Abstract
Condensation and segregation of mitotic chromosomes is a critical process for cellular propagation, and, in mammals, mitotic errors can contribute to the pathogenesis of cancer. In this report, we demonstrate that the retinoblastoma protein (pRB), a well-known regulator of progression through the G1 phase of the cell cycle, plays a critical role in mitotic chromosome condensation that is independent of G1-to-S-phase regulation. Using gene targeted mutant mice, we studied this aspect of pRB function in isolation, and demonstrate that it is an essential part of pRB-mediated tumor suppression. Cancer-prone Trp53−/− mice succumb to more aggressive forms of cancer when pRB's ability to condense chromosomes is compromised. Furthermore, we demonstrate that defective mitotic chromosome structure caused by mutant pRB accelerates loss of heterozygosity, leading to earlier tumor formation in Trp53+/− mice. These data reveal a new mechanism of tumor suppression, facilitated by pRB, in which genome stability is maintained by proper condensation of mitotic chromosomes.
Keywords: Cohesion, condensation, chromosome instability, cell cycle, chromatin
The compaction of the mammalian genome into mitotic chromosomes, and their faithful segregation to recipient daughter cells in mitosis, is a critical event for eukaryotic cells. This stage of the cell division cycle carries considerable risk, as there is no opportunity to reverse the effects of missegregated or damaged chromosomes in daughter cells. For these reasons, understanding mitosis is of vital importance, because changes in genome integrity can lead to cancer.
The processes of chromosome condensation and segregation are intimately linked, as insufficient condensation of chromosome arms can prevent their proper separation in anaphase (Belmont 2006). Of chief importance in facilitating proper chromosome structure is the mitotic chromosome scaffold, upon which mitotic chromosomes are condensed. These scaffolds are composed largely of topoisomerase 2 and condensins (Maeshima and Laemmli 2003). While the enzymatic activity of topoisomerases suggests a mechanism by which they participate in chromosome compaction, the precise role of condensins has been less clear. There are two types of condensin complexes (I and II) that are very similar structurally. Each is comprised of an SMC2 and SMC4 subunit that interact to form the coiled-coil arms of their ring-like structures (Losada and Hirano 2005). In addition, each condensin complex also contains distinct subunits: CAP-H, CAP-D2, and CAP-G in condensin I, and CAP-H2, CAP-D3, and CAP-G2 in condensin II (Losada and Hirano 2005). Surprisingly, depletion of individual components of these complexes by RNAi does not prevent outright chromosome condensation (Ono et al. 2003; Hirota et al. 2004). Instead, each complex appears to offer a unique contribution to the architecture of mitotic chromosomes. This may be due in part to differences in the timing of their loading onto chromosomes, with Condensin II being present on chromatin in interphase, and Condensin I being loaded only after nuclear envelope breakdown (Ono et al. 2003; Hirota et al. 2004). Additionally, differences in subunit composition imply that they may have different functional or regulatory properties (Losada and Hirano 2005). Importantly, defects and delays in chromatid condensation are manifested as lagging chromosomes during anaphase that impede mitotic progression, resulting in aneuploidy (Hirota et al. 2004; Ono et al. 2004; Samoshkin et al. 2009). However, there is limited evidence connecting condensin proteins and chromosome condensation to cancer (Ham et al. 2007; Lapointe et al. 2008).
In addition to condensation defects leading to segregation errors, faithful chromosome segregation can also be affected by mitotic spindle abnormalities. This includes the mitotic spindle checkpoint, which serves to detect unattached kinetochores at metaphase (Nasmyth 2005; Musacchio and Salmon 2007). Signals that emanate from a complex containing MAD2 and BUBR1 at unattached kinetochores prevent the E3 ubiquitin ligase complex APC/C from triggering the degradation of cyclin B1 and securin. Once this checkpoint is satisfied, mitotic cyclin-dependent kinase activity drops, and a securin-associated protease called separase is free to cleave cohesins, allowing sister chromatid separation (Nasmyth 2005; Musacchio and Salmon 2007). Experimental models in which spindle assembly checkpoint components are misexpressed show errors in chromosome segregation (Pei and Melmed 1997; Hernando et al. 2004; Vader and Lens 2008). Thus, defects in either chromosome condensation or spindle assembly checkpoints can lead to segregation errors and aneuploidy, and potentially contribute to cancer pathogenesis. A number of examples of spindle assembly checkpoint defects contributing to cancer incidence in genetically modified mice have been reported (for review, see Schvartzman et al. 2010). Conversely, even though defects in chromosome condensation cause similar mitotic errors, there are no reports using gene targeted mouse models to confirm a role for condensins as tumor suppressors.
The retinoblastoma tumor suppressor protein (pRB) is best known for its role in regulating the G1-to-S-phase transition early in the cell cycle (for review, see Burkhart and Sage 2008). Its ability to negatively regulate transcription of DNA replication machinery through E2F transcription factors creates a mechanism by which it can inhibit cell cycle entry. More recently, pRB has been shown to influence both chromosome condensation and mitotic checkpoint function (Longworth and Dyson 2010; Schvartzman et al. 2010). Components of the spindle assembly checkpoint, such as MAD2, are regulated by E2F transcription factors (Ren et al. 2002; Hernando et al. 2004). For this reason, loss of E2F regulation by pRB, which is almost ubiquitous in cancer, leads to genome instability (Mayhew et al. 2007; Schvartzman et al. 2010). Conversely, the ability of pRB to influence mitotic chromosome condensation has emerged as an E2F-independent function, and loss of pRB function can influence chromosome loss irrespective of proliferation (Zheng et al. 2002; Gonzalo et al. 2005; Isaac et al. 2006; Longworth et al. 2008). To date, evidence for pRB's involvement in chromosome condensation has been largely genetic. Mouse fibroblasts deficient for all pRB family proteins or a knock-in mutation that partially inactivates pRB have abnormal centromeric heterochromatin, leading to chromosome fusions and aneuploidy (Gonzalo et al. 2005; Isaac et al. 2006). Further analyses that combine the use of Drosophila genetics and mammalian cell culture suggest that pRB can interact with the condensin II subunit CAP-D3, and that this interaction is necessary for chromosome compaction in mitosis (Longworth et al. 2008). While studies of spindle checkpoint components such as MAD2 have offered explanations for why their deregulation in cancer can be traced back to pRB function (Schvartzman et al. 2010), the importance of pRB's role in chromosome condensation is less well understood, and has not yet been connected to a cancer phenotype.
In this study, we investigate the mechanism used by pRB to facilitate mitotic chromosome condensation. We rely on a viable, gene targeted mouse strain in which pRB is mutated to block LXCXE-dependent interactions, such as those with viral oncoproteins and chromatin remodeling enzymes like histone deacetylases (Isaac et al. 2006). Cells from these mice have limited proliferative control defects, except for the responsiveness to transforming growth factor β (TGF-β) and senescence-inducing stimuli (Francis et al. 2009; Talluri et al. 2010). We demonstrate that pRB interacts with the Condensin II complex to establish proper chromosome structure. Our experiments reveal that condensation defects caused by a deficiency in pRB–LXCXE interactions occur before metaphase, and are unrelated to the ability to regulate G1-to-S-phase progression. We used Rb1ΔL/ΔL; Trp53−/− mice as well as Trp53−/− controls—both of which are uniformly defective in their response to G1 arrest stimuli such as DNA damage- and oncogene-induced senescence—to study pRB's role in chromosome condensation in isolation. Rb1ΔL/ΔL; Trp53−/− mice succumb to much more aggressive forms of cancer than p53-deficient controls, and their tumors are characterized by elevated levels of chromosome instability. Furthermore, we demonstrate that defective chromosome condensation caused by mutant pRB can accelerate loss of heterozygosity and cancer onset in Trp53+/− mice. This study reveals that participation in mitotic chromosome condensation is an integral aspect of pRB's function as a tumor suppressor protein.
Results
Aberrant chromosome condensation and segregation in Rb1ΔL/ΔL mutant cells
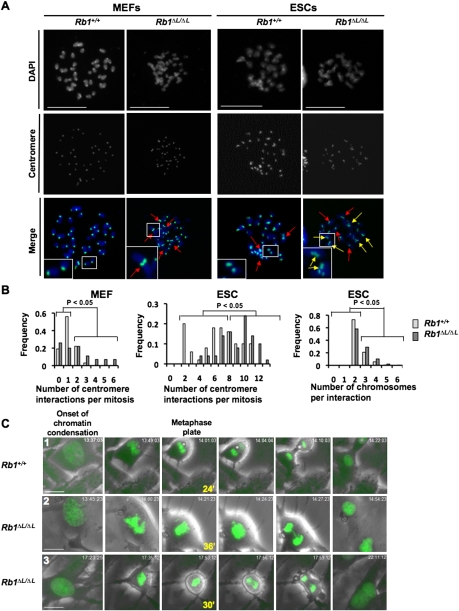
A number of recent reports have indicated that cells deficient for pRB family proteins have chromosomal abnormalities (Gonzalo et al. 2005; Longworth et al. 2008), and we demonstrated centromere fusions and lagging anaphase chromosomes in cells from Rb1ΔL/ΔL mice (Isaac et al. 2006). One interpretation of altered chromosome numbers in these cells is that they are a consequence of deregulated E2F transcriptional control in G1 leading to inappropriate proliferation. In this way, alterations in transcriptional control or commitment to enter S phase early in the cell cycle are manifested in subsequent mitotic errors. To investigate this possibility with Rb1ΔL/ΔL mutant cells, we compared chromosomal abnormalities found in homozygous mouse embryonic fibroblasts (MEFs) with embryonic stem cells (ESCs). Since MEFs have a pronounced G1 phase regulated by pRB (Herrera et al. 1996; Harrington et al. 1998), and ESCs lack the ability to arrest in G1 (Aladjem et al. 1998), we reasoned that chromosomal abnormalities found in both are unlikely to be a consequence of unregulated cell cycle advancement. Figure 1A shows representative chromosome spreads from each cell type for both wild-type and Rb1ΔL/ΔL mutants. Chromosomes were stained with DAPI and a major satellite DNA probe to visualize contacts between centromeres from different chromosomes. In both cell types, a statistically significant increase in centromere interactions (Fig. 1A, inlays) was detected in the mutant Rb1ΔL/ΔL genotype, suggesting an increase in chromosome fusions (χ2 test, P < 0.05 for each comparison in Fig. 1B). Interestingly, there was also an increase in the number of centromeres interacting in Rb1ΔL/ΔL ESCs compared with wild type (χ2 test, P < 0.05) (Fig. 1A [inlay], B [right]). In addition to its contribution to centromere structure, pRB is also known to silence transcription at nearby rDNA repeats (Cavanaugh et al. 1995; Hannan et al. 2000; Ciarmatori et al. 2001). Therefore, we also investigated their involvement in these fusions, as they are found on the p-arms of mouse chromosomes 12, 15, 16, 17, 18, and 19, and are therefore in close proximity to the centromere (Supplemental Fig. 1). These chromosomes are not overrepresented in Rb1ΔL/ΔL fusion events, further suggesting that loss of transcriptional regulation early in the cell cycle does not contribute to this phenotype (Supplemental Fig. 1). These data support a specific role for pRB in regulating chromatin structure at the centromeric repeat sequences of mitotic chromosomes that is independent of pRB's ability to regulate the G1-to-S-phase transition in these cells.
Figure 1.
Rb1ΔL/ΔL cells display aberrant chromosome condensation and segregation. (A) Metaphase chromosome spreads were prepared from MEFs or ESCs of the indicated genotypes. Chromosomes were stained with DAPI (blue) and a probe for major satellite repeats (green) to mark centromeres. Red arrows indicate contact between centromeres from different chromosomes, and yellow arrows indicate centromere contact involving three or more chromosomes. Inlays highlight expanded views of select chromosomes. Bars, 25 μm. (B) The frequency of centromere interactions per mitosis is plotted for each genotype (Rb1ΔL/ΔL and Rb1+/+) and cell type (MEF and ESC). (Right graph) In addition, the number of centromeres involved in each interaction was determined for ESC metaphase spreads, and is displayed as the frequency of multiple chromosome interactions. (C) Video microscopy was performed on MEFs expressing an H2B-GFP reporter by capturing phase-contrast and GFP images every 3 min over a 15-h time course. The images shown begin with the onset of chromatin condensation in prophase as the left-most panel. The last image of the metaphase plate before the onset of anaphase is indicated along with the elapsed time since the onset of prophase (in minutes). The right-most image shows resolved daughter (or binucleated) cells. The numbers in the left-most image correspond to references in the Supplemental Material and Supplemental Movies. Bars, 50 μm.
In order to better understand the origin of defective chromosomal compaction in Rb1ΔL/ΔL mutant cells and its effect on mitosis, we established a video microscopy assay to observe cell division in MEFs. Early passage MEFs were transduced with an H2B-GFP-expressing retrovirus. This lead to equivalent expression of the H2B-GFP fusion in both genotypes. Importantly, expression of this fusion protein was very low and did not lead to a detectable increase in total H2B levels in these cells (Supplemental Fig. 2A). H2B-GFP-expressing cells were monitored microscopically under phase-contrast and fluorescent optics to visualize chromosome condensation, metaphase alignment, anaphase, and cytokinesis. Images from three representative movies are shown in Figure 1C. Typically, mutant cells took longer to progress from the onset of chromosome condensation to the point at which the metaphase plate is most tightly aligned (Table 1). Furthermore, the metaphase plate in mutant mitoses was less compact than wild type (Table 1). To determine whether these phenotypes are associated with defects later in mitosis, we observed sister chromatid separation in anaphase, and found that lagging chromosomes occurred more frequently in mutant cells (Table 1). Lagging chromosomes resulted in a prolonged anaphase that was often resolved abruptly, suggesting either chromosomal breaks or missegregation of whole chromosomes (Fig. 1C, row 2). Alternatively, some cells failed to complete mitosis and became binucleated (Fig. 1C, row 3). In summary, mitosis in Rb1ΔL/ΔL cells is characterized by delayed chromosome condensation, an abnormal metaphase plate, and lagging chromosomes that lead to aneuploidy.
Table 1.
Summary of mitotic phenotypes observed in video microscopy experiments
aFor all statistical tests, Rb1ΔL/ΔL is compared with wild type, and all shRNAs directed against CAP-D3 were compared with shLuc.
bIncludes mitoses where the metaphase plate never visually divided, chromatin decondensed, and cells became tetraploid.
cFive equally spaced cross-sections for each metaphase plate were measured from the last image before the initiation of anaphase.
dA difference from controls that is above 95% confidence interval (χ2 test, P < 0.05).
eAbove 95% confidence interval (t-test, P < 0.05).
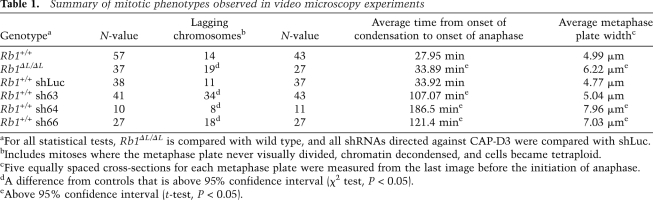
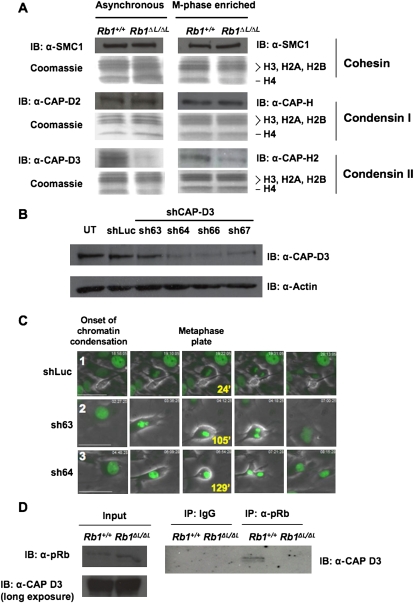
The preceding experiments are consistent with previous reports of defects in condensation and/or cohesion of mitotic chromosomes (Ono et al. 2004). For these reasons, we investigated the levels of the condensin and cohesin protein complexes that were present on chromatin in Rb1ΔL/ΔL cells. Protein extracts were prepared from cell cultures that were synchronized in S phase, released, and harvested at their maximal mitotic index (M-phase-enriched), or that were asynchronous. SDS-PAGE and Western blotting were used to analyze the chromatin fractions from these lysates using histone proteins as a marker for this fraction (Fig. 2A; Suppplemental Fig. 2B). Since Cohesins and Condensins are multiprotein complexes, we chose representative subunits to measure their presence on chromatin. The SMC1 subunit was used as a surrogate for the levels of the Cohesin complex; CAP-H and CAP-D2 were used similarly for Condensin I, and CAP-H2 and CAP-D3 were used to detect Condensin II. This analysis revealed reduced levels of Condensin II on chromatin derived from Rb1ΔL/ΔL cells, while the overall levels of Condensin I and Cohesin were unchanged. To ensure that the reduction in Condensin II loading on chromatin is not due to overall reduction of the protein, we probed for levels in whole-cell extracts and determined that Condensin II protein is expressed at wild-type levels in Rb1ΔL/ΔL fibroblasts (Supplemental Fig. 2C).
Figure 2.
Defective loading of Condensin II complexes on RB1ΔL/ΔL chromosomes. (A) Chromatin fractions were prepared from MEFs that were either proliferating asynchronously or enriched for M-phase cells. Protein content in these fractions was analyzed by SDS-PAGE, followed by Coomassie staining for histone proteins, or Western blotting for the indicated components of the Cohesin and Condensin complexes. (B) Wild-type MEFs were transduced with retroviruses expressing the indicated shRNAs and H2B-GFP. Cell extracts were analyzed by Western blotting for CAP-D3 and Actin. (UT) Untransduced cells. (C) Video microscopy was performed on cells expressing either a control luciferase shRNA, or shRNAs directed against CAP-D3 (sh63 and sh64). Phase-contrast and GFP images were taken every 3 min for 15 h. Representative pictures include the onset of prophase in the left-most panel, and the last view of the metaphase plate before anaphase along with the time elapsed from prophase. The last frame on the right shows the cells after either cytokinesis (shLuc), or failure to resolve as two daughter cells (resulting in binucleation [sh63]) or as persistent anaphase bridges (sh64). The numbers in the left-most image correspond to references in the Supplemental Material and Supplemental Movies. Bars, 50 μm. (D) Extracts were prepared from MEFs of the indicated genotypes. Chromatin fractions from these cells were then subjected to immunoprecipitation with anti-pRB antibodies. The relative amount of CAP-D3, precipitated with wild-type and mutant pRB, was detected by Western blotting. Input levels of relevant proteins from chromatin fractions are shown, and the CAP-D3 blot is overexposed to demonstrate that Condensin II complexes are present in the Rb1 mutant input.
Depletion of the Condensin II subunit CAP-D3 by RNAi delays progression from the onset of condensation to anaphase and results in lagging chromosomes in pRB-deficient HeLa cells (Hirota et al. 2004). Furthermore, a recent study indicates that GST-RB can bind to Condensin II complexes using its LXCXE-binding cleft region (Longworth et al. 2008). For this reason, we investigated the role of CAP-D3 in more detail using video microscopy so that the mitotic defects in Rb1ΔL/ΔL fibroblasts could be directly compared with CAP-D3 deficiency in primary cells. To this end, we generated retroviruses expressing shRNAs against CAP-D3, along with an H2B-GFP reporter, and used them to infect wild-type MEFs. Figure 2B demonstrates the degree of knockdown obtained from each shRNA construct. We used the partial knockdown of sh63, and the more extensive depletion of the others, to investigate the impact of different levels of CAP-D3 expression on mitosis in MEFs. Video microscopy revealed that partial knockdown using sh63 resulted in a delay in reaching anaphase that was longer than in Rb1ΔL/ΔL cells; it also caused lagging chromosomes, but little widening of the metaphase plate (Fig. 2C, row 2; Table 1). Extensive depletion of CAP-D3, shown by sh64 (and sh66) caused a proportionately longer delay from chromosome condensation to anaphase than in sh63, but a similar frequency of lagging chromosomes (Fig. 2C, row 3; Table 1). Interestingly, these depleted cells had a wider metaphase plate, akin to Rb1ΔL/ΔL fibroblasts (Table 1). We interpret this to mean that the earliest defect in mitosis in Rb1ΔL/ΔL MEFs is similar to the delayed condensation that results from CAP-D3 depletion, but is less pronounced. Since RNAi depletion of CAP-D3 has been shown to reduce sister chromosome cohesion (Hirota et al. 2004), this is a likely explanation for the broader metaphase plate in both Rb1ΔL/ΔL and CAP-D3-depleted MEFs. Last, CAP-D3-depleted and Rb1ΔL/ΔL mutant cells both exhibit lagging chromosomes in anaphase as a result of these chromosomal abnormalities. To ensure that the similarity of phenotype between Rb1ΔL/ΔL cells and CAP-D3-depleted cells is not a coincidence, we investigated the physical interaction between pRB and the Condensin II complex. Immunoprecipitation of pRB followed by Western blotting for CAP-D3 reveals that pRB interacts with the Condensin II complex in the chromatin fraction of wild-type but not Rb1ΔL/ΔL MEFs (Fig. 2D). Western blots reveal the input levels of each of the relevant proteins, and that CAP-D3 is present in this fraction. Furthermore, control immunoprecipitations were performed to detect pRB–E2F3 interactions to confirm the specificity of this interaction defect (Supplemental Fig. 2E).
Therefore, these experiments reveal a role for pRB outside of the regulation of E2F target genes in the G1 phase of the cell cycle. We demonstrate that endogenous pRB interacts with Condensin II to compact mitotic chromosomes. Furthermore, a deficiency in this process causes a specific defect in condensation during prophase that manifests as lagging anaphase chromosomes in a primary cell culture system.
The Rb1ΔL mutation exacerbates tumorigenesis in Trp53−/− mice
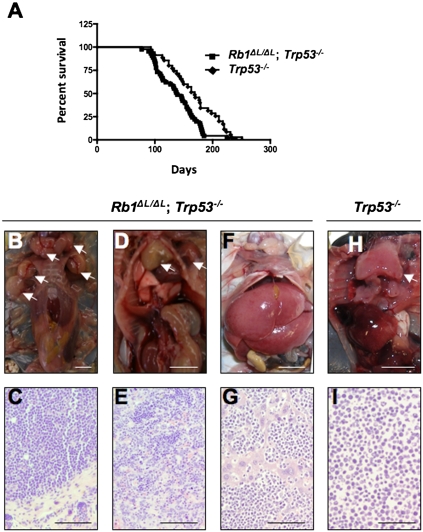
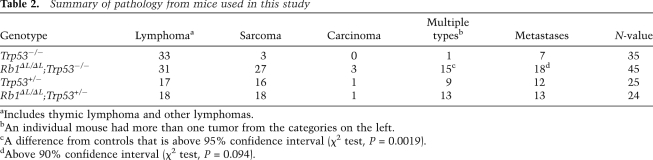
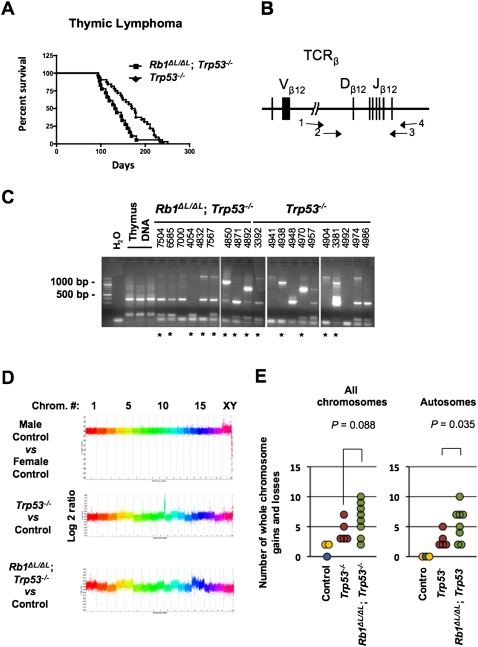
Since chromosome instability is commonly thought to contribute to tumorigenesis by facilitating the acquisition of malignant characteristics, we sought to investigate how the Rb1ΔL mutation impacts cancer pathogenesis. Since we showed that the Rb1ΔL mutation compromises G1 cell cycle arrest in response to negative growth signals from DNA damage and oncogene-induced senescence (Talluri et al. 2010), we chose to cross Rb1ΔL/ΔL mice into a Trp53−/− background, as Trp53−/− mice are known to be defective for the G1 arrest response from DNA breaks and senescence (Lowe et al. 1993; Serrano et al. 1997; Braig et al. 2005; Post et al. 2010). Consequently, comparing Rb1ΔL/ΔL; Trp53−/− mice with Trp53−/− controls eliminates G1-to-S-phase regulation in response to these stimuli in both cohorts of mice, and allows pRB's role in mitosis to be studied in relative isolation. Interestingly, Rb1ΔL/ΔL; Trp53−/− mice succumb to cancer in a significantly shorter time than Trp53−/− controls (log rank test, P = 0.0067) (Fig. 3A). Trp53−/− mice have been reported to develop mainly thymic lymphomas with rare metastases, and our data are no exception (Table 2). However, in addition to earlier tumor onset in Rb1ΔL/ΔL; Trp53−/− mice, we also see a trend toward more aggressive tumors, increased numbers of animals with multiple tumors, and more frequent metastases in Rb1ΔL/ΔL; Trp53−/− compound mutant mice (Table 2). Examples of this aggressive cancer phenotype are shown in Figure 3. Figure 3B shows a mouse with lymphoma affecting at least five major lymph nodes in the thoracic and cervical regions. Figure 3C shows an hematoxylin and eosin (H&E)-stained tissue section from one of these lymph nodes, revealing densely packed cells in immediate contact with the underlying epidermis, with no resemblance of normal lymph node structure. The mouse pictured in Figure 3D had both thymic lymphoma and a sarcoma associated with its left forelimb. The accompanying histology in Figure 3E shows that the sarcoma is being invaded by lymphocytes from the nearby thymic lymphoma. Figure 3F shows striking distension of the liver in a mouse that is caused by metastasis. Histology of this liver in Figure 3G shows islands of hepatocytes amid abundant invading lymphocytes. Last, the image in Figure 3H shows a typical enlarged thymus from a Trp53−/− control. Histology of these lymphomas was typically characterized by larger cells (Fig. 3H), suggesting that Trp53−/− lymphomas are of a lower grade than those in Figure 1, C, E, and G. From this analysis, it is clear that Rb1ΔL/ΔL; Trp53−/− mice succumb to cancer more rapidly than Trp53−/− controls, and that the characteristics of disease in compound mutant animals indicates that these cancers are more aggressive than those found in Trp53−/− mice.
Figure 3.
More aggressive tumors in Rb1ΔL/ΔL; Trp53−/− mice. (A) Kaplan-Meier survival proportions are shown for Rb1ΔL/ΔL; Trp53−/− (n = 45) and Trp53−/− (n = 35) mice. (B–I) Representative images of cancers found in Trp53−/− control and Rb1ΔL/ΔL; Trp53−/− compound mutants. (B) This necropsy reveals lymphoma with multiple affected lymph nodes, as indicated by arrows. (C) H&E staining of a tissue section from one of the affected lymph nodes from B. (D) Mouse with thymic lymphoma and a sarcoma near its left forelimb; both are indicated by arrows. (E) Histological analysis of the sarcoma in D reveals extensive infiltration of this tumor by cells from the neighboring lymphoma. (F) Necropsy demonstrates an enlarged liver in this Rb1ΔL/ΔL; Trp53−/− mutant mouse. (G) Histology of the liver in F reveals extensive infiltration by lymphocytes, indicative of metastasis. (H) Necropsy of a Trp53−/− control mouse shows an enlarged thymus that is typical of these mice. (I) This micrograph shows H&E staining of a thymic lymphoma from a Trp53−/− mouse. Bars: B,D,F,H, 1 cm; C,E,G,I, 100 μm.
Table 2.
Summary of pathology from mice used in this study
aIncludes thymic lymphoma and other lymphomas.
bAn individual mouse had more than one tumor from the categories on the left.
cA difference from controls that is above 95% confidence interval (χ2 test, P = 0.0019).
dAbove 90% confidence interval (χ2 test, P = 0.094).
In order to make direct comparisons between Rb1ΔL/ΔL; Trp53−/−- and Trp53−/−-derived tumors, we decided to focus on thymic lymphomas. These tumors have been studied extensively in Trp53−/− animals, and have been shown to be near diploid with rare chromosomal translocations (Liao et al. 1998; Artandi et al. 2000; Braig et al. 2005). In this way, they offered an ideal starting point for investigating the effects caused by the Rb1ΔL mutation on chromosome instability. To ensure that the comparison between these tumors was appropriate, we sought to investigate whether the Rb1ΔL mutation affects thymic development in a way that could bias this analysis. First, defects caused by the Rb1ΔL mutation alone are not sufficient to cause cancer in this or any other organ in these mice (Supplemental Fig. 3). Second, gross histological analysis of thymi from Rb1ΔL/ΔL and Rb1ΔL/ΔL; Trp53−/− animals did not reveal any obvious abnormalities (Supplemental Fig. 4A). Furthermore, analysis of CD4- and CD8-positive cells revealed no alterations in T-cell development, and rates of proliferation were unaltered by the Rb1ΔL mutation (Supplemental Fig. 4B,C). Finally, E2F target gene expression was not deregulated in thymi from these mice (Supplemental Fig. 4D). From these experiments, we conclude that differences in Rb1ΔL/ΔL; Trp53−/− and Trp53−/− thymic lymphomas are unlikely to be explained by differences in either development, or the basal proliferation rate of cells in this organ.
Examining the survival proportions of the animals that succumbed to thymic lymphoma alone revealed that the Rb1ΔL/ΔL; Trp53−/− mice have a shorter latency before tumor formation compared with Trp53−/− controls (log rank test, P = 0.0198) (Fig. 4A). One possibility to explain the difference in aggressiveness of these tumors is that compound mutant lymphomas may arise from many initiating thymocytes, resulting in a polyclonal tumor, whereas the Trp53−/− controls may be mono- or oligoclonal. To address this question, we used a PCR assay to detect individual T-cell receptor recombination events to estimate the number of individual thymocytes that have become transformed and populate each lymphoma (Fig. 4B,C). This revealed that tumors of both genotypes were rarely monoclonal, and both showed a similar range of clonality, suggesting that clonality does not bias our comparison between the thymic lymphomas found in animals of these two genotypes.
Figure 4.
Increased genomic instability in Rb1ΔL/ΔL; Trp53−/− thymic lymphoma. (A) Kaplan-Meier survival proportions are shown for Rb1ΔL/ΔL; Trp53−/− (n = 18) and Trp53−/− (n = 32) mice that succumbed to thymic lymphoma. (B) Schematic diagram of the T-cell receptor β (TCRβ) locus that was PCR-amplified to assess clonality of thymic lymphomas. Primer pairs 1 and 4 and 2 and 3 were used in a nested strategy to amplify rearranged forms of this gene found in tumor samples, as described in the Materials and Methods. (C) Agarose gel electrophoresis of T-cell receptor β (TCRβ) PCR, including a water-only negative control, and three normal thymus samples as positive controls. Four-digit numbers correspond to ear tag numbers for individual mice, and are present to allow correlations with pathology and array data in Supplemental Tables 1 and 2. The asterisks indicate samples that were used for aCGH analysis. (D) Control, or tumor DNA versus control, was used for hybridization to whole-genome arrays to determine regions of gain or loss in thymic lymphoma samples. Representative graphs depicting log2 ratio values plotted against chromosome number are shown. Data points from individual chromosomes are shown in different colors. (E) Whole-chromosome gains and losses were inferred by differences in an entire chromosome and were compared with controls. The number of whole-chromosome changes for each tumor is plotted against their genotypes. The control male versus control male hybridization is shown in blue, the male versus female hybridizations are shown in yellow, and Trp53−/− and Rb1ΔL/ΔL; Trp53−/− samples are denoted by red and green, respectively. The analysis of all chromosomes, or autosomes alone, are shown. The mean number of changes was compared between genotypes using a t-test.
To investigate the effects of the Rb1ΔL mutation on chromosome instability, we used array comparative genomic hybridization (aCGH) to compare the genomes of thymic lymphoma cells from Rb1ΔL/ΔL; Trp53−/− and Trp53−/− mice. Figure 4D shows representative log2 ratio plots from male versus female control hybridizations, as well as from selected Rb1ΔL/ΔL; Trp53−/− and Trp53−/− tumors hybridized against same sex control DNA. Using male versus male and male versus female control hybridizations as a guide for normal copy number and whole-chromosome gains, we inferred changes in chromosome copy number present in these tumors. Since these lymphomas are polyclonal, the gain or loss of a single chromosome in one clone can be a relatively modest change when the whole thymus is analyzed as one. For this reason, we searched for chromosomes that were statistically different than control, and did not try to distinguish if these represent single or multiple chromosome gains. Satisfyingly, the average number of gains and losses in our Trp53−/− lymphoma controls (4.2) was similar to the frequency of chromosome number changes reported by karyotyping in other studies of Trp53-deficient lymphomas in which these cancers typically have chromosome counts in the low 40s (Liao et al. 1998; Artandi et al. 2000; Braig et al. 2005). Our analysis revealed that whole-chromosome gains or losses were more prevalent in Rb1ΔL/ΔL; Trp53−/− lymphomas than in Trp53−/− controls (Fig. 4E). Because not all tumors originate from mice of the same sex, and this creates unequal opportunities to gain and lose sex chromosomes, we displayed these data in graphs for all chromosomes and for autosomes. This suggests that whole-chromosome instability may be the underlying mechanism that increases cancer susceptibility in Rb1ΔL/ΔL; Trp53−/− animals compared with Trp53−/− mice, an interpretation that is consistent with centromere fusions observed in metaphase spreads (Fig. 1A) and lagging chromosomes in anaphase that were observed in our videos (Fig. 1C). We note that the quantity of copy number segments (local regions of gain or loss relative to adjacent chromosomal sequences) is also elevated in most Rb1ΔL/ΔL; Trp53−/− thymic lymphomas (Supplemental Fig. 5). This implies that smaller genomic rearrangements also take place, and this is consistent with resolution of lagging chromosomes in Figure 1C, row 2, occurring by chromosomal breakage. Therefore, both forms of chromosomal instability may be caused by the Rb1ΔL mutation, and contribute to the increase in cancer susceptibility that we observe.
Our analysis of the Rb1ΔL allele's effects on cancer reveal that it causes a dramatic increase in susceptibility, and Rb1ΔL/ΔL; Trp53−/− mice are characterized by more aggressive tumors than the Trp53−/− controls. Furthermore, characterization of genomic abnormalities found in Rb1ΔL/ΔL tumors demonstrates that they are consistent with the chromosomal and mitotic abnormalities in primary cultures that we observed in Figure 1 by video microscopy. These data strongly suggest that pRB facilitates mitotic chromosome condensation as part of its function as a tumor suppressor.
Accelerated loss of heterozygosity in Rb1ΔL/ΔL mice
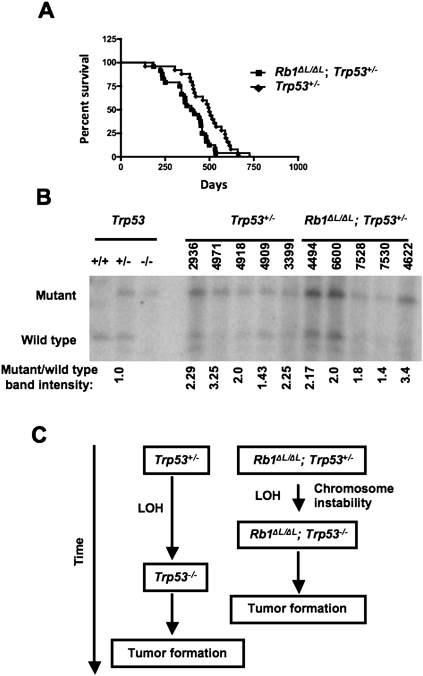
Thus far, our work suggests that failure to condense and properly segregate mitotic chromosomes because of defective pRB–LXCXE interactions leads to chromosome instability and exacerbates cancer pathogenesis. In order to test this more directly, we generated cohorts of Rb1ΔL/ΔL; Trp53+/− and Trp53+/− control animals. It is known that Trp53+/− mice develop a broad spectrum of tumors, including thymic lymphomas, after a considerably longer latency period than Trp53−/− mice (Jacks et al. 1994; Purdie et al. 1994; Donehower et al. 1995). Loss of heterozygosity, which eliminates the remaining wild-type allele, is reported with high frequency in tumors from Trp53+/− mice, strongly suggesting that it is the rate-limiting step for tumor formation, since it can cause the same types of cancer found in Trp53−/− animals (Jacks et al. 1994). For this reason, use of Trp53+/− mice has emerged as an assay for genome instability effects on cancer (Kuperwasser et al. 2000; Smith et al. 2006; Baker et al. 2009). We followed these animals over ∼2 yr and discovered that the Rb1ΔL/ΔL; Trp53+/− compound mutants succumbed to cancer at a significantly younger age than Trp53+/− controls (log rank test, P = 0.0105) (Fig. 5A). Consistent with previous reports, we found that these animals were susceptible to a broad range of cancer types in both Rb1ΔL/ΔL; Trp53+/− and control genotypes (Table 2). In this cross, the best evidence for increased aggressiveness in the compound mutants was the earlier age of cancer incidence. The histopathology of these tumors was similar, reinforcing the validity of comparing these two cohorts for their mechanism of tumor initiation (Supplemental Table 1). To search for loss of heterozygosity, DNA was extracted from tumors and subjected to Southern blot analysis to measure the relative abundance of wild-type and null alleles of Trp53. This revealed that the wild-type allele was reduced in abundance in tumor samples derived from all animals (Fig. 5B); the existence of residual wild-type Trp53 is consistent with the presence of Trp53+/− stroma in these tumor samples. Since these data reveal loss of the wild-type Trp53 locus in all of the tumors we analyzed, and Rb1ΔL/ΔL; Trp53+/− mice develop cancer earlier, this suggests that the rate-limiting step for tumor formation—namely, loss of heterozygosity—has taken place more rapidly and was facilitated by the Rb1ΔL mutation in these mice (Fig. 5C).
Figure 5.
Accelerated loss of heterozygosity in Rb1ΔL/ΔL; Trp53+/− mice. (A) Kaplan-Meier survival proportions are shown for Rb1ΔL/ΔL; Trp53+/− (n = 24) and Trp53+/− (n = 25) mice that succumbed to detectable cancers. (B) Southern blot analysis of tumors from Rb1ΔL/ΔL; Trp53+/− and Trp53+/− mice was performed to assess the relative abundance of wild-type and null Trp53 alleles. Four-digit numbers correspond to ear tags for individual mice to allow correlation with pathology data in Supplemental Table 1. The ratio of mutant to wild-type allele abundance was determined by phosphorimaging, and is displayed below each lane. (C) Model of the Rb1ΔL mutation's role in cancer susceptibility of these mice. Prior reports establish that Trp53+/− mice succumb to cancer after a long latency, and that it is accompanied by loss of heterozygosity at the Trp53 locus. The age at which cancer initiates in Rb1ΔL/ΔL; Trp53+/− mice and the loss of the wild-type Trp53 locus in these tumors suggests that chromosome instability, caused by the Rb1ΔL mutation, induces loss of heterozygosity more rapidly, causing an earlier onset of cancer.
This analysis of loss of heterozygosity in tumors from Rb1ΔL/ΔL; Trp53+/− mice offers evidence that cells bearing the Rb1ΔL mutation are more prone to chromosomal instability. Based on this experiment, and the analysis of Rb1ΔL/ΔL; Trp53−/− lymphomas, our study reveals that chromosome condensation mediated by pRB is likely an important component of its role as a tumor suppressor.
Discussion
This study investigates the role of the pRB in mitotic chromosome condensation. Our data indicate that this is a mechanism by which pRB acts as a tumor suppressor. The novelty of this tumor-suppressive mechanism relies extensively on the ability to separate the mitotic-specific functions of pRB from cell cycle entry control in our cancer-prone mice. The analysis of thymic lymphomas in Rb1ΔL/ΔL; Trp53−/− mice allows us to distinguish between the effects of the Rb1ΔL mutation at these different points in the cell cycle. First, the response to DNA damage, or other stress-inducing stimuli that activate p53, leads to increased p21/CIP1 expression; this in turn inhibits cyclin-dependent kinases, and leads to pRB activation during G1 and cell cycle arrest (Campisi and d'Adda di Fagagna 2007). Since we demonstrated previously that Rb1ΔL/ΔL cells are defective for a G1 arrest in response to γ-irradiation or oncogene-induced senescence (Talluri et al. 2010), including p53 deficiency in both cohorts of our tumor study prevents these Rb1ΔL defects from confounding our interpretations. In addition to these defects, we also determined that Rb1ΔL/ΔL cells are resistant to the growth inhibitory effects of TGF-β (Francis et al. 2009). This growth inhibitory cytokine has been shown to play a key role in peripheral T-cell regulation. In particular, transgenic mice expressing a dominant-negative TGF-β type II receptor in CD8-positive T cells are prone to develop lymphoproliferative disease and, ultimately, lymphoma (Lucas et al. 2000, 2004). Interestingly, the phenotype of these animals is very different from Rb1ΔL/ΔL; Trp53−/− mice, with extensive expansion of T cells in the periphery but not the thymus. In fact, these studies suggest that TGF-β signaling may have very little function in the thymus. Since Rb1ΔL/ΔL mice do not display any lymphoproliferative characteristics in their lifetime (Supplemental Fig. 4), it is unlikely that defective TGF-β growth control can explain the cancer phenotype of Rb1ΔL/ΔL; Trp53−/− mice. There is no evidence of aberrant proliferation or alterations in thymic development or mophology in Rb1ΔL/ΔL; Trp53−/− mice. Furthermore, E2F target genes are regulated normally in this tissue. Because there is evidence of elevated chromosomal instability in these thymic lymphomas, our conclusion that pRB can function as a tumor suppressor by facilitating chromosome condensation is the most appropriate interpretation of these data.
The physical interaction between pRB and Condensin II offers a logical explanation for how pRB can participate in mitotic chromosome condensation. The phenotypes seen in video microscopy experiments of Rb1ΔL/ΔL cells suggest an acute defect in condensation during prophase. However, the extensive reduction in Condensin II levels on chromatin in asynchronously proliferating Rb1ΔL/ΔL MEFs suggests that pRB participates in chromatin loading earlier in the cell cycle, as these cultures contain only a small proportion of mitotic cells. In addition, Condensin II is known to be present on chromatin in interphase nuclei (Hirota et al. 2004). These observations suggest that pRB's role in chromosome condensation may take place earlier in the cell cycle, perhaps in G1 where it is relatively unphosphorylated and already thought to regulate chromatin structure. Immunoprecipitation and Western blotting experiments demonstrate that wild-type pRB interacts with Condensin II in chromatin fractions, but this interaction was absent from Rb1ΔL/ΔL chromatin. Because Condensin II is underrepresented in this fraction in the first place, the lack of interaction may not reflect a need for the pRB–LXCXE-binding cleft to mediate physical contact with Condensin II, but may indicate that this aspect of pRB is more important for Condensin II to be loaded on chromatin. It will be important in future studies to determine precisely how pRB uses LXCXE-type interactions to exert its regulatory role over Condensin II function. At this point, we have no evidence to indicate that it must be direct. Similar studies from the Dyson and te Riele laboratories (Manning et al. 2010; van Harn et al. 2010) show that pRB may also participate in chromosome cohesion at the centromere. In those studies, Cohesin complexes are reduced at centromeres. We cannot rule out that a similar biochemical defect may be present in Rb1ΔL/ΔL cells. However, condensin complexes are also well known to be concentrated at centromeric heterochromatin (Ono et al. 2004; Oliveira et al. 2005; Vagnarelli et al. 2006). Taken together, this suggests that future studies to understand pRB's role in mitosis will need to focus more closely on its ability to regulate chromatin at centromeric regions, as this is likely where it acts to ensure proper chromosome architecture and segregation in mitosis.
This study reveals a novel mechanism of tumor suppression by pRB. While other reports have indicated that defective pRB is associated with chromosomal abnormalities (Hernando et al. 2004; Gonzalo et al. 2005; Iovino et al. 2006; Isaac et al. 2006; Longworth et al. 2008; Amato et al. 2009), our work demonstrates that this manifests in more rapid tumor formation. This raises the question: How important is this aspect of tumor suppression by pRB relative to its well-characterized role in regulating E2F transcription factors and entry into S phase? The lack of spontaneous tumors in our Rb1ΔL/ΔL mice may suggest that it is less important. We favor a more cautious view of this question. The inability to arrest proliferation in G1 because of a pRB deficiency is also accompanied by deregulation of activator E2Fs, and this creates an intrinsic progrowth signal. This is inherently a stronger oncogenic event than diminished chromosome condensation because it combines the loss of negative growth regulation with the gain of a growth-promoting signal. Defective chromosome condensation on the other hand, creates the opportunity for genetic change that can contribute to cancer pathogenesis, but does not provide an inappropriate growth-promoting signal. For these reasons, experiments designed to equalize the loss of safeguards with gain of proliferative advantages will be necessary to appropriately compare these aspects of tumor suppression by pRB. Only through this type of investigation will it be possible to fully comprehend what makes the Rb gene such a critical factor in cell cycle regulation and cancer.
Materials and methods
Cell culture, viral infections, and microscopy
Primary MEFs were prepared and cultured according to standard methods as reported previously (Hurford et al. 1997). ESCs were prepared by intercrossing Rb1ΔL/+ mice and harvesting day 3.5 blastocysts for culture. The inner cell mass was dissociated and colonies were picked and expanded for genotyping as described (Hogan et al. 1994). Mitotic chromosome spreads were prepared from MEFs by treating cells with 50 ng/mL colcemid for 3 h before harvesting, swelling, and fixing. ESC chromosome spreads were generated similarly after treating cultures with 10 μg/mL colchicine for 3 h. Chromosome spreads were stained with a major satellite pericentromeric probe as before (Isaac et al. 2006). Staining of rDNA repeats and the probes used were as described (Grummt et al. 1979; Romanova et al. 2006). Fluorescent microscopic images were captured on a Zeiss Axioskop 40 microscope using a Spotflex camera and EyeImage software.
To introduce H2B-GFP into MEFs, we created a pBABE retroviral vector that expresses H2B-GFP by cloning the gene from pBOS-H2B-GFP. Short hairpin retroviral vectors targeting CAP-D3 were purchased from Open Biosystems, and were cloned into the pLMP plasmid along with the H2B-GFP gene. Viral vectors were packaged into ecotropic retroviruses using Bosc23 cells, and were subsequently used to infect MEFs as described (Pear et al. 1993).
Live cell microscopy was carried out by plating early passage, H2B-GFP-transduced MEFs onto glass-bottom tissue culture dishes (MatTek) containing phenol-free DMEM and 5% FBS supplemented with penicillin, streptomycin, and glutamine. During imaging, cells were maintained at 37°C with 5% CO2 using a stage-mounted environment chamber (Neue Biosciences), and were monitored with an automated inverted microscope (DMI 6000b, Leica). Phase-contrast and fluorescent images were collected every 3 min over a 15-h time course using Openlab Software automation (Ritchie et al. 2008). Full-length movies for each example shown in Figures 1 and 2 are available in the Supplemental Material. Measurements of metaphase plate dimensions were made using Volocity software.
Stained tissue sections were examined microscopically on a Zeiss Axioskop 40 microscope, and were photographed using a Spotflex camera and EyeImage software.
Antibodies and protein detection
The following antibodies were used to detect or precipitate proteins in this study: rabbit anti-histone H2B (07-371, Upstate Biotechnologies), goat anti-GFP (G095, Clontech), rabbit anti-CAP-H (a kind gift of Kyoko Yokomori, University of California at Irvine) (Heale et al. 2006), rabbit anti-CAP-H2 (a generous gift of Tatsuya Hirano, Riken, Japan) (Ono et al. 2003), rabbit anti-SMC1 (A300-055A, Bethyl Laboratories), anti-pRB (G3-245, BD-Pharmingen), anti-BubR1: C-20 (sc-16195, Santa Cruz Biotechnology), anti-PCNA: PC10 (sc-56, Santa Cruz Biotechnology), anti-MCM7: 141.2 (sc-9966, Santa Cruz Biotechnology), anti-E2F3: C-18 (sc-878, Santa Cruz Biotechnology), and anti-E2F3 clone PG37 (05-551, Upstate Biotechnologies). Antibodies to CAP-D3 were raised against a GST fusion protein containing amino acids 1243–1506 of CAP-D3. CAP-D2 antibodies were raised against a GST fusion containing amino acids 943–1132. Rabbits were immunized in a commercial facility (PTG Laboratories). Antibodies were affinity-purified by adsorbing to a His-tagged version of the same protein coupled to a Sulfolink column (Pierce), eluted in low pH, and neutralized.
Cell extracts for Western blotting were prepared by freeze–thaw lysis as described previously (Hurford et al. 1997). Chromatin fractions were prepared by low-salt wash of nuclei followed by nuclease treatment to solubilize chromatin-bound proteins (Mendez and Stillman 2000). MEFs were synchronized using a two-step method of confluence arrest and aphidicolin, followed by washout and collection of cells when the mitotic index is greatest (Isaac et al. 2006). Nuclease-prepared chromatin fractions were used for immunoprecipitation experiments where they were precleared with mouse IgG and protein-G magnetic beads before precipitation using anti-pRB antibodies. Proteins in all experiments were resolved by SDS-PAGE followed by Western blotting using standard techniques.
Mice
The Rb1ΔL mutant strain carries three amino acid substitutions in its Rb1-encoded protein (I746A, N750A, and M754A) that disrupt interactions with LXCXE motif-containing proteins, but not E2F transcription factors (Dick et al. 2000; Isaac et al. 2006); details of its construct and initial characterization have been published previously (Isaac et al. 2006; Francis et al. 2009; Talluri et al. 2010). The Trp53−/− mice were purchased from Jackson Laboratories and were intercrossed with Rb1ΔL mutants to produce the required genotypes for this study. All animals were maintained in a mixed 129/B6 background. Mice were housed and maintained according to the guidelines of the Canadian Council on Animal Care. Animals were followed throughout their lives for signs of tumor burden, and were euthanized when tumors became visible or the animal experienced sudden weight loss or became lethargic. All animals were subjected to a thorough necropsy, and abnormal tissues, organs, or tumors were fixed in formalin and processed for histological assessment. Portions of tumors were also snap-frozen and used to prepare genomic DNA. Tissues were embedded, sectioned, and stained with H&E according to standard methods, and were photographed as described above. See Supplemental Table 1 for synopsis of histopathology for all animals used in this study.
Thymic development
To investigate thymic development and proliferation, we isolated thymuses from 6- to 8-wk-old animals. Tissue was either fixed and frozen for cryosectioning, or dissociated and stained for flow cytometry. Fluorescently labeled antibodies against CD4 (553650) and CD8 (553032) were purchased from BD Pharmingen. CD4/CD8 flow cytometry was carried out essentially as described (Bruins et al. 2004). To detect proliferation, mice were injected with 200 μL of 16 mg/mL BrdU in DMEM 1 h before euthanasia. BrdU-labeled thymuses were fixed, sectioned, and stained with anti-BrdU antibodies (347580, BD Biosciences), and were photographed under fluorescent optics as described above.
PCR, Southern blotting, and aCGH
High-molecular-weight DNA was extracted from frozen tumor samples using standard DNA isolation procedures and were used in the following analyses: DNA from thymic lymphomas was analyzed for T-cell receptor V(D)J recombination patterns using nested PCR. In brief, the Dβ12 and Jβ12 region was characterized using primer pairs and PCR conditions as described by Whitehurst et al. (1999). Loss of heterozygosity of Trp53 in tumors from Trp53+/− mice was determined by Southern blotting. Genomic DNA was digested with EcoRI and StuI, and was resolved, transferred, and hybridized using standard methods. The probe was a genomic DNA clone encompassing intron 7 to intron 9. Band intensities were measured on a Molecular Dynamics Storm scanner PhosphorImager and densitometry analysis was preformed using AlphaEase FC software.
For aCGH experiments, DNA was extracted from livers of five male and female wild-type animals to create pools of control DNA. Same sex control versus control, control male versus control female, and tumor DNA versus the appropriate sex control hybridizations were performed by NimbleGen on a mouse whole-genome array (design 2006-07-26-MM8-WG-CGH). Segmentation analysis described by Olshen et al. (2004) was performed, and was used to infer changes in copy number. A segment was defined as a group of adjacent data points whose log2 ratio values were not statistically different. To determine cutoff values, indicating normal copy number, the mean log2 ratio for all data points of individual chromosomes from the control hybridization was determined. A range of plus or minus one standard deviation from this mean was determined to encompass the log2 ratio from 99.8% of all segments from the control hybridization, suggesting that it was a reliable range that captures the vast majority of normal copy number chromosomal segments. This range was calculated for each individual chromosome from the control hybridization. Segments from individual chromosomes from each tumor hybridization were then compared with the appropriate range for their chromosome to determine if they were of a normal copy number. This allowed us to assess, on a chromosome-by-chromosome basis, the copy number status of each segment. Using this approach, we interpreted whole-chromosome gains or losses based on the chromosomal locations of each constituent segment, and their associated copy number status (see Supplemental Fig. 6 for an illustrated example). See Supplemental Table 2 for raw segment data for aCGH experiments.
Acknowledgments
We are grateful to our colleagues in the Dyson and te Riele laboratories for communicating unpublished results. We thank many members of the London Regional Cancer Program and Children's Health Research Institute for advice during the course of this work, in particular, Francisca Aidoo and Christian Isaac for help during the initial stages of this work. C.C. acknowledges fellowship support from NSERC and OGS, and is a member of the CaRTT training program. K.R. is supported by OGS. S.M.F. is the recipient of a Translational Breast Cancer Research Studentship from the London Regional Cancer Program and acknowledges past support from CBCF Ontario and OGSST. N.G.B. is a CIHR New Investigator. F.A.D. is a Research Scientist of the Canadian Cancer Society, and this work was funded by MOP-64253 from the CIHR.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1917610.
Supplemental material is available at http://www.genesdev.org.
References
- Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM 1998. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol 8: 145–155 [DOI] [PubMed] [Google Scholar]
- Amato A, Lentini L, Schillaci T, Iovino F, Di Leonardo A 2009. RNAi mediated acute depletion of retinoblastoma protein (pRb) promotes aneuploidy in human primary cells via micronuclei formation. BMC Cell Biol 10: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA 2000. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406: 641–645 [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jin F, Jeganathan KB, van Deursen JM 2009. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell 16: 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS 2006. Mitotic chromosome structure and condensation. Curr Opin Cell Biol 18: 632–638 [DOI] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436: 660–665 [DOI] [PubMed] [Google Scholar]
- Bruins W, Zwart E, Attardi LD, Iwakuma T, Hoogervorst EM, Beems RB, Miranda B, van Oostrom CT, van den Berg J, van den Aardweg GJ, et al. 2004. Increased sensitivity to UV radiation in mice with a p53 point mutation at Ser389. Mol Cell Biol 24: 8884–8894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart DL, Sage J 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 8: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F 2007. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol 8: 729–740 [DOI] [PubMed] [Google Scholar]
- Cavanaugh AH, Hempel WM, Taylor LJ, Rogalsky V, Todorov G, Rothblum LI 1995. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 374: 177–180 [DOI] [PubMed] [Google Scholar]
- Ciarmatori S, Scott PH, Sutcliffe JE, McLees A, Alzuherri HM, Dannenberg JH, te Riele H, Grummt I, Voit R, White RJ 2001. Overlapping functions of the pRb family in the regulation of rRNA synthesis. Mol Cell Biol 21: 5806–5814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick FA, Sailhamer E, Dyson NJ 2000. Mutagenesis of the pRB pocket domain reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol Cell Biol 20: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Vogel H, McArthur MJ, Montgomery CAJ, Park SH, Thompson T, Ford RJ, Bradley A 1995. Effects of genetic background on tumorigenesis in p53-deficient mice. Mol Carcinog 14: 16–22 [DOI] [PubMed] [Google Scholar]
- Francis SM, Bergsied J, Isaac CE, Coschi CH, Martens AL, Hojilla CV, Chakrabarti S, Dimattia GE, Khoka R, Wang JY, et al. 2009. A functional connection between pRB and transforming growth factor β in growth inhibition and mammary gland development. Mol Cell Biol 29: 4455–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, et al. 2005. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol 7: 420–428 [DOI] [PubMed] [Google Scholar]
- Grummt I, Soellner C, Scholz I 1979. Characterization of a cloned ribosomal fragment from mouse which contains the 18S coding region and adjacent spacer sequences. Nucleic Acids Res 6: 1351–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham MF, Takakuwa T, Rahadiani N, Tresnasari K, Nakajima H, Aozasa K 2007. Condensin mutations and abnormal chromosomal structures in pyothorax-associated lymphoma. Cancer Sci 98: 1041–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan KM, Hannan RD, Smith SD, Jefferson LS, Lun M, Rothblum LI 2000. Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene 19: 4988–4999 [DOI] [PubMed] [Google Scholar]
- Harrington EA, Bruce JL, Harlow E, Dyson N 1998. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc Natl Acad Sci 95: 11945–11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale JT, Ball AR Jr, Schmiesing JA, Kim JS, Kong X, Zhou S, Hudson DF, Earnshaw WC, Yokomori K 2006. Condensin I interacts with the PARP-1–XRCC1 complex and functions in DNA single-strand break repair. Mol Cell 21: 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, et al. 2004. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430: 797–802 [DOI] [PubMed] [Google Scholar]
- Herrera RE, Sah VP, Williams BO, Makela TP, Weinberg RA, Jacks T 1996. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol 16: 2402–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM 2004. Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci 117: 6435–6445 [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E 1994. Manipulating the mouse embryo: A laboratory manual Cold Spring Harbor Press, Plainview, NY [Google Scholar]
- Hurford R, Cobrinik D, Lee M-H, Dyson N 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev 11: 1447–1463 [DOI] [PubMed] [Google Scholar]
- Iovino F, Lentini L, Amato A, Di Leonardo A 2006. RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Mol Cancer 5: 38 doi: 10.1186/1476-4598-5-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac CE, Francis SM, Martens AL, Julian LM, Seifried LA, Erdmann N, Binne UK, Harrington L, Sicinski P, Berube NG, et al. 2006. The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol 26: 3659–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7 [DOI] [PubMed] [Google Scholar]
- Kuperwasser C, Hurlbut GD, Kittrell FS, Dickinson ES, Laucirica R, Medina D, Naber SP, Jerry DJ 2000. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am J Pathol 157: 2151–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Malhotra S, Higgins JP, Bair E, Thompson M, Salari K, Giacomini CP, Ferrari M, Montgomery K, Tibshirani R, et al. 2008. hCAP-D3 expression marks a prostate cancer subtype with favorable clinical behavior and androgen signaling signature. Am J Surg Pathol 32: 205–209 [DOI] [PubMed] [Google Scholar]
- Liao MJ, Zhang XX, Hill R, Gao J, Qumsiyeh MB, Nichols W, Van Dyke T 1998. No requirement for V(D)J recombination in p53-deficient thymic lymphoma. Mol Cell Biol 18: 3495–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Dyson NJ 2010. pRb, a local chromatin organizer with global possibilities. Chromosoma 119: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longworth MS, Herr A, Ji JY, Dyson NJ 2008. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev 22: 1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano T 2005. Dynamic molecular linkers of the genome: The first decade of SMC proteins. Genes Dev 19: 1269–1287 [DOI] [PubMed] [Google Scholar]
- Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362: 847–849 [DOI] [PubMed] [Google Scholar]
- Lucas PJ, Kim SJ, Melby SJ, Gress RE 2000. Disruption of T cell homeostasis in mice expressing a T cell-specific dominant negative transforming growth factor β II receptor. J Exp Med 191: 1187–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas PJ, McNeil N, Hilgenfeld E, Choudhury B, Kim SJ, Eckhaus MA, Ried T, Gress RE 2004. Transforming growth factor-β pathway serves as a primary tumor suppressor in CD8+ T cell tumorigenesis. Cancer Res 64: 6524–6529 [DOI] [PubMed] [Google Scholar]
- Maeshima K, Laemmli UK 2003. A two-step scaffolding model for mitotic chromosome assembly. Dev Cell 4: 467–480 [DOI] [PubMed] [Google Scholar]
- Manning AL, Longworth MS, Dyson NJ 2010. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev (this issue). doi: 10.1101/gad.1917310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew CN, Carter SL, Fox SR, Sexton CR, Reed CA, Srinivasan SV, Liu X, Wikenheiser-Brokamp K, Boivin GP, Lee JS, et al. 2007. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology 133: 976–984 [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol Cell Biol 20: 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED 2007. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Nasmyth K 2005. How do so few control so many? Cell 120: 739–746 [DOI] [PubMed] [Google Scholar]
- Oliveira RA, Coelho PA, Sunkel CE 2005. The condensin I subunit Barren/CAP-H is essential for the structural integrity of centromeric heterochromatin during mitosis. Mol Cell Biol 25: 8971–8984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M 2004. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5: 557–572 [DOI] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115: 109–121 [DOI] [PubMed] [Google Scholar]
- Ono T, Fang Y, Spector DL, Hirano T 2004. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell 15: 3296–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D 1993. Production of high-titre helper-free retroviuses by transient transfection. Proc Natl Acad Sci 90: 8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Melmed S 1997. Isolation and characterization of a pituitary tumor-transforming gene (PTTG). Mol Endocrinol 11: 433–441 [DOI] [PubMed] [Google Scholar]
- Post SM, Quintas-Cardama A, Terzian T, Smith C, Eischen CM, Lozano G 2010. p53-dependent senescence delays Emu-myc-induced B-cell lymphomagenesis. Oncogene 29: 1260–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdie CA, Harrison DJ, Peter A, Dobbie L, White S, Howie SE, Salter DM, Bird CC, Wyllie AH, Hooper ML, et al. 1994. Tumour incidence, spectrum and ploidy in mice with a large deletion in the p53 gene. Oncogene 9: 603–609 [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 16: 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K, Seah C, Moulin J, Isaac C, Dick F, Berube NG 2008. Loss of ATRX leads to chromosome cohesion and congression defects. J Cell Biol 180: 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova L, Korobova F, Noniashvilli E, Dyban A, Zatsepina O 2006. High resolution mapping of ribosomal DNA in early mouse embryos by fluorescence in situ hybridization. Biol Reprod 74: 807–815 [DOI] [PubMed] [Google Scholar]
- Samoshkin A, Arnaoutov A, Jansen LE, Ouspenski I, Dye L, Karpova T, McNally J, Dasso M, Cleveland DW, Strunnikov A 2009. Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLoS One 4: e6831 doi: 10.1371/journal.pone.0006831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JM, Sotillo R, Benezra R 2010. Mitotic chromosomal instability and cancer: Mouse modelling of the human disease. Nat Rev Cancer 10: 102–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW 1997. Oncogenic ras provokes premature senescence associated with accumulation of p53 and p16. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Smith AP, Henze M, Lee JA, Osborn KG, Keck JM, Tedesco D, Bortner DM, Rosenberg MP, Reed SI 2006. Deregulated cyclin E promotes p53 loss of heterozygosity and tumorigenesis in the mouse mammary gland. Oncogene 25: 7245–7259 [DOI] [PubMed] [Google Scholar]
- Talluri S, Isaac CE, Ahmad M, Henley SA, Francis SM, Martens AL, Bremner R, Dick FA 2010. A G1 checkpoint mediated by the retinoblastoma protein that is dispensable in terminal differentiation but essential for senescence. Mol Cell Biol 30: 948–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G, Lens SM 2008. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta 1786: 60–72 [DOI] [PubMed] [Google Scholar]
- Vagnarelli P, Hudson DF, Ribeiro SA, Trinkle-Mulcahy L, Spence JM, Lai F, Farr CJ, Lamond AI, Earnshaw WC 2006. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat Cell Biol 8: 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Harn T, Foijer F, van Vugt M, Banerjee R, Yang F, Oostra A, Joenje H, te Riele H 2010. Loss of RB proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev (this issue). doi: 10.1101/gad.580710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst CE, Chattopadhyay S, Chen J 1999. Control of V(D)J recombinational accessibility of the D β 1 gene segment at the TCRβ locus by a germline promoter. Immunity 10: 313–322 [DOI] [PubMed] [Google Scholar]
- Zheng L, Flesken-Nikitin A, Chen PL, Lee WH 2002. Deficiency of retinoblastoma gene in mouse embryonic stem cells leads to genetic instability. Cancer Res 62: 2498–2502 [PubMed] [Google Scholar]