Abstract
Chromosome instability (CIN) is a common feature of tumor cells. By monitoring chromosome segregation, we show that depletion of the retinoblastoma protein (pRB) causes rates of missegregation comparable with those seen in CIN tumor cells. The retinoblastoma tumor suppressor is frequently inactivated in human cancers and is best known for its regulation of the G1/S-phase transition. Recent studies have shown that pRB inactivation also slows mitotic progression and promotes aneuploidy, but reasons for these phenotypes are not well understood. Here we describe the underlying mitotic defects of pRB-deficient cells that cause chromosome missegregation. Analysis of mitotic cells reveals that pRB depletion compromises centromeric localization of CAP-D3/condensin II and chromosome cohesion, leading to an increase in intercentromeric distance and deformation of centromeric structure. These defects promote merotelic attachment, resulting in failure of chromosome congression and an increased propensity for lagging chromosomes following mitotic delay. While complete loss of centromere function or chromosome cohesion would have catastrophic consequences, these more moderate defects allow pRB-deficient cells to proliferate but undermine the fidelity of mitosis, leading to whole-chromosome gains and losses. These observations explain an important consequence of RB1 inactivation, and suggest that subtle defects in centromere function are a frequent source of merotely and CIN in cancer.
Keywords: Cohesin, condensin, retinoblastoma, merotely, CIN, aneuploid
High-throughput genomic profiling studies illustrate the fact that most human tumors are aneuploid and display abnormalities in the number of whole chromosomes or chromosome arms (Hanahan and Weinberg 2000; Albertson et al. 2003; Beroukhim et al. 2010). Furthermore, many tumors have been shown to be chromosomally unstable (Lengauer et al. 1997). Chromosome instability (CIN), defined as an elevated rate of gains and losses of whole chromosomes (10–100× more often than stable diploid cells) (Lengauer et al. 1997), has been proposed to promote the evolution of tumor cells. Such genomic changes potentially promote metastasis and chemotherapeutic resistance, and correlate with poorer patient prognosis (Nowell 1976; Kuukasjarvi et al. 1997; Rajagopalan and Lengauer 2004; Gao et al. 2007). Recent studies show that aneuploidy and CIN can have a causal role in tumorigenesis and relapse (Rasnick and Duesberg 1999; Weaver et al. 2007; Baker et al. 2009; Sotillo et al. 2010).
CIN likely results from persistent defects in mitotic fidelity, and several mechanisms have been described that cause cells to missegregate whole chromosomes, including defects in bipolar spindle formation, chromosome–spindle association, chromosome cohesion, and the spindle assembly checkpoint (Cahill et al. 1998; Nigg 2002; Sotillo et al. 2007; Thompson and Compton 2008; Baker et al. 2009; Bakhoum et al. 2009; Ganem et al. 2009; Silkworth et al. 2009).
The retinoblastoma tumor susceptibility gene (RB1) is a key regulator of cell proliferation. RB1 was one of the first tumor suppressor genes to be identified, and its product (pRB) is functionally inactivated in most forms of cancer (Bookstein and Lee 1991; Marshall 1991; Weinberg 1995). Although mutation of RB1 is a key, rate-limiting event in the development of most retinoblastoma, recent studies suggest that homozygous mutation of RB1 causes the appearance of benign retinoma, and these subsequently progress to retinoblastoma (Dimaras et al. 2008). This malignant progression correlates with greatly increased levels of aneuploidy and genomic instability. The idea that mutation of RB1 may cause genomic instability is consistent with studies carried out using cultured cells. Populations of Rb−/− cells, or cells in which pRB is functionally inactivated, have elevated numbers of aneuploid cells, and are prone to increases in ploidy (Hernando et al. 2004; Iovino et al. 2006; Isaac et al. 2006; Mayhew et al. 2007; Srinivasan et al. 2007; Amato et al. 2009). Together, these studies have lead to the hypothesis that much of the aneuploidy seen in tumor cells is a by-product of the inactivation of the pRB pathway. Support for this idea comes from a completely independent line of investigation: Expression profiling has led to the identification of expression signatures that correlate with CIN. Remarkably, 10 of the 20 genes that show the highest correlation with CIN are genes that are known E2F targets, and are surrogate markers of pRB inactivation (Markey et al. 2002; Black et al. 2005; Carter et al. 2006).
Whether loss of pRB function leads to aneuploidy directly (through mechanisms that disrupt normal chromosome segregation) or indirectly (by allowing for the proliferation of spontaneously arising aneuploid cells) remains unclear. Evidence suggesting pRB loss may promote chromosome missegregation directly comes from a number of different observations; these include alterations in centrosome number (Iovino et al. 2006), defects in the spindle assembly checkpoint (Hernando et al. 2004), and formation of micronuclei (Amato et al. 2009), all of which are correlated with abnormal chromosome segregation. In addition, loss of pRB function alters the transcription of several genes with roles during mitosis (such as Aurora A, astrin, Cdc20, Mad2, Nek2, and Ndc80) (Iovino et al. 2006; Chakraborty et al. 2007), and several of these transcriptional changes likely have functional consequences. One potentially important target of pRB is Mad2 (Hernando et al. 2004). Overexpression of Mad2 can cause aneuploidy, and can compromise the regulation of the spindle assembly checkpoint (Sotillo et al. 2007). In contrast, it has also been reported Mad2 levels decrease, rather than increase, in pRB-depleted cells (Amato et al. 2009). Currently, the link between pRB and aneuploidy is poorly understood because there is little data on how the mitotic machinery, which controls chromosome segregation, is altered when pRB is lost.
In this study, we show that the depletion of pRB from nontransformed, primary epithelial cells increases rates of chromosome missegregation to levels comparable with CIN tumor cells. This change occurs despite an intact spindle checkpoint and normal centrosome numbers. We show that the loss of pRB causes defects in centromeric condensation and sister chromatid cohesion, leading to centromere dysfunction during mitosis. The resulting aneuploid cells remain viable and capable of further cell division. Taken together, these observations explain why pRB mutant cells are prone to CIN.
Results
Rb depletion induces CIN
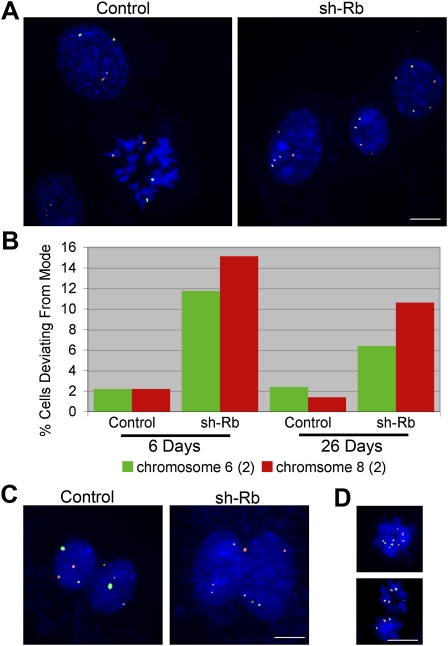
To determine the influence of pRB on chromosome segregation, we used RPE-1 cells, a well-characterized line of h-TERT immortalized human retinal pigment epithelial cells that has a stable, diploid karyotype and is frequently used for studies of mitosis. RPE-1 cells were infected with empty vector, or vector encoding shRNA specific for GFP or pRB (Morris et al. 2008). An efficient and stable knock down of pRB was evident in cells treated with pRB-specific shRNA (sh-Rb) 6 d post-infection (Supplemental Fig. S1A,B). To determine if cells depleted of pRB become aneuploid, we used fluorescence in situ hybridization (FISH) to monitor chromosome numbers. Centromere-specific probes for chromosomes 6 and 8 showed that populations of both control cells and cells depleted of pRB have a modal number of 2. While only 2% of control cells deviated from this mode, 11% of pRB-deficient cells exhibit an abnormal number of chromosome 6, and 15% showed an abnormal number of chromosome 8 (n > 500 cells per population) (Fig. 1A,B). This phenotype was not a transient response to the depletion of pRB. A high level of aneuploidy was maintained over time, and we observed aneuploid cells undergoing mitosis (Fig. 1D, top), indicating that pRB-deficient aneuploid cells are able to proliferate. Within 4 wk of chronic pRB depletion, the vast majority (90%, n = 40 per condition) (Supplemental Fig. S1C) of cells in the sh-Rb population possessed an abnormal karyotype, being near diploid, with the gain or loss of several whole chromosomes.
Figure 1.
Loss of pRB induces CIN. (A,B) FISH analysis with probes for chromosomes 6 and 8 revealed a high degree of aneuploidy in populations of pRB-depleted RPE-1 (sh-Rb) cells that persisted following several weeks of chronic depletion. Both control and sh-Rb populations exhibited a modal copy number of two for both chromosomes 6 and 8. (C) Analysis of individual segregation events revealed high rates of chromosome segregation errors in pRB-depleted cells (0.87% and 0.64% segregation error/division for chromosomes 6 and 8, respectively). (D) Following chronic depletion of pRB, aneuploid cells continued to proliferate (prometaphase cell) (top panel) and to missegregate chromosomes (late anaphase/telophase cell) (bottom panel). Chromosome 6 is shown in green, and chromosome 8 is shown in red throughout. Bar, 5 μm.
These data are consistent with previous studies suggesting that functional loss of pRB leads to aneuploidy (Hernando et al. 2004; Iovino et al. 2006; Isaac et al. 2006; Amato et al. 2009). However, when considered alone, the accumulation of aneuploid cells in a population is not an accurate measure of chromosome missegregation. To measure the frequency of chromosome missegregation, cells were plated at very low densities, and FISH was performed to assess the segregation of chromosomes 6 and 8 in recently divided daughter cells where we could account for each pair of chromosomes. This analysis of individual segregation events showed that cells depleted of pRB missegregate chromosomes 6 and 8 at rates of 0.87% and 0.64% per cell division, respectively (control, n > 1000 segregation events; sh-Rb, n = 462 segregation events) (Fig. 1C). If all chromosomes are similarly affected, this suggests that pRB-depleted cells experience a missegregation event approximately once every six cell divisions. In contrast, we did not observe any chromosome missegregation events in control-treated RPE-1 cells, consistent with previous reports that show chromosome missegregation is extremely rare in RPE-1 cells (one chromosome every 100 divisions) (Thompson and Compton 2008; Ganem et al. 2009).
Taken together, these results demonstrate that the loss of pRB causes the frequent and consistent missegregation of whole chromosomes. Importantly, both the degree of aneuploidy seen in populations of pRB-deficient cells and the rates of chromosome missegregation are similar to well-characterized CIN tumor cell lines (Lengauer et al. 1997; Thompson and Compton 2008). Since pRB is functionally inactivated in many cancer cells, chromosome segregation defects resulting from the loss of pRB are likely to be a major source of the CIN phenotype in human tumors.
Influence of Rb depletion on mitotic progression
To identify defects that can cause the missegregation of whole chromosomes, we examined mitotic progression in pRB-depleted cells. Populations of sh-Rb-treated cells do not show a significant change in overall rate of cell proliferation (Supplemental Fig. S2A; Amato et al. 2009). However, these cells do exhibit a twofold increase in the mitotic index compared with controls (Supplemental Fig. S2B), with a greater proportion of pRB-depleted cells in prometaphase and fewer in metaphase (Supplemental Fig. S2C). This change is consistent with a delay in mitotic progression that has been reported previously (Hernando et al. 2004). Microtubule poisons nocodazole and colcemid induce a strong mitotic arrest in pRB-depleted RPE-1 cells (Supplemental Fig. S2D; data not shown), indicating that the spindle assembly checkpoint is intact. This mitotic arrest was accompanied by the recruitment of BubR1 (a component of the spindle assembly checkpoint) to kinetochores, and analysis of fixed cells showed that BubR1 was depleted from all kinetochores prior to anaphase onset (data not shown), indicating that anaphase does not initiate prematurely in the absence of pRB.
The overduplication of centrosomes has been observed in cells in which the pRB pathway is targeted (Duensing et al. 2001; Iovino et al. 2006), and extra centrosomes have been shown to cause chromosome missegregation by promoting passage through a multipolar spindle intermediate (Iovino et al. 2006; Ganem et al. 2009; Silkworth et al. 2009). To determine whether this phenomenon might underlie chromosome missegregation in pRB-depleted RPE-1 cells, we scored the incidence of multipolar spindles as well as the number of centrioles in these cells. To avoid differences that result indirectly from changes in the mitotic index, we scored nondividing cells and mitotic cells separately. In nondividing cells, the distribution of centriole number was unchanged by pRB depletion (Supplemental Fig. S3A,B). In addition, no increase in multipolar spindle formation and no mitotic cells containing more than two centrioles per spindle pole were observed in pRB-depleted cells (Supplemental Fig. S3C). Nontransformed cells do not tolerate supernumerary centrosomes; consequently, extra centrioles are quickly lost from such cells (Ganem et al. 2009). It is possible that pRB depletion might transiently induce the formation of supernumerary centrosomes. However, at the time during which chromosome segregation events are scored, extra centrosomes are not present and therefore are not the cause of the observed segregation errors.
Centromeric dysfunction in cells lacking pRB
Having excluded defects in the spindle assembly checkpoint and centrosome overduplication as possible sources of the chromosome segregation defects, we noted three clear phenotypes in cells depleted of pRB. Similar changes were seen in cells depleted of pRB by treatment with lentiviral shRNA constructs or with siRNA constructs that target different sequences (see the Materials and Methods).
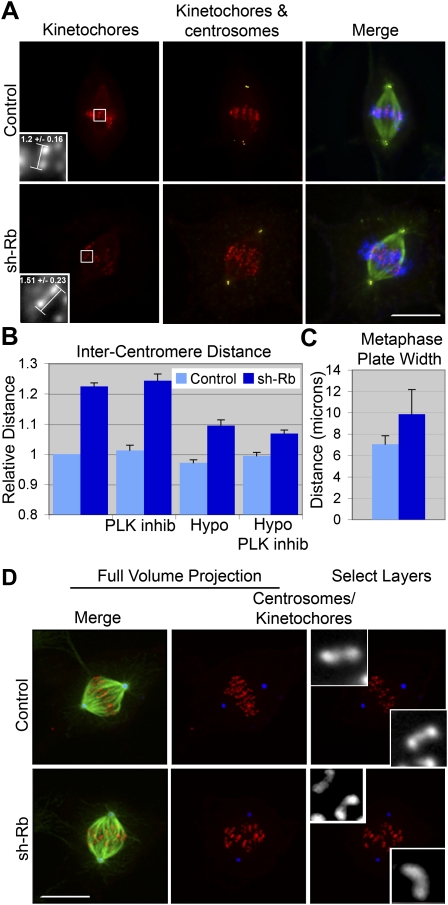
First, pRB-depleted cells have a striking increase (>20%) in intercentromeric distance (1.24 ± 0.09 vs. 1.51 ± 0.07, P < 0.0001) (Fig. 2A,B). This increase is somewhat dependent on the presence of microtubules, since treatments that depolymerize microtubules (hypotonic treatment or addition of nocodazole) partially alleviate this change (Fig. 2B; data not shown). Such treatments failed to restore the intercentromeric distance of pRB-depleted cells to that of control cells, suggesting that the loss of pRB causes a fundamental centromeric defect that is exaggerated by the forces associated with functional kinetochore–microtubule attachments.
Figure 2.
pRB loss causes centromeric dysfunction. (A) RPE-1 cells lacking pRB exhibited >20% increase in intercentromeric distance. (B) This increase was not restored by inhibition of PLK1, but was partially corrected by microtubule depolymerization. (C) Chromosome alignment was also impaired, and the width of the metaphase plate was increased ∼40% in cells lacking pRB. Error bars represent standard error (SEM) of three independent experiments. (D) Centromeres were distorted when pRB was depleted, such that the barbell configuration (staining the kinetochores and associated centromeric region) seen with ACA staining in control cells (insets in top panels) was often bent (insets in bottom panels) in cells lacking pRB. Kinetochores are shown in red, microtubules are shown in green, and centrioles are shown in yellow (A) or blue (D). Similar results were obtained with siRNAs or shRNAs; the data shown were obtained from shRNA-treated cells. Bars, 10 μm. Insets are 4× enlargements.
Second, pRB-depleted cells exhibit a defect in chromosome congression, and chromosomes have difficulty aligning in a tight metaphase plate. Quantitation of this defect showed that the width of the metaphase plate increased 40% in pRB-depleted cells compared with that seen in control cells (7.06 ± 0.78 vs. 9.86 ± 2.31, P < 0.03) (Figs. 2C; Supplemental Fig. S4). Live-cell imaging of RPE-1 cells expressing H2B-GFP (to allow visualization of chromosomes) or RFP-CENPA (to allow visualization of centromeres) supported fixed cell analysis and confirmed that chromosomes in cells depleted of pRB congress to a disorganized metaphase plate prior to anaphase onset (data not shown). Lack of kinetochore microtubule attachment and chromosome biorientation could prevent proper chromosome congression. However, pRB-depleted cells form calcium-stable microtubules similarly to control cells, suggesting they are proficient to form stable kinetochore–microtubule interactions, and chromosomes are capable of biorientation (Supplemental Fig. S4B). Recently, kinetochore congression and alignment within the metaphase plate have been shown to be influenced by centromere stiffness (Jaqaman et al. 2010), supporting the idea that centromere structure may be compromised in pRB-depleted cells.
Third, a close analysis of sister kinetochore pairs revealed that, while properly bioriented chromosomes align with sister kinetochores oriented along the pole-to-pole axis, bioriented sister kinetochores in cells lacking pRB often deviate from this axis (Fig. 2D). This phenotype has been reported for merotelically attached chromosomes where one or both sister kinetochores are associated with microtubules emanating from both spindle poles (Draviam et al. 2006). Merotelic attachments are a common feature of CIN cells (Thompson and Compton 2008), and are not sensed by the spindle assembly checkpoint. If left uncorrected, these attachment errors give rise to lagging chromosomes during anaphase and missegregation events (Cimini et al. 2001, 2003). Staining of sister kinetochore pairs with the anti-centromeric antigen (ACA) labels sister kinetochores and the adjoining centromeric region in a barbell pattern. In pRB-depleted cells, the normal barbell pattern is dramatically bent at many kinetochore pairs (Fig. 2D, inset). This type of centromeric distortion is reminiscent of the distortion seen with merotelic kinetochore attachments (Cimini et al. 2003; Draviam et al. 2006).
Taken together, these observations show that there is an underlying defect in centromeric structure in pRB-depleted cells in which decreased centromeric rigidity allows for improper kinetochore–microtubule attachments and defects in congression.
pRB loss influences sister chromatid cohesion
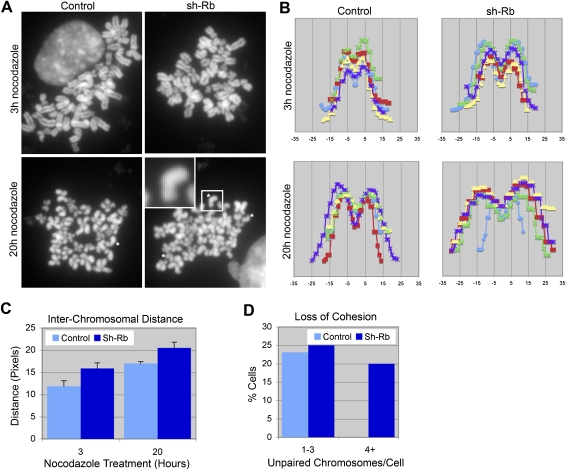
The depletion of cohesin complex components has been shown to cause defects in centromeric structure and function that are similar to the properties of pRB-depleted cells (Kenney and Heald 2006; Jaqaman et al. 2010). To determine whether the chromosomal abnormalities seen in pRB-depleted cells might also lead to defects in chromosome cohesion, we examined metaphase chromosome spreads. Cells treated with shRNA specific for pRB were found to have an increased incidence of premature loss of sister chromatid cohesion (Fig. 3A,D). Furthermore, arms of sister chromatids that maintained cohesion in pRB-depleted cells were found to be further apart than in control cells (Fig. 3B). Strikingly, both of these defects were exacerbated by prolonged mitotic arrest (Fig. 3A [bottom panel], C). These results suggest that chromosome cohesion is impaired in pRB-depleted cells, and that sister chromatids are prone to separate in the absence of pRB, particularly when cells are delayed in mitosis.
Figure 3.
pRB loss disrupts sister chromatid cohesion. (A) Cells depleted of pRB (shRb) have an increased incidence of premature loss of sister chromatid cohesion (examples of separated sister are indicated by asterisks [*]). (B) Line scans through arms of sisters that maintain cohesion revealed an increase in interchromosomal distance in response to pRB loss. Each colored line represents a line scan through the arms of paired sister chromatids, with each chart depicting five line scans within a single representative cell. Prolonged mitotic arrest increased the interchromosomal distance (C) (error bars represent standard error [SEM]; P < 0.05 between control and sh-Rb cells at both 3 and 20 h) and promoted the complete loss of cohesion (D), as measured by the number of cells with one to three or four or more unpaired chromosomes.
Centromeric defects promote chromosome malattachment and segregation errors
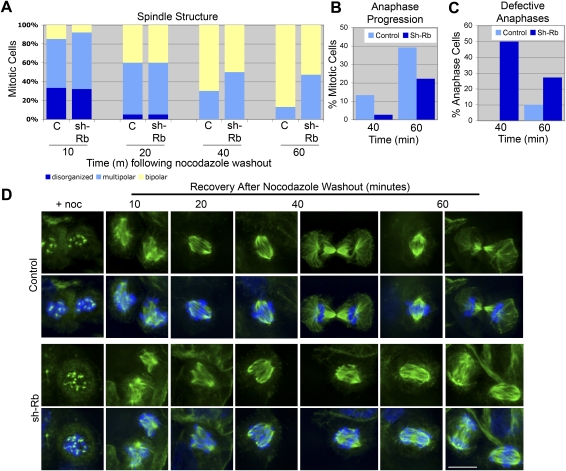
To test the idea that the centromeric defects of pRB-deficient cells can compromise mitotic fidelity, we examined progression through mitosis following release from a mitotic arrest. Completion of mitosis following release from nocodazole-induced microtubule depolymerization promotes transit through a multipolar spindle intermediate, which results in formation of syntelic and merotelic kinetochore attachments (Cimini et al. 2001). Because merotelic attachments are not sensed by the spindle assembly checkpoint, cells may enter anaphase without correcting such erroneous attachments, as evidenced by lagging chromosomes (Cimini et al. 2001). In this sensitized system, cells lacking pRB exhibit a delay in the formation of a bipolar spindle, a delay in anaphase progression, an increased number of anaphase cells exhibiting lagging chromosomes, and an increased number of laggards per cell (Fig. 4).
Figure 4.
pRB loss compromises the fidelity of mitotic progression. Formation of a bipolar spindle (A,D), and progression into anaphase (B) following release from nocodazole-induced mitotic arrest is delayed when pRB is depleted (shRb). (A) The state of spindle organization is characterized as disorganized microtubule arrays (dark blue), multipolar spindles (light blue), and bipolar spindles (yellow). Numbers represent percentage of mitotic cells exhibiting specified spindle structure. P < 0.01 for differences in spindle structure and anaphase progression at time points 40 and 60 min between control and shRb samples. (C) pRB-depleted cells that enter anaphase exhibit an increase in the incidence lagging chromatids over control cells. (D) Examples of representative spindle structure at specified time points following nocodazole washout. Microtubules are shown in green, and DNA is shown in blue. Bar, 10 μm.
As described above, the depletion of pRB promotes premature loss of sister chromatid cohesion, and this defect is enhanced by prolonged mitotic delay (Fig. 3). We therefore expected that individual unpaired kinetochores should be apparent by immunofluorescence following release from nocodazole arrest. Indeed, single kinetochores located near spindle poles (Supplemental Fig. S5A) were evident in pRB-deficient cells but not control cells following nocodazole washout. Under these experimental conditions, spindle formation occurs following drug washout, after chromatid cohesion has been lost between some sister pairs, and lone chromatids are unable to efficiently biorient and align at the spindle equator. However, such lone kinetochores were not routinely observed near spindle poles in control or pRB-depleted cells that had not been subjected to prolonged mitotic arrest. We infer that the reduced level of sister chromatid cohesion seen in pRB-depleted cells is sufficient for chromosome biorientation. Although sister chromatids are more likely to separate when pRB-depleted cells are delayed in mitosis, weakened sister chromatid cohesion also promotes merotelic attachment, and this may allow single chromosomes to maintain alignment that is achieved prior to loss of cohesion (Supplemental Fig. S5B,C). The delays in mitotic progression and the increase in lagging chromosomes following release from a nocodozole arrest indicate that pRB-depleted cells are especially sensitive to conditions that promote merotelic attachments.
pRB loss disrupts centromeric cohesin and condensin II localization
In vertebrate cells, components of both the cohesin and condensin complexes are important for normal centromeric structure and function (Ono et al. 2004; Kenney and Heald 2006; Samoshkin et al. 2009). Cohesin and condensin complexes first associate with DNA during telophase (for review, see Peters et al. 2008). Cohesin is the primary determinant of sister chromatid cohesion, and cohesion is established during S phase concurrent with DNA replication. As cells progress through mitosis, cohesin complexes are removed in two steps. During the prophase-to-metaphase transition, cohesin is first removed from chromosome arms by phosphorylation of cohesin subunits. Subsequently, during the metaphase-to-anaphase transition, cohesin is removed from the centromeric region by cleavage of the Rad21/Scc1 subunit (Peters et al. 2008).
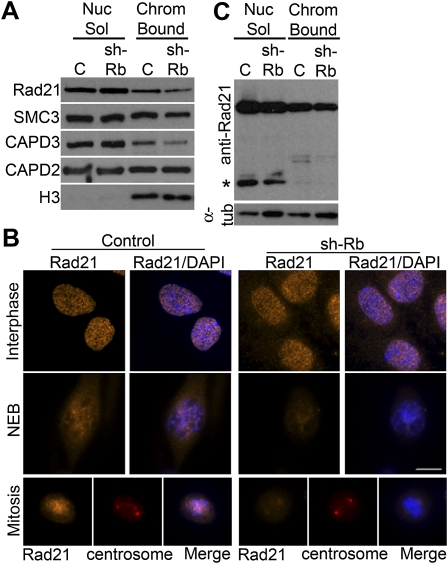
The overall levels of the cohesin proteins Rad21 and SMC3 were unchanged in pRB-depleted cells, and the levels of chromatin-associated Rad21 and SMC3 were only slightly reduced when pRB was depleted (Fig. 5A; Supplemental Fig. S6A). This is consistent with the observation that the degree of chromatid cohesion in these cells is sufficient to maintain cohesion upon mitotic entry. However, centromeric-localized cohesin represents only ∼10% of all cohesin complexes (Peters et al. 2008), and centromere-specific changes in cohesin localization may cause only a minor change in the overall pool of chromatin-associated protein. Using immunofluorescence, we observed that the punctate pattern of Rad21 seen in cells that have undergone nuclear envelope breakdown was specifically reduced in pRB-depleted cells (Fig. 5B).
Figure 5.
pRB depletion impairs cohesin localization. (A) Western blot analysis shows that the chromatin-bound fraction of cohesin components Rad21 and SMC3 and the condensin II component CAP-D3, but not the condensin I component CAP-D2, were reduced in pRB-depleted (shRb) cells. (B) Punctate Rad21 staining was apparent in control cells after nuclear envelope breakdown (NEB) and during mitosis, but was dramatically reduced in cells depleted of pRB. Rad21 is shown in orange, chromosomes are shown in blue, and centrosomes are shown in red. Bar, 10 μm. (C) The levels of cleaved Rad21 (indicated by an asterisk [*]) did not increase in shRb-depleted cells.
Polo-like kinase 1 (PLK1) promotes the removal of cohesin during the transition to metaphase (Lenart et al. 2007). Since PLK1 is an E2F1 target gene and its expression has been shown to increase in the absence of pRB (Amato et al. 2009), we tested whether inhibition of PLK1 could rescue the effects of pRB depletion. Addition of the PLK1 inhibitor BI 2536 (Chemie Tek) induced a strong mitotic arrest and prevented loss of sister chromatid cohesion in both control and pRB-depleted cells. However, PLK1 inhibition did not suppress the increased intercentromeric distance seen in pRB-depleted cells (Fig. 2C), suggesting that this defect is not due to an inappropriate PLK1-dependent removal of cohesin. Because centromeric cohesin is removed by Separase-dependent cleavage of the Rad21 subunit, we compared the levels of cleaved Rad21 in control and pRB-depleted RPE-1 cells. No increase in Rad21 cleavage was observed in pRB-depleted cells (Fig. 5C), suggesting that the reduction in centromeric cohesin is more likely due to a defect in the loading or maintenance of cohesin at the centromere rather than to excessive Rad21 cleavage.
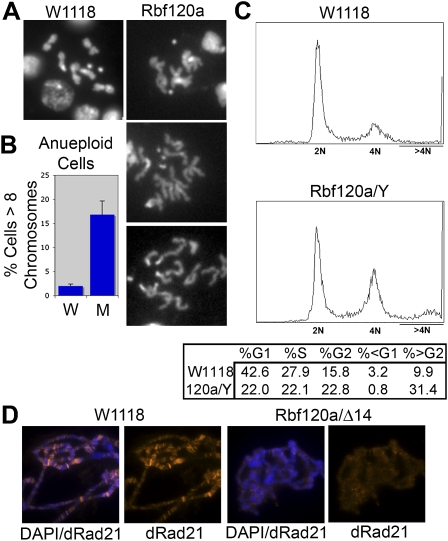
To test whether the link between pRB and sister chromatid cohesion might be conserved during evolution, we compared neuroblasts of wild-type Drosophila larvae with larvae that are strongly hypomorphic for the pRB homolog RBF1. As in pRB-depleted human cells, rbf1120 mutant animals exhibit defects in sister chromatid cohesion and an increase in aneuploidy (Fig. 6A–C). Consistent with the idea that pRB family members promote loading of the cohesin complex onto DNA, immunostaining experiments show reduced dRad21 banding on the polytene chromosomes of rbf1120/Δ14 mutant larvae (Fig. 6D). Interestingly, defects in sister chromatid cohesion have also been observed in mouse embryonic fibroblasts lacking the three Rb family proteins (TKO MEFs) (van Harn et al. 2010).
Figure 6.
Loss of RBF1 in Drosophila causes chromatid cohesion defects and aneuploidy, similar to that seen in pRB-depleted human cells. (A,B) Mitotic chromosome spreads from dissected larval neuroblasts show frequent loss of sister chromatid cohesion (A) and increased incidence of aneuploidy (B). (W) Wild type; (M) rbf1 mutant. Error bars represent standard error (SEM) of three individual animals per condition. (C) FACs analysis of whole larval neuroblasts from wild-type and rbf1 mutant animals similarly reveals that the rbf1 mutants exhibit an increase in aneuploid cells. (D) dRad21 banding on salivary gland polytene chromosomes is decreased in rbf1 mutant animals.
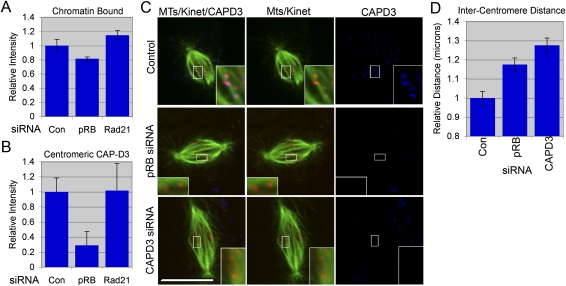
Our previous studies using Drosophila showed that RBF1 interacts physically with the condensin II subunit dCAP-D3 and promotes its association with chromatin (Longworth et al. 2008). This physical interaction is conserved in mammalian cells, and requires the LXCXE-binding cleft of pRB; however, the significance of this interaction in mammalian cells was unclear. Consistent with the published data, fractionation experiments showed that the level of chromatin-associated CAP-D3 (condensin II) was reduced in pRB-depleted cells (Figs. 5A, 7A; Supplemental Fig.S6C). In comparison, the level of chromatin-associated CAP-D2 (condensin I) was unaffected by pRB depletion (Fig. 5A). Intriguingly, condensin II complexes have been shown to be enriched in the centromeric region of mitotic chromosomes (Ono et al. 2004). We therefore tested whether pRB might be required specifically for normal centromeric recruitment of CAP-D3, and whether the loss of CAP-D3 could cause defects in centromere function similar to pRB-depleted cells.
Figure 7.
Centromeric localization of condensin II is compromised in pRB-depleted cells. (A) Chromatin-associated CAP-D3 is reduced in cells depleted of pRB by siRNA, but not in cells depleted of Rad21 (control vs. pRB depletion: P < 0.0001) (quantification of immunofluorescence from Supplemental Fig. 6). (B,C) Centromeric CAP-D3 localization is reduced in pRB-depleted cells to <30% of that seen in control cells (control vs. pRB depletion: P < 0.0001). Microtubules are shown in green, kinetochores are shown in red, and CAP-D3 is shown in blue. (D) The depletion of CAP-D3 causes an increase in intercentromeric distance similar to that observed in pRB-depleted cells. All error bars represent standard error (SEM) of three independent experiments.
Immunofluorescence experiments confirmed that CAP-D3 is strongly enriched at the centromere of control RPE-1 cells. This localization was strikingly reduced in cells treated with pRB-specific siRNA, but not cells depleted of Rad21 (Fig. 7B,C). To assess the functional significance of reduced CAP-D3, RPE-1 cells were treated with siRNAs specific for pRB, CAP-D2, or CAP-D3, and examined for centromeric defects. pRB- and CAP-D3-depleted cells showed a similar increase in intercentromeric distance that was not evident in control-treated cells, nor in cells specifically depleted of CAP-D2 (Fig. 7C,D). Previous studies have shown that the depletion of CAP-D3 enhances the loss of sister chromatid cohesion in a similar manner to that reported here in pRB-depleted cells (Hirota et al. 2004). Taken together, our results show that pRB-depleted cells have a defect in chromosome cohesion, that this defect is most evident at the centromere, and that the defects in centromere function appear to be caused by a reduction in the binding and/or regulation of condensin II and cohesin complexes.
Discussion
The loss of pRB causes a consistent, high rate of chromosome segregation errors, leading to the generation of aneuploid, chromosomally unstable cells. The results described here reveal why this occurs: The loss of pRB compromises centromere function and cohesion, and these changes undermine the fidelity of mitosis.
pRB, CIN, and tumorigenesis
Some cancer cells have a near-tetraploid karyotype, suggesting that missegregation of single chromosomes occurs in conjunction with, or is facilitated by, doubling of the genome (through cell fusion, endoreduplication, or failure of cytokinesis, etc.). However, most human tumor cells are near-diploid (Weaver and Cleveland 2006), and seem unlikely to have progressed through a tetraploid intermediate. We show that loss of pRB results in near-diploid anueploid cells, and that the rate of chromosome missegregation resulting from the inactivation of pRB is remarkably similar to that measured in human tumor cells. Given that the pRB pathway is disrupted in most tumor cells, the idea that pRB inactivation causes CIN provides a simple explanation for the prevalence of chromosome missegregation in human cancer. pRB inactivation may be especially relevant during tumorigenesis because its loss both increases segregation errors and compromises cell cycle arrest. Previous studies have focused on the role of pRB in G1/S-phase progression, but understanding the basis for chromosome segregation defects in pRB-deficient cells is equally important: This information gives insight into the processes and pathways that are misregulated in human tumors. Moreover, the characterization of these defects may reveal ways to restore chromosome stability, thereby reducing metastatic potential. Conversely, treatments that can further exaggerate these changes may cause mitotic catastrophe and eventual death in cells that lack pRB function.
Rb loss: a subtle defect that undermines the fidelity of mitosis
Here we show that pRB loss causes defects in centromere function and reduces sister chromatid cohesion. These changes promote merotelic attachment and increase the incidence of unpaired sister chromatids. While the complete loss of cohesion would have catastrophic consequences, moderate defects in centromeric condensation and cohesion have been shown to cause chromosome segregation errors in tissue culture and various model systems (for review, see Nasmyth and Haering 2009). Hence, the functional inactivation of pRB is a subtle enemy during tumorigenesis because it reduces the fidelity of mitosis without causing more dramatic changes that would compromise cell proliferation.
pRB is necessary for the proper action of condensin II and cohesin components at the centromere. Condensins and cohesins are both required for normal centromere structure and function. Failure to recruit these components leads to a decrease in rigidity between sister centromeres and promotes merotelic kinetochore attachment—defects that are evident in pRB-depleted cells in the increased intercentromeric distance, deformation of centromeric structure, failure of chromosome congression, and increased propensity for lagging chromosomes following mitotic arrest. This combination of centromeric cohesion defects and merotelic kinetochore orientation is highly consistent with studies showing the importance of pericentromeric cohesion in promoting kinetochore biorientation (Ng et al. 2009). Many studies have shown that merotelic attachments are a dominant cause of chromosome missegregation in CIN tumor cell lines (Cimini et al. 2001; Thompson and Compton 2008; Ganem et al. 2009). Given the frequency of lesions in the pRB pathway, and the evidence that pRB loss promotes merotelic attachments, it is likely that much of the CIN in tumors is initiated by functional inactivation of pRB. This underscores an important caveat to the widespread use of human tumor cells in the study of normal mitotic progression and chromosome segregation.
The link between pRB and chromosome segregation
We suggest that the mitotic phenotype of pRB-deficient cells may have multiple components. First, as illustrated here, are the changes resulting from the physical connection between pRB and condensin II. pRB is required for the concentration of CAP-D3 at the centromere, and the depletion of either pRB or CAP-D3 gives similar centromeric defects. These observations are consistent with evidence that condensin II proteins are necessary for centromere function (Hirota et al. 2004; Samoshkin et al. 2009), that pRB associates physically with CAP-D3, and that pRB can promote the association of CAP-D3 with chromatin as cells exit mitosis and enter G1 (Longworth et al. 2008). Taken together, these results suggest that the interaction between pRB and CAP-D3 is important for normal centromere function.
Second, the centromeric defects of pRB-depleted cells are associated with reduced cohesion. Since condensation and cohesion are interrelated processes, it is possible that the cohesion defects are secondary to reduced condensin II activity. Indeed, the depletion of CAP-D3 has been shown to enhance the loss of sister chromatid cohesion in a manner similar to that reported here (Hirota et al. 2004). Additionally, we note that the tight association of cohesin with DNA and the establishment of sister chromatid cohesion are intimately linked to the initiation of DNA replication (Terret et al. 2009; for review, see Nasmyth and Haering 2009). The loss of pRB not only alters the expression of replication proteins, but also changes the spatial organization of DNA synthesis within the nucleus (Barbie et al. 2004), a change that may also impact the loading of cohesin and the maintenance of condensin II complexes. The changes described here add to the emerging view that defects in chromosome cohesion are important in human disease. Mutations in cohesin components and mutations in regulators of cohesin association with DNA have been identified in various human disorders, including Cornelia de Lange syndrome (CdLS) and Roberts syndrome (RBS) (Mannini et al. 2009), as well as colorectoral cancer (Barber et al. 2008). Interestingly, many of the mutations identified in human disease do not alter cohesin protein levels, but instead affect the loading of the cohesin complex or change its affinity for DNA (Mannini et al. 2009; Revenkova et al. 2009).
Third, it is important to remember that the centromere defects resulting from the loss of pRB do not occur in a normal cell, but in a cell in which the deregulation of E2F has caused numerous mitotic proteins to be misexpressed. Unlike the change seen when pRB or CAP-D3 are depleted, the overexpression of E2F1 did not increase the intercentromeric distance, even though it increased the expression of several E2F-regulated genes (Supplemental Fig. S6F–I). This suggests that changes seen in centromere structure and function are not directly caused by elevated E2F activity. However, we note that the deregulation of E2F may enhance the importance of these structural changes. Several groups have shown that the length of mitosis increases in pRB-deficient cells (Hernando et al. 2004; Amato et al. 2009). This mitotic delay has been suggested to be due to an E2F-dependent up-regulation of Mad2 and hyperactivity of the spindle assembly checkpoint (Hernando et al. 2004). Although the centromeric defects described are not apparently caused by the deregulation of E2F, and therefore are unlikely to be closely connected to the up-regulation of Mad2, the finding that pRB-deficient cells have defects in cohesion that are exaggerated by prolonged mitosis may help to explain why such a delay can have important consequences. One could argue that the inactivation of pRB has two synergistic effects: a condensation defect that weakens sister chromatid cohesion and promotes merotelic attachment, and a mitotic delay that enhances these defects.
In our previous studies, we found that all three pRB family members are able to interact with CAP-D3 (Longworth et al. 2008). The up-regulation of p107 and p130 has been shown to allow them to compensate for the absence of pRB, although these effects are often tissue- or context-specific (Pacal and Bremner 2006; Burkhart and Sage 2008). We know that the depletion of pRB causes centromeric defects and aneuploidy in nontransformed cells as diverse as RPE1 epithelial cells and IMR90 fibroblasts (data not shown); however, it remains to be determined whether p107 and/or p130 also influence centromere function and the fidelity of chromosome segregation, and whether there are specific tissues or contexts in which these family members impact the degree of chromosome missegregation resulting from the inactivation of pRB.
Loss of pRB promotes genomic instability
Genomic instability has been implicated recently in the development of retinoblastoma (Dimaras et al. 2008), and the results reported here outline one way in which the inactivation of pRB promotes genetic change. Consistent with these observations, Coschi et al. (2010) found that mutation of the LXCXE cleft, which reduces the ability of pRB to interact with CAP-D3, enhances tumorigenesis and genomic instability in mouse tumor models. It is tempting to speculate that the elevated levels of DNA damage observed in pRB-deficient cells (Pickering and Kowalik 2006) and in pRB, p107, p130 TKO cells (van Harn et al. 2010)) may further increase the likelihood that cells will enter mitosis with damaged DNA and activate mitotic checkpoints (Mikhailov et al. 2002). While further studies are clearly needed to characterize the full extent of the changes resulting from reduced condensin II and cohesin function in cells lacking pRB, work presented here delineates an important and clinically relevant mechanistic role for pRB in the maintenance of genome stability.
Materials and methods
Cell culture and RNAi
hTERT-RPE-1 cells were grown in Dulbecco's Modified Essential Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were infected with LLP empty vector, LLP-shGFP, or LLP-shRbCD (targeting sequence GGTTGTGTCGAAATTGGATCA) as described previously (Morris et al. 2008); placed under puromycin selection for 4–5 d; and analyzed at day 6 or later as specified. For all experiments involving the shRNA depletion of pRB (indicated as shRb), cells infected with empty vector and cells infected with a shGFP-containing vector were both used as controls.
For experiments in which the short-term depletion of targets was achieved using siRNAs, cells were transfected with RNAi MAX (Invitrogen) according to the manufacturer's directions, with 50 nM pool of four pRB-specific siRNA constructs (Dharmacon Smartpool; target sequence 1, CGAAAUCAGUGUCCAUAAA; target sequence 2, GAGUUGACCUAGAUGAGAU; target sequence 3, AAACUACGCUUUGAUAUUG; target sequence 4, GAAUCUGCUUGUCCUCUUA), a CAPD2-specific siRNA construct (CCAUAUGCUCAGUGCUACA) (Hirota et al. 2004), a CAPD3-specific siRNA construct (CAUGGAUCUAUGGAGAGUA) (Hirota et al. 2004), or a Rad21-specific siRNA construct (GGUGAAAAUGGCAUUACGG) (Watrin et al. 2006). mRNA samples were collected and cells were fixed and stained for analysis as described below, 36–40 h after siRNA transfection. For consistency, in the experiments where cohesin and condensin components were depleted by siRNA, we also used siRNAs to deplete pRB (indicated as siRb in the text). As described for the shRNA treatments, the treatment with siRb strongly decreased the level of pRB mRNA and the level of pRB protein detected by immunofluorescence. As expected, the depletion of pRB increased the levels of pRB/E2F-regulated mRNAs p107 and SMC2 (Supplemental Fig. S4A). To test the effects of E2F1 overexpression, RPE-1 cells were transfected with HA-E2F1 and examined 36 h later. Real-time quantitative PCR was performed to determine relative mRNA levels as published previously (Morris et al. 2008).
Drosophila stocks
All fly stocks and crosses were maintained at 25°C. Female progeny of the Rbf1120a/Δ14 and male progeny of the Rbf1120a stocks were analyzed for mitotic defects, chromosome structure, and aneuploidy.
FISH, chromosome spreads, drug treatment, and cell cycle arrest
Cells were prepared and fixed, and α-satellite-specific probes for chromosomes 6 and 8 (Cytocell) were hybridized according to protocols in Thompson and Compton (2008). Chromosome spreads were prepared as described in Ganem et al. (2009). Cells were treated with 100 ng/mL nocodazole or 100 ng/mL colcemid for 5 h for karyotype analysis, or for indicated times for analysis of sister chromatid cohesion. To measure interchromosomal distances, line scans to measure pixel intensity were performed on five chromosomes in each of 10 cells per condition. Centriole numbers were counted in nonproliferating cells that were grown to confluency. The lack of proliferation was confirmed by the appearance of confluency and the lack of mitotic cells, as determined by phase-contrast imaging prior to fixation, and by microtubule and chromatin structure following fixation. For nocodazole washout experiments, cells were treated with 100 ng/mL nocodazole for 16 h, then washed three times in drug-free medium and fixed as described below at the indicated time points. Mitotic cells were staged and spindle structure was analyzed according to DAPI and α-tubulin staining. More than 50 mitotic cells were scored for spindle structure and mitotic progression for each condition and at each time point in three independent experiments. Dissected third instar larval Drosophila neuroblasts were treated in 1 mM colchicine for 2 h prior to fixation in 4% paraformaldehyde for 30 min. Brains were then incubated sequentially in ice-cold methanol for 2 min and 45% acetic acid for 3 min, squashed between slide and coverslip in 60% acetic acid, and frozen in liquid nitrogen. Chromosome numbers were scored for at least 100 mitotic cells for each of three neuroblasts of w1118 and rbf1 mutant animals.
Immunofluorescence microscopy
Cells were extracted in either microtubule-stabilizing buffer (4 M glycerol, 100 mM PIPES at pH 6.9, 1 mM EGTA, 5 mM MgCl2, 0.5% Triton X-100) or calcium-containing buffer (100 mM PIPES at pH 6.8, 1 mM MgCl2, 0.1% Triton X-100, 1 mM CaCl2), followed by fixation in 1% gluteraldehyde (microtubule staining: dm1α [Sigma]) or cold methanol (ACA [Antibodies, Inc.], BubR1 [Abcam], CAPD2 [Bethyl Laboratories], CAPD3 [Bethyl Laboratories], CENP-A [Cell Signaling], Centrin-2 [Santa Cruz Biotechnology], Rad21 [Abcam], and pRB [Cell Signaling and BD Pharmigen]). Alternatively, cells were fixed in cold methanol without additional pre-extraction. Subsequent antibody incubations and washes were done in TBS-BSA (10 mM Tris at pH 7.5, 150 mM NaCl, 1% bovine serum albumin). DNA was detected with 0.2 μg/mL DAPI (Sigma-Aldrich). Coverslips were mounted with ProLong Antifade mounting medium (Molecular Probes). Fluorescent images of fixed and live cells were captured with a Hamamatsu Orca AG cooled CCD camera mounted on a Nikon TI/Yokagawa CSU-10 spinning-disk confocal microscope with a 100×, 1.4 NA objective or a Hamamatsu EM CCD camera mounted on an Olympus IX81 microscope with a 100×, 1.4 NA objective. A series of 0.25-μm optical sections were collected in the Z-axis for each channel (DAPI, fluorescein, Cy5, and/or Texas red). Iterative restoration was performed on images using NIS elements for images acquired on the Nikon. Selected planes from the Z-series were then overlaid to generate the final image. Mitotic index and mitotic progression were quantified by immunofluorescence using chromatin compaction (DAPI stain) and spindle structure (α-tubulin) as indicators of mitotic state. For calculation of the mitotic index, n > 5000 cells per condition were used. For evaluation of mitotic progression, n > 300 mitotic cells were examined per condition. Measurements of intercentromere distances and metaphase plate width were made with Slidebook analysis software; line scans measuring interchromosomal distances were acquired with NIS elements software. Measurements of chromatin-bound or centromere-localized cohesin and condensin components by immunofluorescence were made with Slidebook software by selecting nuclei based on DAPI staining or kinetochores/centromeres based on ACA staining, measuring pixel intensities in >50 selected areas for each condition, and normalizing all intensities to average intensities in control cells. All measurements were performed for three independent experiments. All error bars represent standard errors (SEM). The Student's t-test was used to calculate the significance of differences between samples.
Acknowledgments
We thank David Pellman for RPE-1 cell lines, Terry Orr-Weaver for the dRad21 antibodies, Anders Naar for help with microscopy, Neil Ganem for critical reading of the manuscript, and Mathew Meyerson, Tanja van Harn, Hein te Riele, and Fred Dick for sharing their unpublished results. A.L.M. is supported by a post-doctoral fellowship from the American Cancer Society; M.S.L. is supported by fellowships from the Leukemia and Lymphoma Society and the Charles King Trust. This work was supported by NIH grants R01CA064402 and R01GM081607 to N.J.D.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1917310.
Supplemental material is available at http://www.genesdev.org.
References
- Albertson DG, Collins C, McCormick F, Gray JW 2003. Chromosome aberrations in solid tumors. Nat Genet 34: 369–376 [DOI] [PubMed] [Google Scholar]
- Amato A, Lentini L, Schillaci T, Iovino F, Di Leonardo A 2009. RNAi mediated acute depletion of retinoblastoma protein (pRb) promotes aneuploidy in human primary cells via micronuclei formation. BMC Cell Biol 10: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jin F, Jeganathan KB, van Deursen JM 2009. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell 16: 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA 2009. Genome stability is ensured by temporal control of kinetochore–microtubule dynamics. Nat Cell Biol 11: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, et al. 2008. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci 105: 3443–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Kudlow BA, Frock R, Zhao J, Johnson BR, Dyson N, Harlow E, Kennedy BK 2004. Nuclear reorganization of mammalian DNA synthesis prior to cell cycle exit. Mol Cell Biol 24: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black EP, Hallstrom T, Dressman HK, West M, Nevins JR 2005. Distinctions in the specificity of E2F function revealed by gene expression signatures. Proc Natl Acad Sci 102: 15948–15953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookstein R, Lee WH 1991. Molecular genetics of the retinoblastoma suppressor gene. Crit Rev Oncog 2: 211–227 [PubMed] [Google Scholar]
- Burkhart DL, Sage J 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 8: 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B 1998. Mutations of mitotic checkpoint genes in human cancers. Nature 392: 300–303 [DOI] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z 2006. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 38: 1043–1048 [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Khare S, Dorairaj SK, Prabhakaran VC, Prakash DR, Kumar A 2007. Identification of genes associated with tumorigenesis of retinoblastoma by microarray analysis. Genomics 90: 344–353 [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED 2001. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol 153: 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Moree B, Canman JC, Salmon ED 2003. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci 116: 4213–4225 [DOI] [PubMed] [Google Scholar]
- Coschi CH, Martens AL, Ritchie K, Francis SM, Chakrabarti S, Berube NG, Dick FA 2010. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev (this issue). doi: 10.1101/gad.1917610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaras H, Khetan V, Halliday W, Orlic M, Prigoda NL, Piovesan B, Marrano P, Corson TW, Eagle RC Jr, Squire JA, et al. 2008. Loss of RB1 induces non-proliferative retinoma: Increasing genomic instability correlates with progression to retinoblastoma. Hum Mol Genet 17: 1363–1372 [DOI] [PubMed] [Google Scholar]
- Draviam VM, Shapiro I, Aldridge B, Sorger PK 2006. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC-depleted cells. EMBO J 25: 2814–2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duensing S, Duensing A, Crum CP, Munger K 2001. Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res 61: 2356–2360 [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature 460: 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Furge K, Koeman J, Dykema K, Su Y, Cutler ML, Werts A, Haak P, Vande Woude GF 2007. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci 104: 8995–9000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000. The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, et al. 2004. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430: 797–802 [DOI] [PubMed] [Google Scholar]
- Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM 2004. Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci 117: 6435–6445 [DOI] [PubMed] [Google Scholar]
- Iovino F, Lentini L, Amato A, Di Leonardo A 2006. RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Mol Cancer 5: 38 doi: 10.1186/1476-4598-5-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac CE, Francis SM, Martens AL, Julian LM, Seifried LA, Erdmann N, Binne UK, Harrington L, Sicinski P, Berube NG, et al. 2006. The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol 26: 3659–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman K, King EM, Amaro AC, Winter JR, Dorn JF, Elliott HL, McHedlishvili N, McClelland SE, Porter IM, Posch M, et al. 2010. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J Cell Biol 188: 665–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney RD, Heald R 2006. Essential roles for cohesin in kinetochore and spindle function in Xenopus egg extracts. J Cell Sci 119: 5057–5066 [DOI] [PubMed] [Google Scholar]
- Kuukasjarvi T, Karhu R, Tanner M, Kahkonen M, Schaffer A, Nupponen N, Pennanen S, Kallioniemi A, Kallioniemi OP, Isola J 1997. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res 57: 1597–1604 [PubMed] [Google Scholar]
- Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM 2007. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol 17: 304–315 [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B 1997. Genetic instability in colorectal cancers. Nature 386: 623–627 [DOI] [PubMed] [Google Scholar]
- Longworth MS, Herr A, Ji JY, Dyson NJ 2008. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev 22: 1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannini L, Liu J, Krantz ID, Musio A 2009. Spectrum and consequences of SMC1A mutations: The unexpected involvement of a core component of cohesin in human disease. Hum Mutat 31: 5–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markey MP, Angus SP, Strobeck MW, Williams SL, Gunawardena RW, Aronow BJ, Knudsen ES 2002. Unbiased analysis of RB-mediated transcriptional repression identifies novel targets and distinctions from E2F action. Cancer Res 62: 6587–6597 [PubMed] [Google Scholar]
- Marshall CJ 1991. Tumor suppressor genes. Cell 64: 313–326 [DOI] [PubMed] [Google Scholar]
- Mayhew CN, Carter SL, Fox SR, Sexton CR, Reed CA, Srinivasan SV, Liu X, Wikenheiser-Brokamp K, Boivin GP, Lee JS, et al. 2007. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology 133: 976–984 [DOI] [PubMed] [Google Scholar]
- Mikhailov A, Cole RW, Rieder CL 2002. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr Biol 12: 1797–1806 [DOI] [PubMed] [Google Scholar]
- Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, Kwon EJ, Haigis KM, Naar AM, Dyson NJ 2008. E2F1 represses β-catenin transcription and is antagonized by both pRB and CDK8. Nature 455: 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH 2009. Cohesin: Its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Ng TM, Waples WG, Lavoie BD, Biggins S 2009. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol Biol Cell 20: 3818–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA 2002. Centrosome aberrations: Cause or consequence of cancer progression? Nat Rev Cancer 2: 815–825 [DOI] [PubMed] [Google Scholar]
- Nowell PC 1976. The clonal evolution of tumor cell populations. Science 194: 23–28 [DOI] [PubMed] [Google Scholar]
- Ono T, Fang Y, Spector DL, Hirano T 2004. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell 15: 3296–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacal M, Bremner R 2006. Insights from animal models on the origins and progression of retinoblastoma. Curr Mol Med 6: 759–781 [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J 2008. The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114 [DOI] [PubMed] [Google Scholar]
- Pickering MT, Kowalik TF 2006. Rb inactivation leads to E2F1-mediated DNA double-strand break accumulation. Oncogene 25: 746–755 [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Lengauer C 2004. Aneuploidy and cancer. Nature 432: 338–341 [DOI] [PubMed] [Google Scholar]
- Rasnick D, Duesberg PH 1999. How aneuploidy affects metabolic control and causes cancer. Biochem J 340: 621–630 [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Focarelli ML, Susani L, Paulis M, Bassi MT, Mannini L, Frattini A, Delia D, Krantz I, Vezzoni P, et al. 2009. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum Mol Genet 18: 418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samoshkin A, Arnaoutov A, Jansen LE, Ouspenski I, Dye L, Karpova T, McNally J, Dasso M, Cleveland DW, Strunnikov A 2009. Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLoS One 4: e6831 doi: 10.1371/journal.pone.0006831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Scholl LM, Cimini D 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One 4: e6564 doi: 10.1371/journal.pone.0006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R 2007. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 11: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman JM, Socci ND, Benezra R 2010. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 464: 436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan SV, Mayhew CN, Schwemberger S, Zagorski W, Knudsen ES 2007. RB loss promotes aberrant ploidy by deregulating levels and activity of DNA replication factors. J Biol Chem 282: 23867–23877 [DOI] [PubMed] [Google Scholar]
- Terret ME, Sherwood R, Rahman S, Qin J, Jallepalli PV 2009. Cohesin acetylation speeds the replication fork. Nature 462: 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol 180: 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Harn T, Foijer F, van Vugt M, Banerjee R, Yang F, Oostra A, Joenje H, te Riele H 2010. Loss of RB proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev (this issue). doi: 10.1101/gad.580710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM 2006. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol 16: 863–874 [DOI] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW 2006. Does aneuploidy cause cancer? Curr Opin Cell Biol 18: 658–667 [DOI] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11: 25–36 [DOI] [PubMed] [Google Scholar]
- Weinberg RA 1995. The retinoblastoma protein and cell cycle control. Cell 81: 323–330 [DOI] [PubMed] [Google Scholar]