Abstract
The metazoan circadian clock mechanism involves cyclic transcriptional activation and repression by proteins whose degradation is highly regulated via the ubiquitin-proteasome pathway. The heme receptor Rev-erbα, a core negative component of the circadian network, controls circadian oscillation of several clock genes, including Bmal1 Rev-erbα protein degradation can be triggered by inhibitors of glycogen synthase kinase 3β, such as lithium, and also by serum shock, which synchronizes circadian rhythms in cultured cells. Here we report that two E3 ligases, Arf-bp1 and Pam (Myc-bp2), are copurified with Rev-erbα and required for its ubiquitination. RNA-interference–mediated depletion of Arf-bp1 and Pam stabilizes the Rev-erbα protein and protects Rev-erbα from degradation triggered by either lithium or serum shock treatment. This degradation pathway modulates the expression of Rev-erbα–regulated Clock gene and circadian function in mouse hepatoma cells. Thus, Arf-bp1 and Pam are novel regulators of circadian gene expression that target Rev-erbα for degradation.
Keywords: ubiquitin, circadian rhythm
The physiology and behavior of mammals are subject to daily oscillation driven by endogenous circadian rhythm systems (1–3). In mammals, the core circadian network is generated and maintained via a tightly regulated transcriptional-translational feedback loop (4–6). BMAL1 and CLOCK, the positive components of the circadian clock, activate many circadian output genes, as well as the negative limbs of the circadian loops (2, 7–9). The core negative limb is constituted by the PERIOD (PER) and CRYPTOCHROME (CRY) proteins, which bind to and inhibit BMAL1/CLOCK (4). The other negative limb is mediated by the nuclear receptor Rev-erbα (10), which directly represses Bmal1 gene expression and plays a crucial role in circadian oscillation of Bmal1 mRNA (11).
Emerging evidence has demonstrated that the negative components of the circadian clock are subjected to ubiquitination and subsequent proteasome-mediated degradation (12, 13). For example, the F-box protein FBXL3 regulates the stability of CRY proteins (14–16), whereas another F-box protein containing E3 ligase SCF-βTrcp1 targets phosphorylated PER2 protein for degradation in both Drosophila and mammals (17–20). During a normal circadian cycle, hepatic Rev-erbα protein peaks during the circadian day and becomes undetectable during the circadian night (10). Similar to CRY and PER proteins, Rev-erbα proteins also undergo ubiquitination and proteasome degradation (21), but the mechanism controlling this temporal rhythm of Rev-erbα protein remains unknown.
Lithium, a common treatment for bipolar disorder, can lengthen the period of circadian rhythm in a wide range of experimental systems including insects, mice, and humans (22, 23). In vitro studies suggest that lithium might target multiple clock proteins via its inhibition of GSK3β (21, 24), although whether GSK3β is the sole circadian target of lithium remains unclear. We previously reported that Rev-erbα degradation is accelerated by lithium and stabilized by GSK3β, suggesting the existence of a lithium-induced degradation pathway for controlling Rev-erbα protein cycling (21). In the present work, we used an affinity purification-based proteomic approach to identify two E3 ligases that play an important role in regulating Rev-erbα protein turnover in response to lithium and in antagonizing Rev-erbα–mediated clock gene regulation. These E3 ligases thus regulate clock gene expression by controlling the degradation of Rev-erbα protein.
Results
Rev-erbα Copurifies with E3 Ligases Arf-bp1 and Pam.
We were interested in identifying the degradation pathway, particularly the ubiquitin E3 ligase, for Rev-erbα that is relevant to its clock function. During the ubiquitination process, E3 ligases often form transient protein–protein interactions with their substrates. We hypothesized that the interaction between Rev-erbα and its putative E3 ligases would be stabilized when the degradation pathway was initiated under conditions in which proteasome function was blocked. Flag epitope-tagged Rev-erbα (f-Rev-erbα) was found to have a half-life of about 30 min (Fig. 1A), whereas a Rev-erbα mutant that mimics phosphorylation on serines 55 and 59 (55/59SD) (21) was stable, with a half-life of >2 h (Fig. S1A). Furthermore, lithium dramatically reduced f-Rev-erbα protein, opposite of its effect on stabilization of β-catenin in this cell line, indicating preservation of the lithium-mediated degradation pathway (Fig. 1B). The lithium effect likely occurs through inhibition of GSK3β, given that a specific GSK3β inhibitor also caused the degradation of f-Rev-erbα protein (Fig. S1B).
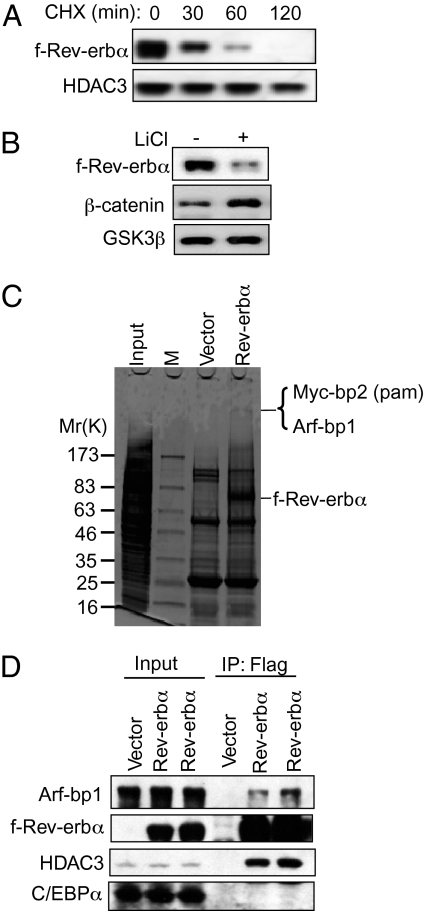
Fig. 1.
The E3 ligase proteins Arf-bp1 and Pam (Myc-bp2) copurify with f-Rev-erbα. (A) The protein half-life of f-Rev-erbα in the presence of cycloheximide was determined in a 293T cell line stably transfected with HA-f-Rev-erbα (HFR). The protein levels of HDAC3 and of an internal loading control were measured. (B) The protein levels of f-Rev-erbα in HFR cells treated with lithium chloride (20 mM) were determined by immunoblot analysis. The β-catenin level serves as a positive control for the action of lithium. (C) Affinity purification of f-Rev-erbα interacting complex from HFR cells. Flag affinity-purification products were run on a polyacrylamide gel and stained with Coomassie blue. The ~400-kDa band was subjected to MALDI-TOF mass spectrometry, which identified two proteins: Pam (Myc-bp2) and Arf-bp1. (D) The interaction of f-Rev-erbα and Arf-bp1 was confirmed by Flag immunoprecipitation (IP), followed by immunoblotting with an antibody against Arf-bp1. Two independent HFR clones (HFR3 and HFR8) were examined for this purpose. The HDAC3 immunoblot was used as a positive control for the f-Rev-erbα IP, and the C/EBPα immunoblot served as a negative control.
The f-Rev-erbα was affinity-purified in the presence of lithium and MG132, a proteasome inhibitor. Coomassie blue staining revealed a high molecular weight component (Fig. 1C). MALDI-TOF mass spectrometry identified two proteins corresponding to this ~400-kDa species: Arf-bp1 and PAM (Myc-bp2) (Fig. 1C). The interaction between f-Rev-erbα and Arf-bp1 was confirmed by immunoblotting (Fig. 1D). Endogenous Rev-erbα bound to Arf-bp1 in HepG2 cells pretreated with lithium and MG132 was detected as well (Fig. S2). The addition of the Rev-erbα ligand heme had no significant effects on the steady-state level of Rev-erbα protein (Fig. S3A) or on the protein half-life of Rev-erbα (Fig. S3B).
Arf-bp1 and Pam Regulate Rev-erbα Protein Levels.
Intriguingly, both Arf-bp1 and Pam are E3 ligases. Arf-bp1 (also known as Ureb1, Lasu1, or Huwe1) belongs to the HECT (homolog of E6AP) family of E3 ligases and is thought to target the ubiquitination and degradation of Mcl, p53, N-myc, and CDC6 (25–27). Pam (also known as Myc-bp2, Phr, or Highwire) belongs to the family of RING-domain E3 ligases and has been shown to regulate mTOR signaling by targeting TSC2 for ubiquitination and degradation (28). Consequently, we tested whether Arf-bp1 and Pam regulate the protein turnover of Rev-erbα. We found that f-Rev-erbα levels were markedly reduced by ectopic expression of both full-length proteins of Arf-bp1 and Pam (Fig. 2A). We then used small interfering RNAs to deplete Arf-bp1 and Pam mRNA and protein (Fig. 2 B and C and Fig. S4). (Note that only antibody against Arf-bp1 is commercially available.) Depletion of Arf-bp1 and Pam markedly stabilized both f-Rev-erbα in HFR cells (Fig. 2B), as well as the endogenous Rev-erbα in Hepa1c1c-7 hepatocytes (Fig. 2C). Furthermore, both Arf-bp1 and Pam are required for the Rev-erbα protein turnover; depletion of either by siRNA was sufficient to stabilize Rev-erbα protein (Fig. S5), suggesting that these two E3 ligases might function together in a complex to degrade the Rev-erbα protein.
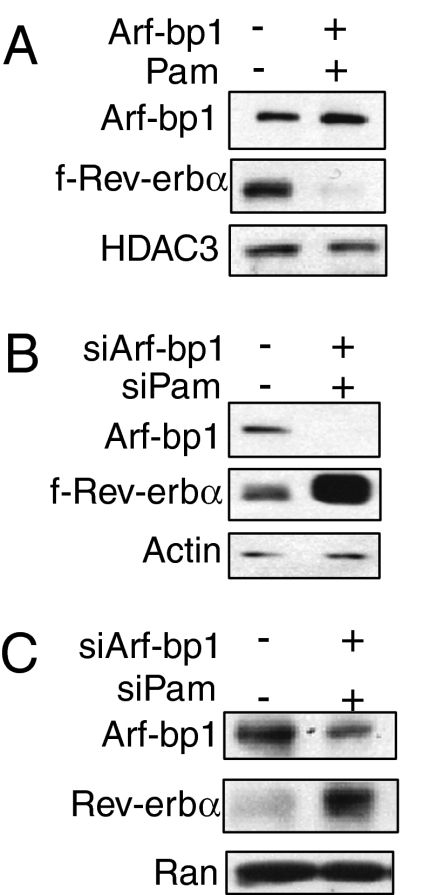
Fig. 2.
The E3 ligase proteins Arf-bp1 and Pam control Rev-erbα protein stability. (A) Effect of Arf-bp1 and Pam overexpression on f-Rev-erbα proteins. HFR cells were transfected with the empty control vector or the expression vectors of Arf-bp1 (2 μg) and Pam (2 μg). The levels of f-Rev-erbα proteins were determined by immunoblot analysis. (B) Effect of Arf-bp1 and Pam knockdown on f-Rev-erbα proteins. HFR cells were transfected with control siRNA or Arf-bp1 and Pam siRNA oligos (300 pmol/well). Cell lysates were analyzed by immunoblotting for the levels of Arf-bp1, f-Rev-erbα, and β-actin as a loading control. Knockdown efficiency for both Arf-bp1 and Pam were determined by RT-qPCR (Fig. S2). (C) Hepa1c1c-7 cells were transfected with the control or Arf-bp1 and PAM siRNA oligos (300 pmol/well) for 60 h before harvest. The endogenous Rev-erbα protein levels were examined by immunoblot analysis.
Arf-bp1 and Pam Promote Ubiquitination and Degradation of Rev-erbα.
An in vivo ubiquitination assay showed that Arf-bp1 and Pam promoted the formation of ubiquitinated Rev-erbα (Fig. 3A). The magnitude of the molecular weight shift suggested that this was polyubiquitination. Indeed, whereas both WT and a K63R mutant form of ubiquitin were conjugated to Rev-erbα, a K48R ubiquitin mutant defective in the formation of polyubiquitin chain (29–31) failed to show the same ubiquitin–Rev-erbα conjugates (Fig. 3B). The ubiquitination process was defective when either Arf-bp1 or Pam was expressed alone (Fig. 3C), indicating that both of these E3 ligases are required. Indeed, ubiquination appeared less when only one was overexpressed, suggesting that the ratio of Arf-bp1 and Pam is important as well. To address whether Arf-bp1 and Pam promote ubiquitination and proteasome degradation of Rev-erbα, we determined the half life of the f-Rev-erbα protein in the presence of the protein synthesis inhibitor cycloheximide. Depletion of both Arf-bp1 and Pam markedly prolonged the half-life of f-Rev-erbα protein (Fig. 3D).
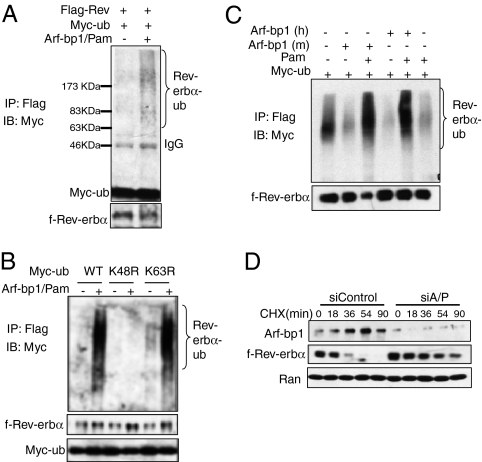
Fig. 3.
The E3 ligase proteins Arf-bp1 and Pam promote ubiquitination and degradation of Rev-erbα protein. (A) In vivo ubiquitination of f-Rev-erbα protein by Arf-bp1 and Pam. HFR cells were transfected with expression vectors encoding Myc-ubiquitin with either GFP or Arf-bp1 and Pam. The Rev-erbα-Myc-ubiquitin conjugates were detected by immunoblotting with anti-Myc antibody. (B) Arf-bp1 and Pam promotes polyubiquitination solely via lysine 48 linkage. Two ubiquitin mutants (K48R and K63R) were used along with the WT ubiquitin. All ubiquitin expression vectors were Myc-tagged, and anti-Myc antibody was used to detect Rev-erbα-myc-ubiquitin conjugates. (C) Either Arf-bp1 or Pam alone is not sufficient to promote ubiquitination of Rev-erbα. In this in vivo ubiquitination assay, the Arf-bp1 and Pam plasmids were added individually or together into the transfection mixture. (D) HFR cells were transfected with the control siRNA or siRNA oligos for Arf-bp1 and Pam for 60 h before being treated with cycloheximide (20 μg/mL) for the indicated times. The f-Rev-erbα protein levels were examined by immunoblot analysis.
Regulation of the Degradation of Rev-erbα by Arf-bp1 and Pam.
Because substrate ubiquitination is usually signal-dependent, and Rev-erbα is a component of the circadian clock network, we investigated the role of Arf-bp1 and Pam in the regulation of Rev-erbα protein stability by lithium or serum shock, a treatment aimed at synchronizing circadian rhythms in cultured cells. Depletion of Arf-bp1 and Pam protected Rev-erbα from lithium-induced degradation (Fig. 4A), consistent with the identification of these E3 ligases in the presence of lithium. Moreover, serum shock, which destabilizes Rev-erbα and induces circadian synchronization (21, 32), did not lead to Rev-erbα degradation when Arf-bp1 and Pam were depleted (Fig. 4B). To explore whether Arf-bp1 and Pam are also necessary for Rev-erbα protein oscillation during a circadian cycle, we monitored Rev-erbα protein levels in Hepa1C1C-7 cells for a period of 56 h after synchronization (Fig. 4C and Fig. S6). Depletion of both Arf-bp1 and Pam significantly affected the oscillation amplitude of Rev-erbα protein without changing its phase. Moreover, Rev-erbα protein oscillation was even more robust when both Arf-bp1 and Pam were knocked down, suggesting that those two proteins may normally play an inhibitory role in Rev-erbα protein oscillation during circadian rhythm.
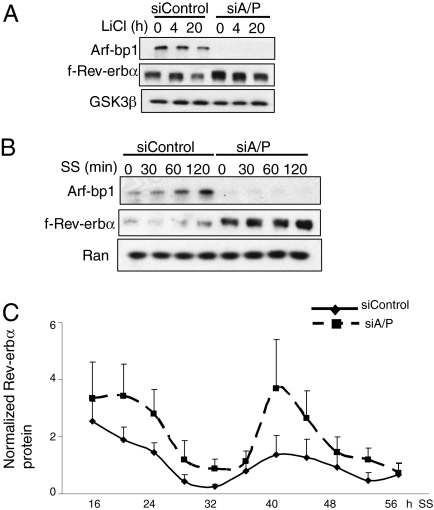
Fig. 4.
Arf-bp1– and Pam-mediated Rev-erbα degradation is circadian-relevant. (A) HFR cells were transfected with the control siRNA or siRNA oligos for Arf-bp1 and PAM for 60 h and then treated with lithium chloride (20 mM) for the indicated periods. The f-Rev-erbα protein levels were evaluated by immunoblot analysis. (B) HFR cells were transfected with the control siRNA or siRNA oligos for Arf-bp1 and PAM for 60 h and then synchronized with serum shock (50% horse serum diluted in DMEM) for the indicated periods. The f-Rev-erbα protein levels were evaluated by immunoblot analysis. (C) Effect of Arf-bp1 and Pam on Rev-erbα protein oscillation in synchronized Hepa1c1c-7 cells. Hepa1c1c-7 cells were transfected with either control or Arf-bp1 and Pam siRNA and synchronized by serum shock treatment. Protein levels of Rev-erbα were measured by immunoblot analysis and quantified by densitometry. Plotted data are mean ± SEM of three replicates.
Arf-bp1 and Pam Regulate Bmal1 Gene Expression.
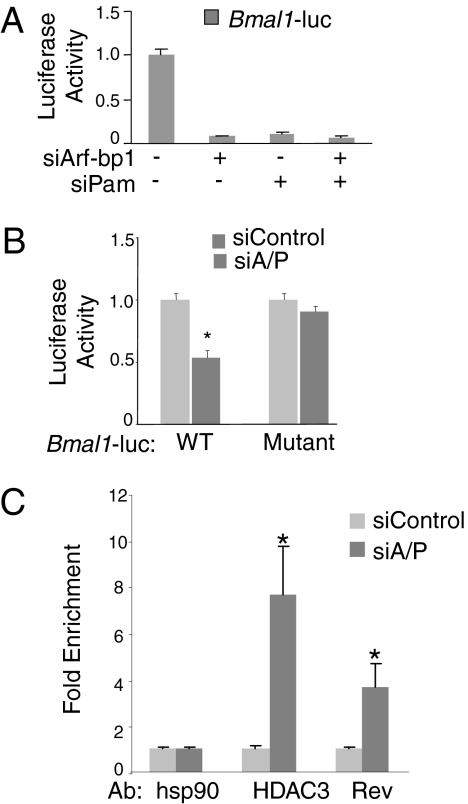
Rev-erbα directly binds and represses the Bmal1 gene, and mice lacking Rev-erbα exhibit dampened oscillation of Bmal1 (10). Therefore, we hypothesized that depletion of Arf-bp1 and Pam, which stabilizes Rev-erbα, would repress Bmal1 transcription. Indeed, depletion of either Pam or Arf-bp1 strongly suppressed the expression of a luciferase reporter gene under the control of the Bmal1 promoter (Fig. 5A). This repression required the Rev-erbα–binding sites; a non–Rev-erbα binding Bmal1 luciferase mutant lost its response to depletion of both Arf-bp1 and Pam (Fig. 5B). Furthermore, chromatin immunoprecipitation (ChIP) experiments demonstrated increased binding of Rev-erbα, along with its repression partner HDAC3, to the Bmal1 promoter when both E3 ligases were knocked down (Fig. 5C).
Fig. 5.
E3 ligases Arf-bp1 and Pam control Rev-erbα–mediated circadian function. (A) Repression of Bmal1 luciferase reporter (pGL4-Bmal1-luc) in 293T cells transfected with the control siRNA or siRNA oligos for Arf-bp1 or PAM alone, or both. Results are expressed as mean ± SD of at least three independent experiments. (B) Repression of either WT (pGL3-SV40-Bmal1-wt-luc) or mutant Bmal1 luciferase reporter (pGL3-SV40-Bmal1-ROREm-luc) in 293T cells transfected with either the control siRNA or siRNA oligos for both Arf-bp1 and Pam. Results are expressed as mean ± SD of at least three independent experiments. *P < 0.05. (C) ChIP for occupancy of Rev-erbα and HDAC3 on the Bmal1 promoter in 293T cells transfected with control siRNA or siRNA oligos for both Arf-bp1 and Pam. Antibodies against Rev-erbα (1:100) and HDAC3 (1:100) were used to immunoprecipitate the DNA-protein complex. Anti-HSP90 was used as a nonspecific antibody control. Data are plotted as mean ± SD of three replicates. *P < 0.05 by Student's t test. The results shown are representative of four separate experiments.
Arf-bp1 and Pam Modulate the Circadian Clock.
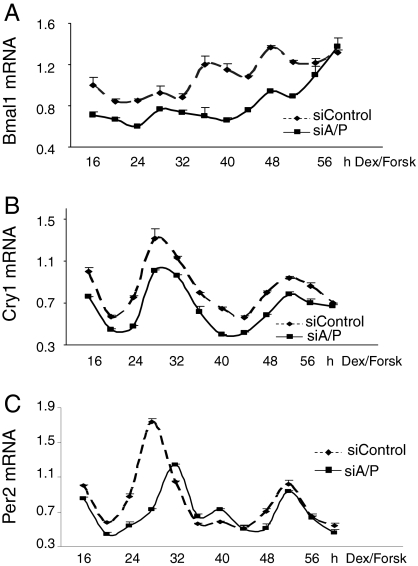
Because Arf-bp1 and Pam are involved in the Rev-erbα–mediated Bmal1 regulation, we sought to determine the role of Arf-bp1 and Pam in regulation of the cell-autonomous clock. Treatment of mouse hepa1c1c-7 liver cells with dexamethasome and forskolin induced circadian oscillation of the Bmal1 gene expression (Fig. 6A). Depletion of Arf-bp1 and Pam (Fig. S4) not only produced a decrease in the expression levels of Bmal1, but also dampened its oscillating amplitude (Fig. 6A). A recent study showed that mRNA oscillation of Bmal1 gene is not essential for core clock function (33). To test whether a persistent inhibition of the Bmal1 expression caused by dysregulated Rev-erbα protein could affect the BMAL1-dependent clock function, we examined the circadian oscillation of Cry1 and Per2 genes, which are both direct targets of the BMAL1/CLOCK complex. As expected, Cry1 oscillation was significantly repressed [P < 0.05, area under the curve (AUC)] on the depletion of Arf-bp1 and Pam, although its circadian period (τ) remained the same (Fig. 6B). In contrast, Per2 oscillation was repressed only within the first cycle, but fully recovered within the second cycle (Fig. 6C), suggesting that Per2 cycling is more resilient to inhibition of Bmal1 expression. Together, these data demonstrate that Arf-bp1 and Pam are novel endogenous modulators of circadian clock gene expression.
Fig. 6.
E3 ligases Arf-bp1 and Pam regulate the cell-autonomous circadian clock. Hepa1c1c-7 cells were transfected with either the control siRNA or siRNA oligos for both Arf-bp1 and Pam. Then, 48 h later, cells were treated with the synchronizing medium containing dexamethasone (100 nM) and forskolin (10 μM) and collected at the indicated times after synchronization. Knockdown of Arf-bp1 and Pam was confirmed by RT-qPCR (Fig. S7A). The Rev-erbα protein in those knockdown conditions was determined by Western blot analysis (Fig. S7B). The value for the amount of mRNA present at the 16 h time point was set at 1. Error bars represent ± range (n = 2). The mRNA levels of Bmal1 (A), Cry1 (B), and Per2 (C) are shown. P for Bmal1 AUC = 0.0073; P for Cry1 AUC = 0.044.
Discussion
Circadian rhythms are generated via an interlocked transcription-translational feedback loop driven by appropriate functions of clock proteins. Timely removal of the negative-feedback loop proteins is critical for the propagation of circadian cycles (13, 34). Our finding that Rev-erbα ubiquitination and protein stability are controlled by Arf-bp1 and Pam implicates these E3 ligases as novel regulators of the circadian network that mediates lithium-stimulated destabilization of Rev-erbα.
An essential role for ubiquitin E3 ligases in circadian clock regulation was initially demonstrated in Drosophila Slimb mutants (17, 18). In mammals, FBXL3, an F-box protein containing E3 ligase, targets CRY for degradation, and FBXL3 mutant mice display a 26-h circadian rhythm because of inappropriate stabilization of CRY protein within each circadian cycle (14–16). A recent study revealed a role of AMP-activated protein kinase–mediated phosphorylation in degrading CRY proteins during the circadian day (35), suggesting that degradation of clock proteins can be triggered by nutritional signals. GSK3β-mediated phosphorylation of circadian proteins, including CRY2 (24) and Rev-erbα (21), is particularly relevant in this context, where the inhibitory effect of lithium on GSK3β activity mimics endogenous signals that inhibit GSK3β, such as the Wnt-signaling pathway.
BMAL1/CLOCK-mediated transcription activation is necessary for the circadian oscillation of Rev-erbα mRNA (36), but until the present study, very little was known about the molecular pathways mediating protein oscillation of Rev-erbα. Interestingly, the ~30-min half-life of Rev-erbα protein appears to be less than that of CRY and PER proteins (14, 18, 20, 37). The finding that depletion of Arf-bp1 and Pam decreases the overall level of Bmal1 mRNA and negatively affects the amplitude of Cry1 and Per2 gene oscillations suggests that altered degradation of Rev-erbα protein is essential for normal clock function. Consistent with this, earlier work has shown that Bmal1 cycling is not essential for its circadian function, although the absolute level of Bmal1 is the rate-limiting factor for a functional clock (33). Our findings are also consistent with recent work suggesting that the amplitude of molecular clock is highly sensitive to disruption of the Rev-erbα/BMAL1 axis (38). However, knockdown of Arf-bp1 and Pam did not completely abolish the oscillation of Rev-erbα protein, suggesting that other mechanisms also contribute to Rev-erbα protein oscillation.
Arf-bp1 and Pam are each required for efficient ubiquitination and degradation of Rev-erbα. This suggests that both are involved in transferring the ubiquitin moiety to the Rev-erbα substrate via the formation of a functional E3 ligase complex. In this case, one E3 ligase might serve as a scaffold protein to assemble a highly efficient ubiquitination complex while the other catalyzes the reaction. An important question that remains to be addressed is how these two E3 ligases function together to promote ubiquitination of the Rev-erbα protein.
The repressive activity of Rev-erbα is regulated posttranslationally at several levels in addition to the degradation pathway reported here. The nuclear receptor corepressor (NCoR)–histone deacetylase 3 (HDAC3) complex is critically important for Rev-erbα repression (11), and its disruption alters cell-autonomous circadian rhythms (39). The interaction between Rev-erbα and the NCoR-HDAC3 complex is enhanced by heme, which functions as a ligand for Rev-erbα (40, 41). The NCoR-HDAC3 complex and heme both directly bind to Rev-erbα, underscoring the importance of the regulation of Rev-erbα protein levels by Arf-bp1 and Pam.
Heme synthesis also exhibits a circadian rhythm (42), which is modulated by the ability of Rev-erbα to repress the coactivator PGC-1α (43). In addition to regulating heme metabolism, Rev-erbα has been shown to regulate the metabolism of glucose, lipid, and bile acid metabolism, particularly in the liver (41, 44, 45). Thus, Rev-erbα serves as a link between metabolic and circadian pathways, and its distinct modes of posttranslational regulation, including the Arf-bp1/Pam–mediated degradation pathway targeted by lithium, might facilitate the fine-tuning of complex physiological processes.
Methods
Plasmids and Reagents.
pQCXIP-HA-f-Rev-erbα was generated using a standard subcloning strategy. pcDNA-Arf-bp1 was provided by Wei Gu (University of Columbia), and pCMX-Pam was provided by Vijaya Ramesh (Harvard University). Additional details are provided in SI Methods.
Generation of Stable Cell Lines.
A 293T-derived stable cell clone expressing HFR cells was generated for the purpose of large-scale protein purification. Here 293T cells were transiently transfected with pQCIPX-HA-FLAG (HF; control vector) or pQCIPX-HFR-f-Rev-erbα expression vector and grown in a selection medium containing puromycin (1.5 μg/mL) for 3 wk. The positive clones were confirmed by both RT-PCR and immunoblot analysis for the presence of transgene. Rev-erbα–expressing clones (HFR) were also tested for their response to lithium treatment and their repression functions on Rev-erbα target genes.
Purification of Rev-erbα from HFR Cells.
Both control (HF) and HFR cells were expanded and treated with lithium chloride (20 mM) and MG132 (2 μM) before harvest. Cell pellets were lysed in ice-cold Flag immunoprecipitation (IP) buffer [50 mM Tris-HCl (pH 7.5), 120 mM NaCl, 0.5 mM EDTA, 1% Triton X-100, and 1× protease inhibitor) for 30 min, after which cell lysates were centrifuged at 19,000 × g for 30 min. Then the supernatant was transferred into a fresh tube containing 100 μL of Flag M2 agarose beads preequilibrated with PBS. After overnight incubation at 4 °C with Flag M2 agarose beads, the immunoprecipitated complexes were further cleared by washing the beads at least five times with Flag IP buffer. Precipitates were eluted with 2× SDS loading buffer, boiled at 95 °C for 10 min to release protein complex, and separated on 4–20% SDS/PAGE gel for Coomassie blue staining (Pierce). Specific bands were cut out from the gel and subjected to mass spectrometry peptide sequencing. The presence of Arf-bp1 in the immunoprecipitated complex was also evaluated by SDS/PAGE gel and immunoblot analysis.
In Vivo Ubiquitination Assay.
HFR cells were cotransfected with pcDNA-Arf-bp1 and pCMV-Pam along with either WT pCMV-myc-ubiquitin or ubiquitin mutants (K48R or K63R). At 36 h after transfection, cells were treated with 20 mM lithium and 2 μM MG132 for 16 h and then lysed in Flag IP buffer. About 1 mg of protein lysates after centrifugation were used in immunoprecipitation assays with Flag M2 agarose beads. Immunocomplexes were resolved on 4–20% gradient gels, and immunoblotting was performed with anti-Myc antibody (Covance). About 5 μL of IP eluent was used for immunoblot analysis to evaluate the expression of f-Rev-erbα and Myc-ubiquitin.
Quantitative RT-PCR.
Total cellular RNA was prepared using the RNeasy Kit (Qiagen). cDNA was synthesized and subjected to the real-time PCR using SYBR Q-PCR mastermix on a Prism 7900 thermal cycler (Applied Biosystems). The value of each cDNA was calculated using the standard curve method and normalized to the value of the housekeeping gene control (arbp). The data were plotted as fold change. AUC analysis was used to calculate the difference of the circadian oscillation of target genes between control and treatment conditions. The primer sequences used in this study are listed in SI Materials and Methods.
Serum Shock and Synchronization Study.
Either 293T or Hepa1c1c-7 liver-derived cells were transfected with siRNA oligos before the synchronization study. In general, at 48 h after transfection, cells were incubated in 0.5% FBS–containing medium for 16 h and then synchronized by various treatments. For the experiment presented in Fig. 4 B and C, the cells were exposed to 50% horse serum diluted in DMEM for the indicated time and then harvested for protein analysis. For the experiment presented in Fig. 6, the cells were synchronized by a medium containing dexamethasome (100 nM) and forskolin (10 μM). Total RNA was extracted at the indicated time points and processed for RT-qPCR analysis using arbp as a normalization control.
Immunoblotting.
Western blot analysis was performed as described previously (21). Cell pellets were lysed in the ice-cold RIPA buffer supplemented with 1× protease inhibitor and 50 mM NaF and then incubated on ice for 20 min. The protein lysates were cleared by centrifugation at 20,800 × g at 4 °C for another 10 min. The resulting supernatants were collected and quantified using a BioRad protein assay kit. Blots also were probed with the following primary antibodies: anti-Flag (Sigma-Aldrich), anti–Rev-erbα and anti-GSK3β (Cell Signaling), anti-Ran (BD Bioscience), anti-HDAC3 (Santa Cruz Biotechnology), and anti–Arf-bp1 and anti-Myc (Covance). For the experiment shown in Fig. 4C and Fig. S6, the protein lysates were measured using with a bicinchoninic acid (BCA) protein assay (Pierce) with BSA as the standard. All of the samples were processed at the same time. For this, 30 μg of protein was loaded on a SDS-PAGE gel with 26 wells. The lowest-exposure film was then used to scan and to perform densitometry analysis with ImageJ software. The film was scanned in color and then converted to gray scale to subtract the background. The area of the box was maintained constant for all of the samples for a particular protein. The integrated density of each band was obtained and normalized with the respective integrated density of Ran.
ChIP Assay.
The ChIP assay was performed as described previously (19). First, 293T cells were transfected with either control siRNA or Arf-bp1/Pam siRNA mixture. Then, 72 h later, the cells were harvested for the ChIP assay. The sonicated chromatin materials were immunoprecipitated with the following antibodies at 4 °C overnight: anti-HDAC3, anti–acetyl histone H3 (Upstate Biotechnology), anti–Rev-erbα, and anti-Hsp90. The resulting DNA fragments were purified and subjected to PCR analysis using primers encompassing the RORE region of the human Bmal1 promoter. The PCR primer sequences were as follows: for hBmal1: forward, 5′-ttggtcagcggcctctcta-3′; reverse, 5′-cccggatacctgcctctt -3′; for h36B4: forward, 5′- acgctgctgaacatgctcaa-3′; reverse, 5′-gatgctgccattgtcgagca-3′.
Supplementary Material
Acknowledgments
We thank W. Gu (Columbia University, New York) for the Arf-bp1 expression vector, R. Ramesh (Harvard University, Cambridge, MA) for the PAM expression vector, C. X. Yuan (University of Pennsylvania proteomics core facility) for mass spectrometry peptide sequencing, and Z. D. Liu for assistance with the AUC analysis. This work was supported by National Institutes of Health Grants DK45586 and DK062434 (to M.A.L.) and Grant DK077449 (to L.Y) and the Penn Endocrinology and Diabetes Research Center (Grant DK19525).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000438107/-/DCSupplemental.
References
- 1.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levi F, Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 3.Schibler U. The 2008 Pittendrigh/Aschoff lecture: Peripheral phase coordination in the mammalian circadian timing system. J Biol Rhythms. 2009;24:3–15. doi: 10.1177/0748730408329383. [DOI] [PubMed] [Google Scholar]
- 4.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 5.Ptácek LJ, Jones CR, Fu YH. Novel insights from genetic and molecular characterization of the human clock. Cold Spring Harb Symp Quant Biol. 2007;72:273–277. doi: 10.1101/sqb.2007.72.017. [DOI] [PubMed] [Google Scholar]
- 6.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 7.Antoch MP, et al. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunger MK, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King DP, et al. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 11.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 12.Sehgal A, et al. Molecular analysis of sleep: wake cycles in Drosophila. Cold Spring Harb Symp Quant Biol. 2007;72:557–564. doi: 10.1101/sqb.2007.72.018. [DOI] [PubMed] [Google Scholar]
- 13.Virshup DM, Eide EJ, Forger DB, Gallego M, Harnish EV. Reversible protein phosphorylation regulates circadian rhythms. Cold Spring Harb Symp Quant Biol. 2007;72:413–420. doi: 10.1101/sqb.2007.72.048. [DOI] [PubMed] [Google Scholar]
- 14.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 15.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 16.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of Cryptochrome and Period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grima B, et al. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- 18.Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- 19.Reischl S, et al. Beta-TrCP1–mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007;22:375–386. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 20.Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls Clock-dependent transcription via casein kinase 1–dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 21.Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- 22.Gould TD, Manji HK. Glycogen synthase kinase-3: A putative molecular target for lithium mimetic drugs. Neuropsychopharmacology. 2005;30:1223–1237. doi: 10.1038/sj.npp.1300731. [DOI] [PubMed] [Google Scholar]
- 23.Iwahana E, et al. Effect of lithium on the circadian rhythms of locomotor activity and glycogen synthase kinase-3 protein expression in the mouse suprachiasmatic nuclei. Eur J Neurosci. 2004;19:2281–2287. doi: 10.1111/j.0953-816X.2004.03322.x. [DOI] [PubMed] [Google Scholar]
- 24.Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. Ser-557–phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J Biol Chem. 2005;280:31714–31721. doi: 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- 25.Chen D, et al. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 26.Warr MR, et al. BH3-ligand regulates access of MCL-1 to its E3 ligase. FEBS Lett. 2005;579:5603–5608. doi: 10.1016/j.febslet.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han S, et al. Pam (protein associated with Myc) functions as an E3 ubiquitin ligase and regulates TSC/mTOR signaling. Cell Signal. 2008;20:1084–1091. doi: 10.1016/j.cellsig.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Ye Y. Polyubiquitin chains: Functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: The apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 33.Liu AC, et al. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanselow K, Kramer A. Role of phosphorylation in the mammalian circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:167–176. doi: 10.1101/sqb.2007.72.036. [DOI] [PubMed] [Google Scholar]
- 35.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siepka SM, Yoo SH, Park J, Lee C, Takahashi JS. Genetics and neurobiology of circadian clocks in mammals. Cold Spring Harb Symp Quant Biol. 2007;72:251–259. doi: 10.1101/sqb.2007.72.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang EE, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alenghat T, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raghuram S, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin L, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 42.Rogers PM, Ying L, Burris TP. Relationship between circadian oscillations of Rev-erbalpha expression and intracellular levels of its ligand, heme. Biochem Biophys Res Commun. 2008;368:955–958. doi: 10.1016/j.bbrc.2008.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu N, Yin L, Hanniman EA, Joshi S, Lazar MA. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbalpha. Genes Dev. 2009;23:2201–2209. doi: 10.1101/gad.1825809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duez H, Staels B. The nuclear receptors Rev-erbs and RORs integrate circadian rhythms and metabolism. Diab Vasc Dis Res. 2008;5:82–88. doi: 10.3132/dvdr.2008.0014. [DOI] [PubMed] [Google Scholar]
- 45.Duez H, et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.