Abstract
Most current assays of HIV antiviral resistance are based on either sequencing of viral genes (genotypic assays) or amplification and insertion of these genes into standardized virus backbones and culture. These latter are called phenotypic assays. But the only generally accepted phenotypic assay is based upon culture of intact patient virus, performed in phytohemagglutinin-activated peripheral blood mononuclear cells (PHA blasts) in the presence of differing drug concentrations. However, PHA blast culture is difficult and not always reproducible. Therefore we have sought cell lines that may produce more predictable results, yet faithfully mirror results in PHA blasts. We have compared 10 different cell lines for receptor and coreceptor expression, growth of laboratory-adapted strains of HIV, growth by direct inoculation of PBMC from infected patients, and in assays of antiviral drug effects. One of these cell lines, C8166-R5, is statistically not inferior to CD8-depleted PHA blasts for culturing HIV from the peripheral blood cells of patients. The effective concentrations of antiviral drugs of all classes were similar when assayed in C8166-R5 or PHA blasts. Known drug-resistant isolates grown in C8166-R5 demonstrated the predicted effects. We followed a patient longitudinally and demonstrated that resistance testing in C8166-R5 was predictive of clinical outcome. These experiments represent the first steps in developing a clinically useful phenotypic drug resistance assay based upon culturing the patient's own virus.
Introduction
The rise of drug-resistant HIV has led to the use of resistance testing in the clinical management of HIV-infected individuals.1,2 Currently used tests are described as either genotypic or phenotypic. Genotyping refers to obtaining the sequence of the genes encoding the targets of therapy: polymerase, protease, and envelope. These sequences are compared to libraries of sequences from isolates of known drug sensitivity and resistance profile, and predictions are made.3–5 Phenotyping is more complex. Supposedly this assays functional virus in the patient at the time the sample was obtained. The only accepted phenotypic drug resistance assay that studies patient-derived infectious HIV is cultivation in phytohemagglutinin-activated peripheral blood mononuclear cells (PHA blasts), utilizing an ACTG-approved protocol.6 Because this assay is difficult, tedious, and hard to standardize, other experimentally simpler assays have been developed and are offered commercially.7,8 But these assays do not start with infectious virus; rather they use viral nucleic acids as their connection to the patient, and insert target genes into well-characterized constructs, whose drug susceptibility is then tested.
The number of infectious particles in a sample of HIV is greatly outnumbered by the copies of viral RNA and pseudovirions that are present. This is especially so in samples directly obtained from patients, in whom as few as 0.01% of nucleic acid copies may be associated with infectivity.9 Therefore drug resistance testing based upon nucleic acid amplification may not reflect the most important population of virus in the patient, the virions capable of infecting other cells. In addition, virus genes that are not examined may influence the rates of replication, transcription, or other infective process that might be missed if only a portion of the virus genome is analyzed.10 Minor populations of virus (<10%) may be missed with sequence-based techniques.
To address the development of a clinically useful antiviral assay based upon the infectious virus present in patients, rather than viral nucleic acids, we have investigated the use of cell lines to replace PHA blasts. Although there are various cell lines that have been used to cultivate HIV, we have chosen a subset of these cells and expressed CCR5 in those that did not do so naturally. We have compared the growth of laboratory and patient-derived virus in the different cell lines and in PHA blasts. We then utilized the best of these cell lines in a series of antiviral resistance assays to show that they provided results similar to those obtained with PHA blast cultures. The goal of this study was to demonstrate that cell lines yield results equivalent to those obtained with PHA blasts, not necessarily to compare all cell lines that have been used to culture HIV. If cell lines can substitute for PHA blasts, the resistance testing of patient-derived infectious HIV may become feasible. Ultimately clinical trials will define whether resistance testing based upon patient-derived infectious virus or amplified nucleic acids yields more relevant answers for patient management.
Materials and Methods
Cells
H9 and PM-1 cells were the gift of Dr. M. Reitz, then at NCI, Bethesda, MD.11,12 1B4 is a subclone of PM-1 chosen for high CCR5 expression, as measured with antibody 2D7 by flow cytometry (see below). H1-J.C53 are HeLa cells transduced to express high levels of CD4 and CCR5; they naturally express CXCR4.13 MT2-R5, MT4-R5, C8166-R5, CEM-R5, VB-R5, and ST-R5 were produced by expressing CCR5 in the CD4+ T cell lines MT2, MT4, C8166, CEMx174, VB, and SupT1, respectively (AIDS Research and Reference Reagent Program, ARRRP, Rockville, MD) with a CCR5-encoding vector pBABE-puro-CCR5,14 a gift of David Dorsky to James Robinson. Following transduction, cells positive for CCR5 were detected by their ability to form syncytia with cells infected with vaccinia expressing ADA gp160, a functional R5 viral envelope (ARRRP). Cells were cloned by limiting dilution. Clones competent for syncytium formation with R5 viruses were maintained in RPMI with 10% fetal calf serum (FCS) and 1 μg/ml puromycin (Sigma Chemical, St. Louis, MO). CEMx174 cells expressing CCR5 have been described previously.15
Peripheral blood cells were density gradient (LSM Lymphocyte Separation Medium, MP Biomedical, Solon, OH) purified. To make PHA blasts, they were cultured at 2 × 106 cells/ml in RPMI 1640 medium plus 10% FCS in the presence of 10 μg/ml PHA. After 2 days the medium was changed without the addition of fresh PHA, and the following day interleukin-2 (IL-2) (ARRRP) was added at a concentration of 200 U/ml. Fresh medium and IL-2 were added every 3–4 days. In many cases, the PHA blasts were made using cells depleted of CD8+ cells by magnetic separation (Dynal Biotech LLC, Invitrogen, Carlsbad, CA) prior to activation.
Flow cytometry
The expression of HIV receptors and coreceptors on the surface of infected cells was studied using indirect immunofluorescence and flow cytometry, as described elsewhere.16,17 Cells were stained with monoclonal antibodies (mAbs) (obtained from ARRRP): Sim.2 (anti-CD4), 2D7 (anti-CCR5), and 12G5 (anti-CXCR4) at 10 μg/ml. Binding was detected with fluorescein isothyocyanate (FITC)-conjugated antimouse Ig (Zymed, South San Francisco, CA). Two thousand cells were analyzed using a FACSstar flow cytometer (Becton Dickinson, Mountain View, CA).
Virus stocks
Virus stocks were grown in PHA blast cultures, aliquoted, and frozen at −70°C until use. Two laboratory strains were used, the X4-tropic molecularly cloned virus NL4-318 and the R5-tropic isolate Ba-L.19 Two clinical isolates from drug-naive patients were also used: 208K8, R5 tropic and 208K10, X4 tropic.20 Drug-resistant virus isolates and their parental drug-sensitive strains were obtained from the ARRRP: AO18 (pre-AZT and post-AZT), AO12 (pre-AZT and post-AZT),21 1495-2 (AZT intermediate),22–24 HIVRTMC,24 xxHIV-LAI-M184V (3TC-resistant),25 RF, RF/V82F/I84V, RF/L-323-12-3, RF/L-323-9-1,26,27 and HIV-1 L10R/M46I/L63P/V82T/I84V.28
Antiretrovirals
Antiviral drugs were obtained in an uncompounded form either from ARRRP—didanosine (ddI), abacavir succinate (ABC), tenofovir disoproxil fumarate (TDF), and lopinavir (LPV), or directly from the manufacturer—nevirapine (NVP) (Boehringer Ingelheim Pharmaceuticals, Inc. Ridgefield, CT), delavirdine (DLV) (Pharmacia & Upjohn, Kalamazoo, MI), efavirenz (EFV) (DuPont Merck Pharmaceutical Company, Willmington, DE), zidovudine (AZT), zalcitabine (ddC), stavudine (d4T) (Sigma Chemical, St. Louis, MO), lamivudine (3TC), amprenavir (APV) (Glaxo Wellcome Inc., Research Triangle Park, NC), saquinavir (SQV) (Hoffmann-La Roche Inc., Nutley, NJ), indinavir (IDV) (Merck & Co., Inc., Whitehouse Station, NJ), ritonavir (RTV) (Abbott, Abbott Park, IL), and nelfinavir (NFV) (Agouron Pharmaceuticals, San Diego, CA).
Measurement of HIV infection
We used an Ag capture enzyme-linked immunosorbent assay (ELISA) for the measurement of the HIV core Ag, p24. The sensitivity of this assay is 5 pg/ml, and experimental details are provided elsewhere.29 Wells were coated with the capture anti-p24 mAb, 183-H12-5C. An appropriate dilution of tissue culture medium treated with 1% Triton X-100 (Sigma) was then added and incubated overnight at 4°C. Following washing, the plates were incubated with biotin-conjugated HIV-immune globulin (ARRRP) as the detecting Ab and then with Amdex-streptavidin HRP (Amersham Pharmacia Biotech, Piscataway, NJ). Signal was detected with the colorimetric HRP substrate tetramethylbenzidine (Sigma) and measured at A450. All samples were run in duplicate or triplicate.
Patients
Patients were recruited from the Medical Center of Louisiana at New Orleans HIV Outpatient (HOP) Clinic using IRB-approved protocols and informed consent. Patients received remuneration for participating. Patients were selected on the basis of plasma viral load greater than > 10,000 copies/ml using the HIV-1 COBAS Amplicor Monitor Assay (Roche Diagnostic Systems, Branchburg, NJ) with a lower limit quantitation of 400 RNA copies/ml. Patients had a viral load assay the visit before the sample was taken or coincident with the sample. Viral load ranged from 27,300 to >750,000 copies/ml. Two patients had two or more samples, often with a therapeutic alteration intervening. One patient, designated 054, was assayed four times over 12 months. On this patient, viral load was performed seven times over the same interval, and commercial resistance testing using a sequence-based system (Trugene HIV-1 Genotyping Kit, Bayer HealthCare LLC, Tarrytown, NY) performed three times (Tulane Retrovirology Laboratory, New Orleans, LA). A total of 57 patients were used in the studies reported here.
Results
Comparison of cell lines for cell-surface expression of receptors and coreceptors and for susceptibility to HIV infection
It has previously been shown in HeLa cells transduced to express CD4 and CCR5 that susceptibility to HIV infection is most closely associated with the level of CD4 expression, provided there is a threshold level of the appropriate corecptor.13,30 To determine the suitability of cells to detect patient-derived isolates of both R5 and X4 tropism, we examined a panel of cell lines for binding by monoclonal antibodies to the virus receptor CD4 and to the coreceptors CXCR4 and CCR5 (Table 1). The mean fluorescent intensity of anti-CD4 ranged from a low of 43.5 on H9 cells to a high of 251.4 on C8166-R5. Expression of both coreceptors, CXCR4 and CCR5, is also high on C8166-R5.
Table 1.
Cell Surface Ag Expression on Different Cell Linesa
| Cell line | Secondary Ab only | Anti-CD4 | Anti-CCR5 | Anti-CXCR4 |
|---|---|---|---|---|
| H9 | 3.6 | 43.5 | 10.6 | 321.5 |
| PM1 | 3.6 | 121.0 | 12.7 | 201.6 |
| IB4 | 7.0 | 68.9 | 69.9 | 113.7 |
| H1-J.C53 | 3.8 | 64.8 | 166.3 | 19.8 |
| MT2-R5 | 3.0 | 66.7 | 42.1 | 160.7 |
| MT4-R5 | 4.3 | 109.5 | 32.1 | 41.4 |
| C8166-R5 | 5.1 | 251.4 | 52.3 | 161.1 |
| CEM-R5 | 5.0 | 61.4 | 53.0 | 29.2 |
| VB-R5 | 3.5 | 238.1 | 25.2 | 42.1 |
| ST-R5 | 4.1 | 245.8 | 131.3 | 117.8 |
Cell surface expression of CD4 receptor and CCR5 and CXCR4 coreceptor on cell lines was measured using indirect immunofluorescence and flow cytometry. Results are reported as mean fluorescence of cells gated by appropriate side and forward scatter.
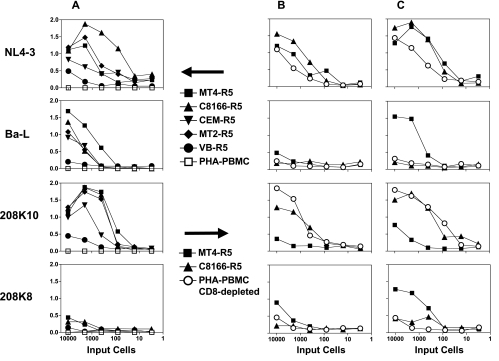
As we envision a cell-line-based phenotypic drug resistance assay, we propose that patient-derived PHA blasts would be coincubated with the indicator cell lines. To mimic this, we first infected PHA blasts with cell-free virus from both laboratory and clinical isolates. Then graded numbers of infected PHA blasts were added to the uninfected indicator cell lines and p24 was measured 3 or 5 days later (Fig. 1). In Fig. 1A different cell lines were compared to PHA blasts that did not have CD8+ cells removed. The production HIV p24 was detected in most cell lines with all virus isolates 3days after infection, but not in PHA blasts. Both C8166-R5 and MT4-R5 have excellent cell growth characteristics in tissue culture when compared to the others (not shown), and produce measurable levels of p24 within 3 days. Therefore we used these cell lines in a second experiment (Fig. 1B and C) and compared them to CD8-depleted PHA blasts. On both 3 and 5 days postinoculation, each of the cell lines became infected and produced p24 at a rate that was greater than or equal to that seen with the PHA blasts. PM-1 cells were tested in these studies and were shown to have infectivity at levels comparable to C8166-R5 and MT4-R5 (data not shown).
FIG. 1.
Spread of HIV-1 infection from HIV-infected PHA blasts to indicator cells. PHA blasts were infected with HIV-1 isolates: molecularly cloned laboratory strains: Ba-L (R5) and NL4-3 (X4), and clinical isolates: 208K8 (R5) and 208K10 (X4). Three days postinfection, blasts were added to uninfected indicator cells: 104 cells from cell lines or 4 × 105 PHA blasts. The number of cells from the infected culture is indicated on the horizontal axis. Infected and uninfected cells were incubated for 3 days (A and B) or 5 days (C) and then supernatant p24 was measured (optical density is shown on the vertical axis).
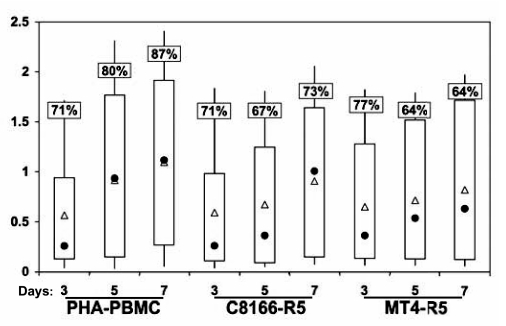
We next tested the ability of the cell lines to detect patient-derived virus, and compared the results to those obtained with CD8-depleted PHA blasts. Patients were selected for study on the basis of moderate to high viral load (>10,000), similar to those expected in the most likely patient population that might be undergoing intensive resistance testing, i.e., patients who are not doing well under their current therapeutic regimen. As a preliminary experiment, all cell lines tested in Fig. 1, along with PM-1 and 1B4, were compared to PHA blasts in cultivating virus from 10 different patients, at times ranging from 2 to 9 days postcultivation. Only C8166-R5 and the MT4-R5 became infected in a manner comparable to the PHA blasts. Therefore only these cells were used in further studies. PHA blasts from 17 different patients were activated for 3 days and then added to indicator cells; production of p24 was measured 3, 5, and 7 days later. Figure 2 shows the results. Two different outcomes were measured: percentage of patients in whom HIV can be detected and optical density obtained on p24 ELISA. Not unexpectedly, CD8-depleted PHA blasts appear to set the standard for cultivating virus derived from patients.
FIG. 2.
Detection of HIV-1 in microcultures after cultivation of PHA-PBMC from infected patients with primary cells and cell lines. A total of 5 × 105 PHA-stimulated, CD8-depleted PBMCs from HIV-1-infected patients were cocultured with 5 × 105 cells from the cell lines or 2 × 106 PHA-stimulated, CD8-depleted PBMCs from a seronegative donor for 7 days. During this time supernatant p24 was measured by ELISA. Results are expressed as optical density obtained in ELISA. For each patient, duplicate experimental wells were assayed in triplicate by p24 ELISA. The percents in the boxes indicate the percentage of patient cultures in which HIV infection could be detected, as defined by p24 levels 2 SD greater than plate mean background. Open triangles represent the mean value of all positive cultures and filled circles represent the median. The top of the thick bar is the 75% percentile and the bottom is the 25% percentile. The lines demonstrate the full range of values.
To determine if cell lines may be substituted for PHA blasts, we used statistical tests of noninferiority. We made comparisons between the cell lines and the blast culture both according to the log-transformed optical densities on p24 ELISA and according to virus growth (Table 2). For the log-transformed optical density measure, mixed effects models were used to generate one-sided 95% confidence intervals for the lower limit of the difference in p24 production in the two cell types; the antilog of the lower limit reflects the lower 95% limit of the ratio of the geometric means.31 Comparisons of the proportion of cultures exhibiting virus growth were performed via the Mantel–Haenszel strategy;32 using repeated measures to analyze the relative risk, one-sided 95% confidence intervals for the ratio of the proportion of samples exhibiting growth for the two treatments were calculated. Virus growth in the cell line, as compared to growth in the blast culture, is expected to be at least the percentage indicated by the lower bound (with 95% confidence). The results indicate that C8166-R5 cells are not inferior to both un-depleted and CD8-depleted PHA blasts.
Table 2.
Comparison of Cell Lines to PHA Blasts for Cultivation of Patient-Derived HIV
| |
C8166-R5 |
MT4-R5 |
||||||
|---|---|---|---|---|---|---|---|---|
| Day 3 | Day5 | Day7 | Overall | Day 3 | Day5 | Day7 | Overall | |
| PBMC (not depleted of CD8+ cells) | ||||||||
| Ratio of geometric mean optical densities (cell line vs. blast culture) | ||||||||
| Observed ratio | 1.02 | 0.78 | 0.80 | 0.87 | 1.10 | 0.58 | 0.33 | 0.58a |
| Lower bound of one-sided 95% CI | 0.74 | 0.47 | 0.46 | 0.69 | 0.72 | 0.29 | 0.16 | 0.44a |
| Ratio of proportion of samples exhibiting virus growth (cell line vs. blast culture) | ||||||||
| Observed ratio | 1.11 | 0.91 | 1.20 | 1.07 | 0.81 | 0.56 | 0.50 | 0.62 |
| Lower bound of one-sided 95% CI | 0.75 | 0.60 | 0.89 | 0.85 | 0.43 | 0.31 | 0.25 | 0.44 |
| PBMC (CD8 not depleted) | ||||||||
| Ratio of geometric mean optical densities (cell line vs. blast culture) | ||||||||
| Observed ratio | 1.03 | 0.74 | 0.79 | 0.85 | 1.26 | 0.90 | 0.66 | 0.93a |
| Lower bound of one-sided 95% CI | 0.77 | 0.52 | 0.50 | 0.70 | 0.88 | 0.54 | 0.41 | 0.74a |
| Ratio of proportion of samples exhibiting virus growth (cell line vs. blast culture) | ||||||||
| Observed ratio | 1.00 | 0.83 | 0.85 | 0.89 | 1.09 | 0.80 | 0.73 | 0.87 |
| Lower bound of one-sided 95% CI | 0.76 | 0.67 | 0.64 | 0.77 | 0.76 | 0.52 | 0.49 | 0.70 |
The relative growth of HIV (as measured by optical density on p24 ELISA) in MT4-R5 cells, compared to the blast culture, decrease significantly over time (p = 0.0048 compared to the unseparated blasts; p = 0.0446 compared to CD8-depleted blasts).
To determine whether the ability to cultivate virus correlates with in vivo physiology, we sought correlations with plasma viral load and CD4 counts (Table 3). Spearman's rank correlation coefficient was used to assess the direction and strength of the associations between ELISA optical density on different days and the patients' CD4 counts and viral loads at the time of sample collection. Surprisingly, HIV growth in C8166-R5 correlates better with patient plasma viral load and CD4 count than growth in CD8-depleted PHA blasts.
Table 3.
Correlation of HIV Cultivation in Different Cells with CD4 Count and Viral Load in Patientsa
| |
Correlation (p-value) between optical density and CD4 count |
Correlation (p-value) between optical density and VL |
||||
|---|---|---|---|---|---|---|
| Cell type | Day 3 | Day 5 | Day 7 | Day 3 | Day 5 | Day 7 |
| Blast (CD8 depleted) | −0.46 (0.0639) | −0.44 (0.995) | −0.33 (0.2260) | 0.61 (0.0098) | 0.52 (0.0462) | 0.47 (0.0761) |
| C8166-R5 | −0.53 (0.0295) | −0.60 (0.0175) | −0.79 (0.0005) | 0.70 (0.0017) | 0.64 (0.0103) | 0.64 (0.0097) |
| MT4-R5 | −0.31 (0.3050) | −0.55 (0.0818) | −0.52 (0.0981) | 0.68 (0.0112) | 0.75 (0.0085) | 0.67 (0.0233) |
Optical density of p24 ELISA obtained on the indicated day of culture was correlated with either patient CD4 count or plasma viral load (VL). Spearman's rank correlation coefficient and p-value are shown; n varies from 11 to 17 per comparison.
Measurement of effects of antiviral agents
The antiviral effects observed in a functional assay of HIV infectivity are dependent upon drug uptake, metabolism, and inactivation by the indicator cells. Therefore it is crucial for us to demonstrate that the same effects with antiviral drugs are observed in the indicator cell lines and the PHA blasts. In an initial assay of this, we infected PHA blasts with four different virus isolates. Infected blasts were then mixed with indicator cells in the presence of different concentrations of antiviral drugs. The results (Table 4) show that antiviral drugs have similar, although not identical, effects in the conventional cultures (CD8-depleted blasts) and in both MT4-R5 and C8166-R5 cell lines.
Table 4.
IC50 Concentration of Antiviral Drugs in Different Cells against Four Different Drug-Sensitive HIV Isolates
| |
|
HIV-1 Strain |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
Ba-L |
NL4-3 |
208K10 |
208K8 |
||||||||
| |
|
Cell type |
|||||||||||
| Drug | Dose rangea(μM) | PHA-blasts | C8166-Rt | MT4-R5 | PHA-blasts | C8166-Rt | MT4-R5 | PHA-blasts | C8166-Rt | MT4-R5 | PHA-blasts | C8166-Rt | MT4-R5 |
| NRTI | |||||||||||||
| AZT | 0.08–10 | <0.08b | 2 | NTc | <0.08 | <0.08 | <0.08 | <0.08 | <0.08 | <0.08 | >10 | <0.08 | NT |
| d4T | 0.08–10 | 0.4 | 2 | NT | 0.4 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.08 | 0.4 | 2 |
| ddI | 0.08–10 | 10 | 10 | NT | 10 | 2 | 0.4 | 2 | 0.4 | 0.08 | 10 | 2 | 10 |
| ddC | 0.008–1 | 0.04 | 0.008 | NT | 0.2 | 0.2 | 0.2 | 0.008 | 0.04 | 0.008 | 0.2 | 0.2 | 0.2 |
| 3TC | 0.04–5 | 0.2 | 0.04 | NT | 0.2 | 0.2 | 0.2 | 0.04 | 0.04 | 0.2 | 0.04 | 0.2 | 1 |
| ABC | 0.5–62.5 | 2.5 | 12.5 | NT | 12.5 | 62.5 | NT | 12.5 | 62.5 | NT | 12.5 | 2.5 | NT |
| NNRTI | |||||||||||||
| DLV | 0.001–0.180 | 0.180 | 0.007 | 0.0070 | 0.001 | 0.007 | 0.04 | 0.007 | 0.007 | 0.001 | 0.007 | 0.001 | 0.001 |
| EFV | 0.003–0.317 | 0.0025 | 0.0025 | 0.0025 | 0.0025 | 0.0025 | 0.0025 | 0.0025 | 0.0025 | 0.0025 | 0.0025 | 0.0025 | 0.0127 |
| NVP | 0.003–0.376 | 0.015 | 0.376 | 0.3760 | 0.075 | 0.003 | 0.376 | 0.075 | 0.015 | 0.003 | 0.376 | 0.003 | 0.075 |
| PI | |||||||||||||
| IDV | 0.001–0.140 | 0.140 | 0.028 | NT | 0.006 | 0.028 | 0.028 | 0.028 | 0.140 | 0.028 | 0.028 | 0.006 | NT |
| NFV | 0.001–0.15 | 0.03 | 0.03 | NT | 0.006 | 0.001 | 0.006 | 0.151 | 0.151 | 0.006 | 0.03 | 0.006 | 0.03 |
| RTV | 0.002–0.28 | 0.277 | 0.277 | NT | 0.011 | 0.011 | 0.277 | 0.277 | 0.055 | 0.055 | 0.055 | 0.055 | 0.055 |
| SQV | 0.001–0.13 | 0.026 | 0.026 | 0.0260 | 0.005 | 0.026 | 0.026 | 0.026 | 0.026 | 0.001 | 0.005 | 0.005 | 0.005 |
| APV | 0.001–0.16 | 0.001 | 0.03 | 0.007 | 0.007 | 0.03 | 0.007 | 0.007 | 0.007 | 0.007 | 0.03 | 0.03 | 0.03 |
Dose range indicates the range of concentrations of antiviral drugs tested.
Inhibitory drug concentration (IC50) is defined as the lowest drug concentration tested giving 50% or greater inhibition of p24 production compared to drug-free control wells.
NT, not tested.
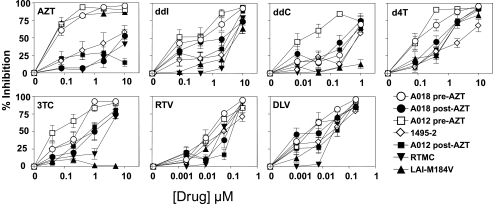
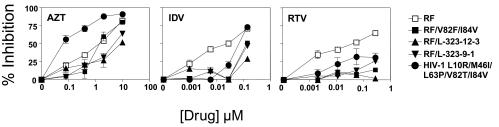
We next tested the ability of the cell lines to detect the presence of drug resistance, using known antiviral-resistant isolates that were defined in conventional PHA blast cultures. Figure 3 shows results obtained with C8166-R5 cells with reverse transcriptase inhibitors (RTIs) and virus isolates, obtained from ARRRP, with known resistance patterns to RTIs.21–25 When tested against AZT, the expected resistances were seen: post-treatment isolates (AO18 and AO12) were orders of magnitude more resistant than pre-treatment isolates, and isolate 1495-2 had intermediate status, as had been reported. The RTMC were resistant to AZT, and also resistant to ddI, 3TC, and delaviridine. Although the M184V strain was sensitive to AZT, it was resistant to ddI, ddC, and especially 3TC. The post-treatment AZT-resistant isolates had intermediate levels of resistance to other RTIs. Figure 4 shows a similar analysis for mutations known to cause resistance to protease inhibitors (PIs). The isolates26–28 were tested against the PIs indinivir and ritonavir, and against AZT, the latter as a control for virus growth. The one virus not derived from the RF isolate was the only isolate with differing resistance to AZT. Compared to the “wild-type” isolate RF, all of the mutants showed high level resistance to ritonavir at all doses tested, and at all but the highest dose of indinivir. These results demonstrate the ability of C8166-R5 indicator cells to identify known drug-resistant isolates.
FIG. 3.
Effect of anti-HIV drugs on RTI-resistant isolates. C8166-R5 cells were infected with viruses having known sensitivity and resistance to RTI drugs. A total of 400 infected cells were mixed with 104 uninfected C8166-R5 and cultured in the presence of the indicated drug at 5-fold dilutions. Supernatant was collected for p24 ELISA. Results are expressed as the percent inhibition compared to cultures in the absence of drug. Viruses A018 and A012 post-AZT have been defined as AZT resistant (the pre-AZT isolates are sensitive) and 1495-2 has been described as having intermediate resistance to AZT.
FIG. 4.
Effect of anti-HIV drugs on PI-resistant isolates. C8166-R5 cells were infected with HIV containing protease mutations. Once infection was established, 400 infected cells were mixed with 104 uninfected C8166-R5 cells and cultured in the presence of the indicated drug at 5-fold dilutions. Supernatant was collected for p24 ELISA. Results are expressed as the percent inhibition compared to cultures in the absence of drug.
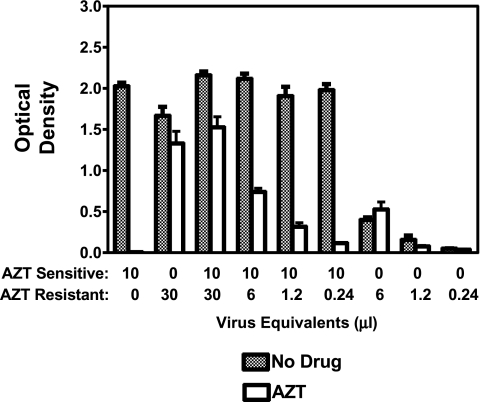
We have proposed that a phenotypic drug resistance assay may have greater sensitivity for detecting minority variants than sequence-based assays, which fail to detect drug-resistant variants in less than 10% of the population.10 To test this, we mixed various amounts of AZT-resistant HIV with drug-sensitive virus, and measured the effect of AZT on virus growth in C8166-R5 cells (Fig. 5). In a 7-day assay, we were able to detect drug-resistant HIV representing less than 1% of the population of infectious HIV.
FIG. 5.
AZT treatment of pC8166 cells infected with the mixture of AZT-sensitive and AZT-resistant isolates. AZT-sensitive (A018 pre-AZT) and AZT-resistant (A018 post-AZT) cultures were premixed. The amount of virus to add to obtain the defined ratios was based upon preliminary titration results for each isolate. C8166-R5 cells (106 in 500 μl) were infected with cell-free virus for 3 h. Following the infection, cells were washed and set up at 105 in 2-ml wells in triplicate. One day postinfection 0 or 5 μM AZT was added to the infected culture. Supernatant p24 was measured on day 7. Optical density at 450 nm is shown on the vertical axis.
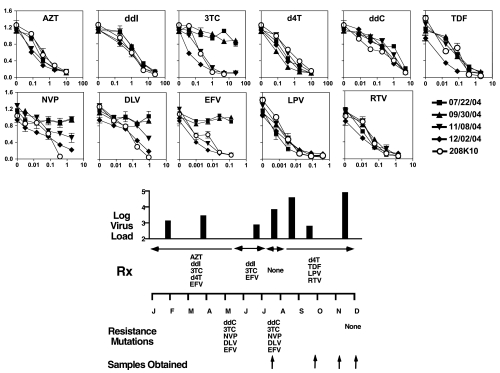
Having demonstrated that C8166-R5 cells have characteristics substantially similar to CD8-depleted PHA blasts for phenotypic drug resistance testing, we next used them to measure the sensitivity and resistance to drugs in patient samples. Patients were usually undergoing alterations in therapy at the time our samples were drawn. We tested drugs that were either used to treat the patients or that would be newly initiated. The patient whose results are shown in Fig. 6 was followed serially, had multiple viral load assays to assess the success of the therapy, and underwent genotypic drug resistance testing. Samples taken in May and September show multiple drug-resistant genotypes and poorly controlled viremia. Phenotypic drug resistance testing confirms the resistance to 3TC, nevirapine, delaverdine, and efavirenz present in July and September. Both genotypic and phenotypic testing demonstrate that these drug-resistant populations disappeared following a therapeutic holiday. Despite surprising concordance between the two types of resistance testing, there was a discrepancy for ddC, with the genotype predicting resistance, but not the phenotype. Since the patient did not receive ddC, we do not know which is correct. Neither assay explains the failure of therapy in December.
FIG. 6.
Antiretroviral phenotypic susceptibility testing in a patient showing drug resistance. CD8-depleted PHA blasts from serial samples obtained from a single patient were cultured for 5–8 days. Then 5 × 105 blasts were mixed with 2 × 105 uninfected C8166-R5 in 2-ml wells. HIV-1 isolate 208-K10, a clinical isolate obtained prior to the use of antiretroviral therapy, is a drug-sensitive control. Once infection was established, as measured by detection of the viral p24 antigen, drug-susceptibility testing was performed by incubating 4 × 102 infected pC8166 cells with 104 uninfected C8166-R5 cells in the presence or absence of the indicated drugs. Supernatant was collected on day 3 for p24 ELISA. The patient's viral load, drug resistance mutations determined by Trugene HIV-1 Genotyping, and treatment are shown in the lower portion of the figure.
Discussion
Antiviral resistance testing has become a standard of care for treating HIV infection, particularly for those whose viremia cannot be controlled by standard antiviral regimens. Multiple different assays exist and it is not yet known which assay best predicts patient response to therapy.2,10,33–37 Genotypic assays rely upon gene sequence and comparison to known resistance mutations. Because sequencing is usually performed on the predominant population, minority populations of resistant virus may be missed.10 Under drug selection, such populations can emerge as dominant. Molecular methods to detect minority populations do exist, but require sequencing multiple clones or amplicons, selective hybridization, or gel analyses, such as denaturing gradient gel electropheresis. Such analyses are not used in routine clinical care. Thus, genotypic assays are believed to be good at demonstrating existing drug-resistance, but may have less predictive value.10 In an attempt to improve predictability, the “virtual phenotype” assay compares genotypic sequences derived from patients to databases of virus sequences whose drug response and clinical outcome are known.37–39 These informatic analyses are sophisticated and evolving, but suffer all the biological limitations of any sequence-based assay.
Phenotypic assays culture patient-derived HIV in the presence of antiviral drugs and measure the ability of the drugs to suppress virus growth. The argument for superiority of these assays is that they test infectious HIV, the virus that is propagating and spreading within the patient. These assays are difficult to standardize, because of differences in growth rates of patient-derived virus and the need for primary cells, PHA blasts, in the most accepted form of this assay.6 The blast cells are tedious to culture, difficult to obtain in large homogeneous batches, and grow slowly.
To avoid these problems, the commercially available phenotypic assays express patient-derived amplicons in a framework of an HIV molecular clone whose growth kinetics are well established.3–5,37,40 This assay is simpler and more reproducible than standard phenotypic assays, and does have the ability to detect minor subpopulations, giving greater predictive value. But because this method is based on using amplified nucleic acids, rather than infectious virus present in the patient, it loses some important theoretical advantages. The result may be influenced by the presence of pseudovirions that do not encode functional HIV.9 Replication and pathogenicity may be influenced by virus genes outside of the amplified sequences. These factors define the importance of a population of virus in a patient, and yet are deliberately eliminated from study when testing by amplification-based phenotypic assays.
A “true” phenotypic assay is one in which patient-derived virus is cultivated in the presence of different concentrations of antiviral drugs. The need for PHA blast cultures, the long period of cultivation necessary to obtain results, and the large amount of patient-to-patient variability in rates of virus replication all combine to limit the clinical development of such an assay. The use of cell lines that faithfully culture patient-derived virus could obviate the first two difficulties. Such cells would likely express high levels of the HIV receptor CD4 and coreceptors CXCR4 and CCR5.13,30 Because it is well known that the method of cultivation of HIV can influence the virus that emerges,41,42 there is hesitation to accept the clinical utility of results derived from cell lines rather than from primary lymphocyte cultures. Other concerns are that cell lines might metabolize antiviral agents differently than primary cells, or that the virus functions that are the target of antiviral activity may differ when virus is grown in different cell types. To propose the substitution of a particular cell line for primary cells, it is critical to demonstrate the equivalency of the cells for these functions. The studies reported here are the first steps in the process of developing useful cell lines for this function.
We began by comparing nine different cell lines for receptor and coreceptor expression (Table 1), susceptibility to infection with virus isolates maintained in our laboratory (Fig. 1), and tissue culture characteristics. Based upon these criteria, we selected two cell lines, each expressing CD4 and CXCR4 naturally and CCR5 by transfection. Using well-characterized virus stocks we found that comparative rates of virus replication were the same in both these cell lines as in PHA blasts (data not shown). These two cell lines were then compared to PHA blasts, and then CD8-depleted PHA blasts, for the ability to culture patient-derived HIV within 1 week. The results indicated that the cell line C8166-R5 was not inferior to CD8-depleted PHA blasts in the ability to cultivate patient-derived HIV (Fig. 2, Table 2). The results of HIV cultivation in C8166-R5 cells correlated with patient CD4 counts and viral loads as well, or better, than those in CD8-depleted blast cells (Table 3). The cell lines were then compared with PHA blast cultures for their ability to demonstrate effects of antiviral drugs on standardized HIV isolates and were found to be equivalent (Table 4). In Figs. 3 and 4, the C8166-R5 cells were shown to be able to detect known resistant virus and to detect cross-resistance. The utility of C8166-R5 cells in detecting small populations of resistant virus (Fig. 5) and in following a single patient (Fig. 6) was then demonstrated. Thus by many of the parameters we deem important, C8166-R5 functions for assay purposes in a manner indistinguishable from CD8-depleted PHA blasts. The C8166-R5 cells are easy to maintain and have a doubling time of <16 h. In regard to the key characteristics tested here, the C8166-R5 cells are stable in tissue culture for long periods of time when maintained under puromycin selection.
A variety of different cell lines have been used to cultivate HIV. The purpose of these studies is to demonstrate the possibility that cell lines may substitute for PHA blasts in the determination of antiviral susceptibility from patient-derived HIV. It was not our purpose to perform an exhaustive analysis to compare every available cell line and identify the best. Our results demonstrate that C8166-R5 cells appear to be equivalent to PHA blasts; other cell lines not compared in these studies may function as well, or even better, than C8166-R5.
The studies reported here are only the first that are necessary to determine if C8166-R5 can substitute for PHA blasts in phenotypic resistance testing, and whether the cell line can be adapted to a clinically useful assay. It will next be essential to compare the cell line and primary cells in a number of patients, and determine whether each yields the same data. If so, and other characteristics are amenable to scale up, it would ultimately be necessary to perform clinical trials in which patient management using genotypic, expression-based phenotypic, and true phenotypic assays is compared. Although we may tout theoretical or commercial advantages of one assay over the other, it is only through carefully designed clinical trials that the most appropriate tests of drug resistance can be determined. Such an assay should allow the physician to choose the antiretroviral drugs that will provide the best long-term control of viremia for the patient.
Acknowledgments
We would like to thank Marcel Charbonnet, Emily Moran, and Megan Weydert for expert technical assistance. This work was supported by NIH Grant CA83756 (S.H.P.) and by the Research Institute for Children.
References
- 1.Hirsch MS. Conway B. D'Aquila RT. Johnson VA. Brun-Vezinet F. Clotet B. Demeter LM. Hammer SM. Jacobsen DM. Kuritzkes DR. Loveday C. Mellors JW. Vella V. Richman DD. Antiretroviral drug resistance testing in adults with HIV infection: Implications for clinical management. International AIDS Society-USA Panel. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 2.Hirsch MS. Brun-Vezinet F. Clotet B. Conway B. Kuritzkes DR. D'Aquila RT. Demeter LM. Hammer SM. Johnson VA. Loveday C. Mellors JW. Jacobsen DM. Richman DD. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA Panel. Clin Infect Dis. 2003;37:113–128. doi: 10.1086/375597. [DOI] [PubMed] [Google Scholar]
- 3.Petropoulos CJ. Parkin NT. Limoli KL. Lie YS. Wrin T. Huang W. Tian H. Smith D. Winslow GA. Capon DJ. Whitcomb JM. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkin NT. Hellmann NS. Whitcomb JM. Kiss L. Chappey C. Petropoulos CJ. Natural variation of drug susceptibility in wild-type human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2004;48:437–443. doi: 10.1128/AAC.48.2.437-443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkin NT. Gupta S. Chappey C. Petropoulos CJ. The K101P and K103R/V179D mutations in human immunodeficiency virus type 1 reverse transcriptase confer resistance to nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother. 2006;50:351–354. doi: 10.1128/AAC.50.1.351-354.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Japour AJ. Mayers DL. Johnson VA. Kuritzkes DR. Beckett LA. Arduino JM. Lane J. Black RJ. Reichelderfer PS. D'Aquila RT. Crumpacker CS. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. The RV-43 Study Group, the AIDS Clinical Trials Group Virology Committee Resistance Working Group. Antimicrob Agents Chemother. 1993;37:1095–1101. doi: 10.1128/aac.37.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertogs K. de Bethune MP. Miller V. Ivens T. Schel P. Van Cauwenberge A. Van Den Eynde C. Van Gerwen V. Azijn H. Van Houtte M. Peeters F. Staszewski S. Conant M. Bloor S. Kemp S. Larder B. Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deeks SG. Hellmann NS. Grant RM. Parkin NT. Petropoulos CJ. Becker M. Symonds W. Chesney M. Volberding PA. Novel four-drug salvage treatment regimens after failure of a human immunodeficiency virus type 1 protease inhibitor-containing regimen: Antiviral activity and correlation of baseline phenotypic drug susceptibility with virologic outcome. J Infect Dis. 1999;179:1375–1381. doi: 10.1086/314775. [DOI] [PubMed] [Google Scholar]
- 9.Piatak M., Jr Saag MS. Yang LC. Clark SJ. Kappes JC. Luk KC. Hahn BH. Shaw GM. Lifson JD. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 10.Shafer RW. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin Microbiol Rev. 2002;15:247–277. doi: 10.1128/CMR.15.2.247-277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lusso P. Cocchi F. Balotta C. Markham PD. Louie A. Farci P. Pal R. Gallo RC. Reitz MS. Growth of macrophage-tropic and primary HIV-1 isolates in a unique CD4+ T-cell clone (PM1): Failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Folks T. Benn S. Rabson A. Theodore T. Hoggan MD. Martin M. Lightfoote M. Sell K. Characterization of a continuous T-cell line susceptible to the cytopathic effects of the acquired immunodeficiency syndrome (AIDS)-associated retrovirus. Proc Natl Acad Sci USA. 1985;82:4539–4543. doi: 10.1073/pnas.82.13.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Platt EJ. Wehrly K. Kuhmann SR. Chesebro B. Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophage-tropic isolates of HIV-1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorsky DI. Harrington RD. An indicator cell assay for T-cell tropic, macrophage-tropic, and primary isolates of HIV-1 based on green fluorescent protein. J Acquir Immune Defic Syndr. 1999;22:213–220. doi: 10.1097/00126334-199911010-00001. [DOI] [PubMed] [Google Scholar]
- 15.Ellis BR. Munene E. Elliott D. Robinson J. Otsyula MG. Michael SF. Seroprevalence of simian immunodeficiency virus in wild and captive born Sykes' monkeys (Cerco-pithecus mitis) in Kenya. Retrovirology. 2004;1:34. doi: 10.1186/1742-4690-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pincus SH. Messer KG. Cole R. Ireland R. VanCott TC. Pinter A. Schwartz DH. Graham BS. Gorse GJ. Vaccine-specific antibody responses induced by HIV-1 envelope subunit vaccines. J Immunol. 1997;158:3511–3520. [PubMed] [Google Scholar]
- 17.Pincus SH. Wehrly K. Cole R. Fang H. Lewis GK. McClure J. Conley AJ. Wahren B. Posner MR. Notkins AL. Tilley SA. Pinter A. Eiden L. Teintze M. Dorward D. Tolstikov VV. In vitro effects of anti-HIV immunotoxins directed against multiple epitopes on the HIV-1 envelope glycoprotein gp160. AIDS Res Hum Retroviruses. 1996;12:1041–1051. doi: 10.1089/aid.1996.12.1041. [DOI] [PubMed] [Google Scholar]
- 18.Adachi A. Gendelman HE. Koenig S. Folks T. Willey R. Rabson A. Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and non-human cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gartner S. Markovits P. Markovitz DM. Kaplan MH. Gallo RC. Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 20.Chesebro B. Wehrly K. Nishio J. Perryman S. Macrophage-tropic HIV isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: Definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larder BA. Darby G. Richman DD. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 22.Gingeras TR. Prodanovich P. Latimer T. Guatelli JC. Richman DD. Barringer KJ. Use of self-sustained sequence replication amplification reaction to analyze and detect mutations in zidovudine-resistant human immunodeficiency virus. J Infect Dis. 1991;164:1066–1074. doi: 10.1093/infdis/164.6.1066. [DOI] [PubMed] [Google Scholar]
- 23.Richman DD. Guatelli JC. Grimes J. Tsiatis A. Gingeras T. Detection of mutations associated with zidovudine resistance in human immunodeficiency virus by use of the polymerase chain reaction. J Infect Dis. 1991;164:1075–1081. doi: 10.1093/infdis/164.6.1075. [DOI] [PubMed] [Google Scholar]
- 24.Larder BA. Kemp SD. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 25.Schinazi RF. Lloyd RM., Jr Nguyen MH. Cannon DL. McMillan A. Ilksoy N. Chu CK. Liotta DC. Bazmi HZ. Mellors JW. Characterization of human immunodeficiency viruses resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto MJ. Garber S. Winslow DL. Reid CD. Aldrich P. Jadhav PK. Patterson CE. Hodge CN. Cheng YS. In vitro isolation and identification of human immunodeficiency virus (HIV) variants with reduced sensitivity to C-2 symmetrical inhibitors of HIV type 1 protease. Proc Natl Acad Sci USA. 1993;15:7543–7547. doi: 10.1073/pnas.90.16.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King RW. Winslow DL. Garber S. Scarnati HT. Bachelor L. Stack S. Otto MJ. Identification of a clinical isolate of HIV-1 with an isoleucine at position 82 of the protease which retains susceptibility to protease inhibitors. Antiviral Res. 1995;28:13–24. doi: 10.1016/0166-3542(95)00033-i. [DOI] [PubMed] [Google Scholar]
- 28.Condra JH. Schleif WA. Blahy OM. Gabryelski LJ. Graham DJ. Quintero JC. Rhodes A. Squires KE. Deutsch PJ. Emini EA. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 29.Pincus SH. Marcotte TK. Forsyth BM. Fang H. In vivo testing of anti-HIV immunotoxins. In: W. A. Hall., editor. Immunotoxin Methods and Protocols. Humana Press; Totowa, NJ: 2001. pp. 277–294. [DOI] [PubMed] [Google Scholar]
- 30.Kabat D. Kozak SL. Wehrly K. Chesebro B. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of HIV. J Virol. 1994;68:2570–2577. doi: 10.1128/jvi.68.4.2570-2577.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Center for Drug Evaluation Research. Food and Drug Administration; Rockville, MD: 2001. Guidance for industry: Statistical approaches to establishing bioequivalence. [Google Scholar]
- 32.Stokes ME. Davis CS. Koch GG. Categorical Data Analysis Using the SAS System. SAS Institute Inc.; Cary, NC: 1995. [Google Scholar]
- 33.Siliciano RF. Scientific rationale for antiretroviral therapy in 2005: Viral reservoirs and resistance evolution. Top HIV Med. 2005;13:96–100. [PubMed] [Google Scholar]
- 34.Clavel F. Hance AJ. HIV drug resistance. N Engl J Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 35.Gallant JE. Antiretroviral drug resistance and resistance testing. Top HIV Med. 2005;13:138–142. [PubMed] [Google Scholar]
- 36.Hammer SM. Saag MS. Schechter M. Montaner JS. Schooley RT. Jacobsen DM. Thompson MA. Carpenter CC. Fischl MA. Gazzard BG. Gatell JM. Hirsch MS. Katzenstein DA. Richman DD. Vella S. Yeni PG. Volberding PA. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society-USA panel. JAMA. 2006;296:827–843. doi: 10.1001/jama.296.7.827. [DOI] [PubMed] [Google Scholar]
- 37.Haubric RH. Resistance and replication capacity assays: Clinical utility and interpretation. Topics HIV Med. 2004;12:52–56. [PubMed] [Google Scholar]
- 38.Parkin NT. Chappey C. Petropoulos CJ. Improving lopinavir genotype algorithm through phenotype correlations: Novel mutation patterns and amprenavir cross-resistance. AIDS. 2003;17:955–961. doi: 10.1097/00002030-200305020-00003. [DOI] [PubMed] [Google Scholar]
- 39.Ross L. Boulme R. Fisher R. Hernandez J. Florance A. Schmit JC. Williams V. A direct comparison of drug susceptibility to HIV type 1 from antiretroviral experienced subjects as assessed by the antivirogram and PhenoSense assays and by seven resistance algorithms. AIDS Res Hum Retroviruses. 2005;21:933–939. doi: 10.1089/aid.2005.21.933. [DOI] [PubMed] [Google Scholar]
- 40.Harrigan PR. Montaner JS. Wegner SA. Verbiest W. Miller V. Wood R. Larder BA. World-wide variation in HIV-1 phenotypic susceptibility in untreated individuals: Biologically relevant values for resistance testing. AIDS. 2001;15:1671–1677. doi: 10.1097/00002030-200109070-00010. [DOI] [PubMed] [Google Scholar]
- 41.Sawyer LSW. Wrin WT. Crawford-Miksza L. Potts B. Wu Y. Weber PA. Alfonso RD. Hanson CV. Neutralization sensitivity of HIV-1 is determined in part by the cell in which the virus is propagated. J Virol. 1994;68:1342–1349. doi: 10.1128/jvi.68.3.1342-1349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang YJ. Fredriksson R. McKeating JA. Fenyö EM. Passage of HIV-1 molecular clones into different cell lines confers differential sensitivity to neutralization. Virology. 1997;238:254–264. doi: 10.1006/viro.1997.8812. [DOI] [PubMed] [Google Scholar]