonstrates that RNA viruses can produce functional miRNAs and suggests that this level of transcriptional regulation may extend beyond DNA viruses.
Keywords: influenza, virus, miRNA, synthetic biology, vector
The discovery of noncoding small RNAs and the specificity by which they can be made to control gene expression has revolutionized our ideas of transcriptional and translational regulation (1). The biogenesis of microRNAs (miRNAs) begins with the transcription of an RNA polymerase II primary miRNA transcript (pri-miRNA) containing an approximately 70- to 80-nt hairpin (1). Formation of the RNA hairpin results in subsequent processing at the base of the double-stranded stem, a process mediated by the cellular complex RNase III enzyme Drosha and a double-stranded RNA-binding protein called DGCR8 (1). Cleavage of the hairpin liberates the miRNA precursor (pre-miRNA) from the larger transcript, permitting nuclear export in an Exportin 5-dependent manner (1). Following translocation, the resulting hairpin is further processed by a second RNase III enzyme called Dicer, which cleaves the bulge of the pre-miRNA and generates an imperfect 22-nt RNA duplex featuring 2-nt 3′ overhangs on each strand. Duplex RNA is subsequently loaded into the RNA-induced silencing complex (RISC), wherein the thermal stability of each end of the duplex is thought to determine which of the strands is used as the guide strand, the basis of posttranscriptional gene silencing (PTGS) specificity (1). The miRNA, loaded into the RISC, is thought to mediate PTGS by binding to 3′ UTRs or the ORFs of mRNA and cause deadenylation and/or translational inhibition (2). In addition to the many well-conserved cellular miRNAs (3), viruses have been shown to synthesize small RNA products (4). Thus far, viral miRNAs have been identified in a number of DNA viruses, including members of the herpesvirus, polyomavirus, and adenovirus families (3–5). These viral miRNAs have been described to regulate their replication cycles (6, 7) and the cellular transcriptome (5, 8–10), and even to mimic endogenous miRNAs (11, 12).
For a virus to produce miRNAs, a hairpin recognizable by Drosha and DGCR8 must be made accessible. Because these cellular components are found in the nucleus, virally produced pri-miRNA would presumably need to be derived from a nuclear virus, because synthesis in the cytoplasm would require nuclear localization or nonconventional processing. Consistent with this idea, cytoplasmic viruses have yet to demonstrate functional miRNA synthesis, despite the identification of small RNA species collectively referred to as viral-derived small RNAs (vsRNAs) (13). Furthermore, the lack of viral miRNAs produced from nuclear RNA virus infections has led many to speculate that viruses with RNA genomes are not amenable to the exploitation of this pathway, because processing of the hairpin would result in the degradation of genomic RNA (14).
In an effort to determine whether viral production of miRNAs is limited to DNA viruses, we incorporated a pri-miRNA into the genome of an RNA virus and characterized its replication properties. Here, we demonstrate that influenza A virus can be engineered to produce a functional miRNA without loss of viral growth. Plasmid-based rescue of influenza A virus, encoding the endogenous neuron-specific microRNA-124 (miR-124) (15), permits normal processing and function of this cellular miRNA while maintaining WT replication levels. A potential obstacle for RNA virus production of miRNAs is that the viral genome will encode a perfect complementary target of the miRNA. In assessing the extent of this limitation, we have ascertained that miRNA targeting of influenza A virus is only effective on the level of mRNA, presumably because of the nuclear localization and molecular organization of the viral ribonucleoprotein (RNP) complex. Taken together, this suggests that an RNA virus can synthesize miRNA without loss of genomic material or self-induced PTGS. Despite the artificial nature of this system, this work demonstrates that RNA viruses are capable of usurping the cell's small RNA machinery as a means of manipulating host and/or viral gene expression and may expand the available molecular vectors that can be used for cellular delivery of small RNAs.
Results
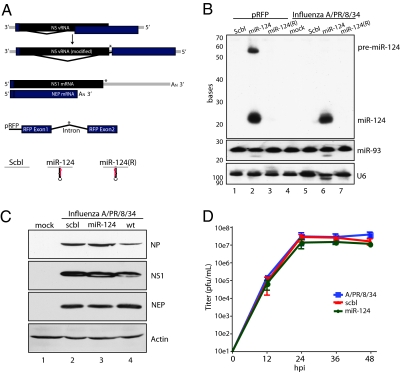
In an effort to determine whether RNA viruses are capable of exploiting the cell's small RNA machinery, we engineered an influenza A virus to encode a known miRNA locus and ascertained how this would affect miRNA processing, PTGS activity, and/or virus replication. Because two of the eight negative-stranded segments that compose the genome of influenza A virus undergo splicing during infection (16), we investigated whether the virus would permit the insertion of a mammalian pri-miRNA in the context of a viral intron, thereby mimicking a number of well-characterized endogenous miRNAs (17). To perform these studies, we chose segment 8, reasoning that being the shorter of two viral transcripts that undergo splicing, it would be more amenable to the addition of genetic material. Segment 8 encodes two proteins, the nonstructural protein 1 (NS1), which confers a block on cellular antiviral activity (18), and the nuclear export protein (NEP, also referred to as NS2), which is responsible for shuttling the mature RNP complexes to the cytoplasm before viral egress and has recently been implicated in controlling virus replication (19, 20). Because the mRNA encoding the N-terminal of NEP/NS2 overlaps with the C-terminal transcript of NS1, we first disrupted the endogenous splice acceptor site and recreated it beyond the stop codon of NS1 (Fig. 1A). Synthesis of this nonoverlapping split ORF created an intergenic region within segment 8 that extended the 3′ UTR of NS1 and the spliced lariat of NEP/NS2. To determine whether the virus would permit insertion of a cellular pri-miRNA, we cloned either a scrambled (scbl) genomic sequence or the murine miR-124-2 locus [in both 5′ to 3′ (miR-124) and 3′ to 5′ [miR-124(R)] orientations] into the intergenic region of segment 8 and generated virus through use of the plasmid-based rescue system (21, 22). Purified virus was propagated in 10-d-old embryonated chicken eggs, growing to titers of ≈10e8 to ≈10e9 pfu/mL. Scbl, miR-124, and miR-124(R) fragments were additionally cloned into the intergenic region of a red fluorescent protein (RFP) expressing plasmid as previously described (23). Virus-dependent miR-124 synthesis was observed at comparable levels to transfected plasmid-based miR-124 production (Fig. 1B). Moreover, miR-124 expression was restricted by the orientation of the pre-miRNA, demonstrating expression only in its endogenous 5′ to 3′ orientation. Furthermore, miR-124 expression required splicing of NEP/NS2, because a construct only expressing NS1 with a miR-124 hairpin in the 3′ UTR failed to produce the small RNA (Fig. S1). In addition, despite the extension of the NS1 3′ UTR and intron length of NEP/NS2, the insertion of neither the scbl sequence nor the pri-miR-124 affected viral protein expression, as demonstrated by robust levels of nucleoprotein (NP) encoded on segment 5 and NS1 or NEP/NS2 encoded on segment 8 (Fig. 1C). To determine the replicative capacity of the NS recombinant viruses, we performed a multicycle growth curve (Fig. 1D). Replication of the recombinant NS viruses demonstrated robust growth and no significant decrease in viral titers compared with WT influenza A/PR/8/34 virus.
Fig. 1.
Engineered split NS1/NEP viruses do not have an impact on viral replication. (A) (Top) Diagram of original NS vRNA segment as compared with the engineered split NS1/NEP construct. (Middle) Diagram of NS1 and NEP mRNA from engineered split vRNA and plasmid-encoding a spliced RFP construct for delivery of exogenous miRNA. (Bottom) Diagram of scbl pri-miR-124 and pri-miR-124(R) inserts. Asterisks show where the inserts were ligated. (B) Small Northern blot of plasmid- and virus-based (MOI = 2) miR-124 expression of scbl, miR-124, and miR-124(R). Levels of miR-93 and U6 were used as loading controls. (C) Western blot analysis of mock or scbl miR-124 or WT A/PR/8/34 influenza (wt) virus infections in MDCK cells (MOI = 2). Blots depict viral NP, NS1, NEP/NS2, and actin. (D) Multicycle growth curve on viruses from C performed in MDCK cells. Error bars depict SD of triplicate samples.
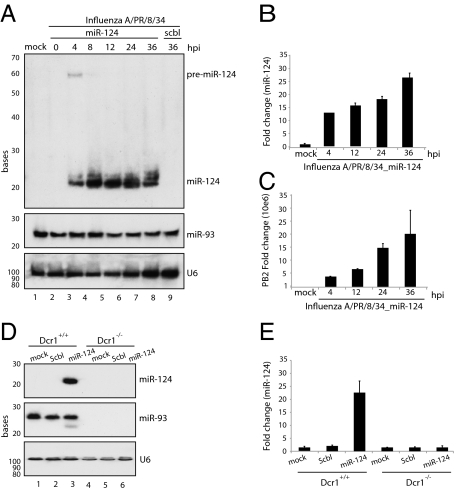
To determine whether the lariat, containing the pri-miRNA generated during NEP/NS2 synthesis, would be continually processed by the endogenous cellular machinery, miR-124–containing influenza A virus infections were performed in Madin–Darby canine kidney (MDCK) cells and were harvested at multiple time points. Small RNA Northern blots for viral-produced miR-124 demonstrated substantial expression of the miRNA as early as 4 h postinfection (Fig. 2A). The robust expression of viral miR-124 was sustained for the duration of infection at levels comparable to those observed for endogenous miR-93. Furthermore, although pre-miR-124 was evident at 4 h postinfection, its absence at later times indicates that viral production of miRNA was not overwhelming the cell's export machinery, a phenomenon previously reported for adenovirus delivery of miRNAs (24). To ensure that the processing of pre-miR-124 mimicked the endogenous Dicer end product, we performed real-time quantification of miR-124 by stem loop-specific RT-PCR (25). Because this assay is specific for the 3′ ends of mature miRNAs and discriminates among related miRNAs that differ by as little as a single nucleotide, the robust 25-fold induction observed in response to the engineered miR-124–containing virus strongly suggests that the mature product is a perfect mimetic of endogenous miR-124 (Fig. 2B). The production of miR-124 also correlated with viral replication as measured by PB2 synthesis (Fig. 2C). To ensure that the production of miR-124 from influenza A virus was processed by the endogenous cell machinery, we performed infections with the scbl control and miR-124–producing viruses in WT- and Dicer-deficient fibroblasts. Total RNA was analyzed by small RNA Northern blotting, demonstrating miR-124 production exclusively in WT cells infected with the miR-124–encoding influenza A virus (Fig. 2D). Loss of miRNA production, as a result of Dicer deficiency, was confirmed by an absence of miR-93 expression. These results were further corroborated through stem loop-specific RT-PCR (Fig. 2E). Taken together, these results suggest that influenza A virus can be engineered to deliver high levels of miR-124 in the context of a de novo virus infection.
Fig. 2.
Engineered viral synthesis of miR-124. (A) Small Northern blot of viral miR-124 at hours postinfection (hpi) indicated (MOI = 1). Levels of miR-93 and U6 were used as loading controls. (B) qRT-PCR analysis of viral miR-124 levels standardized with small nucleolar RNA-202. (C) qRT-PCR analysis of viral PB2 levels standardized with tubulin. (D) Northern blot of viral miR-124 levels in WT and Dcr1−/− fibroblasts. (E) qRT-PCR of samples generated in D. Error bars indicate SD.
One of the RNA viral constraints of encoding a miRNA is that the hairpin itself could form a Drosha substrate during viral replication that would result in genomic slicing, producing two distinct fragments and the miRNA hairpin. This would clearly have an impact on viral progeny output and may induce the formation of defective interfering particles. In the case of influenza A virus, cleavage of the miR-124 hairpin could result in fragmentation of viral cRNA at the base of the miR-124 stem (Fig. 3A). To monitor cRNA levels for cleavage activity, we performed RT on RNA from fibroblasts infected with scbl control or miR-124–containing viruses using an oligo-dT primer or a primer specific for the 3′ cRNA noncoding region, which is absent in both NS1 and NEP/NS2 mRNA (16). Whereas oligo-dT RT synthesized both NS1 and NEP/NS2 mRNA (as well as NS cRNA), 3′ cRNA RT selectively amplified NS cRNA and excluded mRNA, as evident by the lack of NEP/NS2 (Fig. 3B). To determine whether Drosha was capable of processing the miRNA hairpin directly from the genome, we used this discriminating RT reaction to monitor the 5′, 3′, and hairpin regions of the cRNA during de novo virus infection. Quantitative PCR (qPCR) of the NS segment demonstrated that the 5′ and 3′ ends were equally represented between the scbl control and the miR-124–producing influenza A viruses (Fig. 3 C and D). Equal representation of the 5′ and 3′ segment ends suggests that the level of NS synthesis between these two viruses was comparable. To ensure that the qPCR data did not reflect the emergence of a viral revertant, primers specific for the miR-124 NS loop were used to demonstrate that the genomic hairpin was still present (Fig. 3E). Because cleavage of cRNA would result in the inhibition of further viral RNA (vRNA)/cRNA synthesis, the comparable levels of cRNA strongly suggest that viral genomic RNA is not a favorable substrate for Drosha-mediated cleavage. To determine whether genomic RNA was processed by Drosha at any level, we performed 5′ RACE on cRNA (Fig. 3F). In addition to the full-length cRNA product, this analysis amplified a second aberrant cRNA species from the miR-124–producing virus. On sequencing, this ~500-nt species was identified as a heterogenous population of cRNAs. Although some species isolated included 5′ and 3′ cRNA ends with large internal deletions, none of the fragments terminated at the base of the miR-124 hairpin, suggesting random replication intermediates or PCR-mediated splice variants rather than Drosha-mediated activity. Overall, lack of Drosha activity on either NS cRNA (Fig. 3) or the 3′ UTR of NS1 (Fig. S1) suggests that the sole source of miR-124 is the lariat produced during NEP/NS2 synthesis.
Fig. 3.
Viral genomic miRNA hairpins are not substrates for Drosha. (A) Diagram of miR-124 producing segment 8. RNA species include vRNA, cRNA, and mRNA. Primers and reference numbers used in subsequent experiments are depicted. Primers used in RT are as follows: RT1 represents oligo-dT and RT2 is specific to the noncoding region of NS cRNA. (B) RT-PCR products of NEP/NS2 mRNA and NS1 mRNA/3′ NS cRNA. RT1 and RT2 depict primers used in the RT reaction. RNA was derived from mock-infected fibroblasts (−) or cells treated (+) with WT influenza A/PR/8/34. (C) qPCR from mock-treated fibroblasts or cells infected with either scbl or miR-124–producing influenza A viruses. Values depict 5′ NS cRNA levels as compared with tubulin. (D) Same as in C using 3′ cRNA-specific primers. (E) Same as in D using primers specific to the pri-miR-124 insert. (F) 5′ RACE analysis of viral infection. Indicated gel sections were purified and sequenced; representative results are denoted in the table with reference to the numbered diagram in A.
A second hindrance of encoding miRNA in the context of an RNA viral genome is that the genomic strand that encodes the intronic hairpin becomes a perfect inverse complement to the produced miRNA, therefore serving as a potential miRNAt. In the context of influenza, a hairpin produced from mRNA would result in the formation of a miRNAt on the vRNA. This would not occur in the context of cRNA or mRNA because of the imperfect binding along miRNA stem loops. To determine whether this phenomenon causes a significant restriction on RNA virus-produced miRNAs, we engineered additional viruses to determine whether the vRNA could be subject to miRNA-mediated inhibition. For these studies, the segment 8 encoding an intergenic region was used to introduce miR-142 target sites in either the 3′ UTR of NS1 or in the context of vRNA (Fig. 4A). Exogenous expression of miR-142 was achieved by plasmid delivery of the miR-142 hairpin and confirmed by small RNA Northern blotting (Fig. 4B). Because this miRNA has already been demonstrated to induce potent transcriptional inhibition of miR-142 targets (26), we investigated whether the levels of NS1 would be affected when the mRNA or vRNA was targeted (mRNAt and vRNAt, respectively). MDCK cells, or MDCK cells stably expressing miR-142, were infected with a scbl control or with mRNAt or vRNAt recombinant virus at an MOI of 0.1 for 18 h (Fig. 4C). Total protein analysis demonstrated that NS1 levels in control and vRNAt recombinant viruses showed no significant difference regardless of miR-142 expression. In contrast, miR-142 targeting of mRNA (mRNAt) resulted in dramatic loss of NS1 in a miR-142–dependent manner, although viral NP levels remain unaffected. Altogether, these results suggest that the accessibility of genomic RNA to the miRNA/RISC complex is not sufficient to affect the overall transcript levels of the virus.
Fig. 4.
Viral genomic RNA is not targeted by miRNAs. (A) Diagram of recombinant segment 8 encoding an untargeted scbl insert or miR-142 target sites oriented to either the NS vRNA (vRNAt) or the NS1 mRNAt. (B) Small Northern blot probed for miR-142 expression in cells transfected with a miR-142 expression vector. (C) Western blot of MDCK cells and MDCK cells stably expressing miR-142, mock-treated or infected with scbl, vRNAt, or mRNAt viruses (MOI = 0.1). Immunoblots for NS1, NP, and actin are depicted.
Finally, to assess if virus-produced miRNAs are loaded into the RISC complex and capable of mediating PTGS, we determined whether a GFP encoding tandem repeats of miR-124 target elements (GFP_124) could be silenced. Recombinant viral infections and subsequent GFP_124 transfections demonstrated a 47.4% decrease in the number of green fluorescent cells only in the context of the miR-124 expressing influenza A virus (Fig. 5A). Furthermore, to ensure that virus infection could induce PTGS on an endogenous cellular transcript, we used a neuronal precursor cell line (CAD) to determine whether miR-124 expression could stimulate neuron-like differentiation, as previously described (23). To this end, CAD cells were untreated, serum-starved, or infected with the scbl or miR-124–producing influenza A virus strains (Fig. 5B). At 24 h postinfection, or 48 h after serum starvation, cells were fixed and examined by confocal microscopy, demonstrating that serum starvation, or expression of virus-produced miR-124, was sufficient to induce neuron-like morphology. Taken together, these results strongly suggest that influenza A virus can be engineered to encode endogenous and fully functional miRNA.
Fig. 5.
Engineered influenza virus produces functional miR-124. (A) Fibroblasts, transfected with a miR-124 targeted GFP construct were infected with scbl or miR-124–producing (miR-124) influenza A viruses and compared with untreated cells. FACS analysis was used to determine GFP expression (36 hpi). (B) CAD cells were fixed either following 48 h of serum starvation or 24 hpi (MOI = 1) with either scbl or miR-124–producing virus. Cells were stained with β-tubulin before imaging by confocal microscopy. Hoechst dye was used to visualize nuclei.
Discussion
The study of host–pathogen interactions has made considerable progress in characterizing the mechanisms by which cells detect the presence of virus infection and the global transcriptional response that ensues. Virus replication results in the formation of distinct replication intermediates recognized by the cell that lead to the production of IFN-I (27). Secretion of IFN-I communicates a warning message to surrounding cells, prompting a secondary transcriptional response aimed at fortifying their viral defenses through the up-regulation of both antiviral proteins and a small subset of miRNAs (28, 29). Viruses, in turn, counter these defenses through diverse mechanisms. RNA viruses with limiting coding capacity often modulate the cell's antiviral response by inhibiting viral RNA detection, blocking nuclear export, and/or blocking IFN-I signaling all through the direct interaction between viral and host proteins (30). In contrast, larger DNA viruses can perform these functions while additionally modulating the immune response through the production of decoy receptors, chemokines, MHC receptors, and, most recently characterized, miRNAs (31–33). The large coding capacity of DNA viruses, and their predominance for nuclear replication, enables these viruses to usurp the cell's small RNA processing machinery. Although these characteristics are largely limited to DNA viruses, the ability to engineer influenza A virus to encode a functional miRNA suggests that RNA viruses, in general, may also modulate cellular activity in a miRNA-dependent manner.
In this study, we successfully engineered an influenza A virus strain to encode a functional miRNA and we demonstrate that the miRNA is synthesized to levels comparable to those of highly abundant cellular miRNAs. Furthermore, viral generation of miRNAs mimics their endogenous counterparts in their ability to confer PTGS. In addition, PTGS can be achieved without sacrificing the level of virus replication or genome stability. Taken together, this research suggests that future in-depth studies should identify RNA virus-produced miRNAs. It is therefore not surprising that deep sequencing on a number of RNA viruses recently identified large populations of vsRNAs (13). These RNAs were generated from a diverse family of viruses, including poliovirus, hepatitis C virus, Dengue virus, vesicular stomatitis virus, flock house virus, and West Nile virus. Although many vsRNAs demonstrate some aspects of miRNA structure, the physiological function and biogenesis of these heterogeneous vsRNAs remain unknown. Should RNA viruses be capable of inducing PTGS activity, it would undoubtedly serve as a means to suppress cellular immunity or regulate the cellular transcriptome to favor viral replication; however, such an example has yet to be characterized. This may reflect the fact that RNA viruses, although capable, simply do not produce miRNAs. Because the repressive effect of miRNAs on host transcripts rarely exceeds 4-fold (34, 35), viral miRNAs may be inadequate as a strategy to evade the host antiviral response, especially considering the robust induction of many IFN-I–stimulated genes (28). Additionally, the acute nature of RNA virus infections is not well suited to long-term transcriptional modulation by miRNAs, a characteristic not shared with persistent DNA viruses. Should the modest activity of miRNAs outweigh the evolutionary expense of encoding the RNA hairpin, there would be a strong negative selection pressure against RNA virus-encoded miRNAs. Additional studies are required to resolve whether nature has produced such a pathogen.
Finally, regardless of whether RNA viruses produce endogenous miRNAs, the ability to engineer such vectors may have applicational value in small RNA delivery. The issue of effective and nontoxic delivery is a key challenge and serves as the most significant barrier between RNAi technology and its therapeutical application (36). Although lentivirus- and lipid-based delivery models have demonstrated some in vivo success, genomic integration and/or insufficient generation of intracellular miRNAs has limited their applications (36). In contrast, nonintegrating viral vectors have been found to induce ultraphysiological and sustained levels of small RNAs, resulting in toxicity through saturation of the host small RNA cell machinery (24). Although an influenza virus-based delivery method would be confined to the respiratory tract, the extensive clinical data demonstrating the safety of live-attenuated influenza strains and the ability to induce high transient levels of small RNAs may make this an ideal vector for treating viral respiratory infections, asthma, and other acute respiratory diseases by delivering custom-designed miRNA hairpins (37). In conclusion, this work reveals that pathogen-produced miRNAs may extend beyond DNA viruses and suggests that RNA viral vectors may be suitable delivery vehicles for RNA-based therapeutics.
Materials and Methods
Cell Culture.
HEK293, MDCK, CAD, and murine fibroblasts were cultured in DMEM (Mediatech) supplemented with 10% (vol/vol) FBS and 1% penicillin/streptomycin. Dicer-deficient fibroblasts were a kind gift from A. Tarakhovsky (Rockefeller University, New York, NY) and Donal O'Carrol (European Molecular Biology Laboratory, Monterotondo, Italy), and CAD cells were a kind gift from T. Maniatis (Columbia University, New York, NY).
Virus Design and Rescue.
The modified NS segment (A/PR/8/34) was generated by PCR, followed by three-way ligation. Details regarding cloning and generation of the virus can be found in SI Text. Virus rescues using the plasmid-based rescue system are described elsewhere (38).
Virus Infections.
Viral infections were performed at the multiplicity of infections (MOIs) specified. Virus was inoculated into indicated cell lines containing PBS media supplemented with 0.3% BSA (MP Biomedicals) and penicillin/streptomycin for 1 h. Inoculum was then aspirated off and replaced with either fresh complete medium for the indicated times or MEM supplemented with 0.5 or 5% (vol/vol) BSA and L-(tosylamido-2-phenyl) ethyl chloromethyl ketone trypsin.
Northern Blot Analysis.
Northern blots and probe labeling were performed as described previously (39). Probes used include anti-miR-124: 5′-TGGCATTCACCGCGTGCCTTAA-3′, anti-miR-93: 5′-CTACCTGCACGAACAGCACTTTG-3′, miR-142-3p: 5′-TCCATAAAGTAGGAAACACTACA-3′, and anti-U6: 5′-GCCATGCTAATCTTCTCTGTATC-3′.
Western Blot Analysis.
Western blots were generated from total protein separated on a 15% (vol/vol) SDS/PAGE gel. Resolved protein was transferred to nitrocellulose (Bio-Rad), blocked for 1 h with 5% (wt/vol) skim milk at 25 °C, and then incubated with the indicated antibody overnight at 4 °C. Actin (Abcam), NS1, NEP/NS2, and NP (kind gifts from P. Palese, Mount Sinai School of Medicine, New York, NY) antibodies were all used at a concentration of 1 μg/mL in 5% (wt/vol) skim milk. Secondary mouse and rabbit antibodies (GE Healthcare) were used at a 1:5,000 dilution for 1 h at 25 °C. Immobilon Western Chemiluminescent HRP Substrate (Millipore) was used as directed.
Immunofluorescence.
Cells were fixed on glass coverslips by incubating with 4% (vol/vol) formaldehyde overnight at 4 °C. Following two PBS washes, cells were permeabilized with 0.5% octyl phenoxylpolyethoxylethanol detergent in PBS for 10 min and immediately washed two additional times. The cells were then blocked with 0.5% BSA in PBS for 30 min at room temperature. Primary antibody was incubated for 2 h at room temperature at a 1:500 concentration. The monoclonal antibody (E7-β-tubulin) was obtained from the Developmental Studies Hybridoma Bank. Following four washes in 0.5% BSA in PBS, cells were incubated with secondary Rhodamine Red-X (Fisher) at a concentration of 1:750 for 1 h with Hoechst 33342 dye (Invitrogen) added with 15 min remaining. Following four washes, coverslips were mounted on glass slides with Prolong Gold Antifade (Invitrogen). Images were captured with the Leica TCS SP5 DMI microscope at a magnification of ×60.
qPCR.
Conventional qPCR was performed on the indicated cDNA samples using KAPA SYBR FAST qPCR Master Mix (KAPA Biosystems), and miRNA qPCR was performed using TaqMan MicroRNA Assays (Applied Biosystems). Experiments were performed on a Mastercycler ep realplex (Eppendorf). Delta delta cycle threshold (ΔΔCT) values were calculated over replicates using tubulin or small nucleolar RNA 202 as the endogenous housekeeping gene and mock-infected or mock-transfected samples as the calibrator in respective experiments. Values represent the fold difference for each condition compared with mock-infected or mock-transfected samples. Error bars reflect ±SD of fold induction. Primers used for qRT-PCR can be found in SI Materials and Methods.
5′ RACE.
5′ RACE was performed on virally infected samples using the 5′ RACE System for Rapid Amplification of cDNA ends, version 2.0 (Invitrogen).The procedure was carried out according to manufacturer's instructions. In brief, first-strand cDNA synthesis was performed using viral cRNA specific primer 5′-AGTAGAAACAAGGGTGTTTTTTAT-3′. cDNA was purified using SNAP (Invitrogen) purification columns and then tailed with dCTP using terminal deoxynucleotide transferase (TdT) (Invitrogen). The cDNA was then amplified using EconoTaq (Lucigen) with the provided 5′ RACE abridged anchor primer (Invitrogen) and nested NEP primer 5′-AATGGATCCAAACACTGTGTCA-3′. Fragments were then gel-purified using the QIAquick Gel extraction kit (Qiagen) and cloned for sequencing using the TOPO TA Cloning kit (Invitrogen).
Multicycle Growth Curve.
MDCK cells were infected with viruses indicated at an MOI of 0.01. Supernatant (225 μL) was removed at the indicated times. Supernatant was then plaqued in MDCK cells in serial dilutions in triplicate in MEM-agar overlay supplemented with 0.01% DEAE-dextran (Sigma) and 0.1% NaHCO3 (Sigma). Plaques were counted 2 d after infection.
FACS.
GFP_miR-124t was generated by synthesizing and inserting four perfectly complementary miR-124 target sites into the pEGFPC1 plasmid (GenBank accession no. U55763) via HindIII and BamH1. FACS analysis was performed on 2 × 10−6 cells/mL resuspended in PBS with 2% (vol/vol) FBS. GFP expression was quantified through the FL1 channel with the Cytomics Fc 500 (Beckman) instrument.
Supplementary Material
Acknowledgments
We thank Drs. P. Palese (Mount Sinai School of Medicine) and B. Brown (Mount Sinai School of Medicine) for advice, reagents, and comments made during the course of this work. J.T.P. is supported by a Ruth L. Kirschstein National Research Service Award fellowship. A.G.-S. is partly supported by Center for Research on Influenza Pathogenesis (CRIP), a National Institute of Allergy and Infectious Diseases-funded Center of Excellence in Influenza Research and Surveillance (Grant HHSN266200700010C) and by National Institute of Allergy and Infectious Diseases Grants U19AI83025 and U54AI57158. B.R.t. is supported, in part, by Pew Charitable Funds and the U.S. Army Research Office. Confocal laser scanning microscopy was performed at the Mount Sinai School of Medicine Microscopy Shared Resource Facility, supported with funding from National Institutes of Health/National Cancer Institute shared resources Grant 5R24 CA095823-04, National Science Foundation Major Research Instrumentation Grant DBI-9724504, and National Institutes of Health shared instrumentation Grant 1 S10 RR0 9145-01.
Footnotes
Conflict of interest statement: Mount Sinai School of Medicine owns patent positions for reverse genetics of influenza virus, in which A.G.-S. is an inventor.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U55763).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003115107/-/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 7.Bellare P, Ganem D. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: An evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe. 2009;6:570–575. doi: 10.1016/j.chom.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samols MA, et al. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen A, et al. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 2010;24:195–205. doi: 10.1101/gad.553410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnside J, et al. Marek's disease virus encodes MicroRNAs that map to meq and the latency-associated transcript. J Virol. 2006;80:8778–8786. doi: 10.1128/JVI.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skalsky RL, et al. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan R, et al. Sequence conservation and differential expression of Marek's disease virus microRNAs. J Virol. 2008;82:12213–12220. doi: 10.1128/JVI.01722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parameswaran P, et al. Six RNA viruses and forty-one hosts: Viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 2010;6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen BR. Five questions about viruses and microRNAs. PLoS Pathog. 2010;6:e1000787. doi: 10.1371/journal.ppat.1000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 16.Palese P, Shaw M. In: Fields Virology. 5th Ed. Knipe DM, Howley PM, editors. Philadelphia: Raven; 2007. pp. 1648–1698. [Google Scholar]
- 17.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salvatore M, et al. Effects of influenza A virus NS1 protein on protein expression: The NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J Virol. 2002;76:1206–1212. doi: 10.1128/JVI.76.3.1206-1212.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Neill RE, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robb NC, Smith M, Vreede FT, Fodor E. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J Gen Virol. 2009;90:1398–1407. doi: 10.1099/vir.0.009639-0. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fodor E, et al. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:1–9. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 28.de Veer MJ, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 29.Pedersen IM, et al. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.García-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: A lesson in détente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 31.Seet BT, et al. Poxviruses and immune evasion. Annu Rev Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 32.Powers C, DeFilippis V, Malouli D, Früh K. Cytomegalovirus immune evasion. Curr Top Microbiol Immunol. 2008;325:333–359. doi: 10.1007/978-3-540-77349-8_19. [DOI] [PubMed] [Google Scholar]
- 33.Boss IW, Plaisance KB, Renne R. Role of virus-encoded microRNAs in herpesvirus biology. Trends Microbiol. 2009;17:544–553. doi: 10.1016/j.tim.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 35.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mittal V. Improving the efficiency of RNA interference in mammals. Nat Rev Genet. 2004;5:355–365. doi: 10.1038/nrg1323. [DOI] [PubMed] [Google Scholar]
- 37.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 38.Perez JT, et al. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat Biotechnol. 2009;27:572–576. doi: 10.1038/nbt.1542. [DOI] [PubMed] [Google Scholar]
- 39.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.