Abstract
The p53 tumor suppressor limits proliferation in response to cellular stress through several mechanisms. Here, we test whether the recently described ability of p53 to limit stem cell self-renewal suppresses tumorigenesis in acute myeloid leukemia (AML), an aggressive cancer in which p53 mutations are associated with drug resistance and adverse outcome. Our approach combined mosaic mouse models, Cre-lox technology, and in vivo RNAi to disable p53 and simultaneously activate endogenous KrasG12D—a common AML lesion that promotes proliferation but not self-renewal. We show that p53 inactivation strongly cooperates with oncogenic KrasG12D to induce aggressive AML, while both lesions on their own induce T-cell malignancies with long latency. This synergy is based on a pivotal role of p53 in limiting aberrant self-renewal of myeloid progenitor cells, such that loss of p53 counters the deleterious effects of oncogenic Kras on these cells and enables them to self-renew indefinitely. Consequently, myeloid progenitor cells expressing oncogenic Kras and lacking p53 become leukemia-initiating cells, resembling cancer stem cells capable of maintaining AML in vivo. Our results establish an efficient new strategy for interrogating oncogene cooperation, and provide strong evidence that the ability of p53 to limit aberrant self-renewal contributes to its tumor suppressor activity.
Keywords: AML, Ras, cancer stem cells, leukemia, p53, self-renewal, tumor suppressor
p53 mutations are common in human cancers, and are frequently associated with aggressive disease courses and drug resistance (Wattel et al. 1994; Buttitta et al. 1997; Levine and Oren 2009). Germline p53 mutations predispose mice and humans to diverse tumors (Malkin et al. 1990; Donehower 1996), and p53 loss cooperates with a variety of oncogenic lesions—including oncogenic Ras, deregulated Myc, and loss of Pten—to promote tumorigenesis in various experimental models (Hermeking and Eick 1994; Eischen et al. 1999; Johnson et al. 2001; Chen et al. 2005). In addition, various proteins acting upstream of or downstream from p53 in its signaling networks are frequently mutated in human cancers (Bueso-Ramos et al. 1993; Esteller et al. 2001; Lowe and Sherr 2003; Brown et al. 2009). Indeed, mutational disruption of the p53 network may occur in virtually all aggressive end-stage cancers (Brown et al. 2009).
p53 possesses a range of biological activities that may contribute to its role in tumor suppression, including its ability to trigger various cell cycle checkpoints, apoptosis, autophagy, differentiation, and cellular senescence (Meek 2009; Zilfou and Lowe 2009). Moreover, loss of p53 fuels genomic instability to further facilitate tumor evolution. p53 is best understood for its action in response to cellular stress, whereby it elicits one or more of the above biological responses to limit proliferation. As an example, oncogenic Ras can trigger replication stress, which in some instances activates p53 as part of a fail-safe mechanism that ultimately prevents malignant transformation (Serrano et al. 1997; Bartkova et al. 2006; Gonzalez et al. 2006). Consequently, p53 loss cooperates with Ras in many tumor types (Sevignani et al. 1998; Bardeesy et al. 2001; Johnson et al. 2001; Orsulic et al. 2002; Zuber et al. 2009).
Recent studies have linked p53 to the process of stem cell self-renewal (TeKippe et al. 2003; Meletis et al. 2006; Armesilla-Diaz et al. 2009; Liu et al. 2009), which is defined as a biological process in which a stem cell divides to generate one (asymmetric division) or two (symmetric division) daughter stem cells with developmental potentials that are indistinguishable from those of the mother cell (Molofsky et al. 2004). Normal hematopoietic stem cells (HSCs) and mammary stem cells from p53-deficient mice show increased self-renewal in culture and in mice, which, in mammary stem cells, result from enhanced symmetric cell division (Rambhatla et al. 2001; Dumble et al. 2007; Cicalese et al. 2009; Liu et al. 2009). Moreover, suppression of p53 or the p53 pathway enhances the production of induced pluripotent stem (iPS) cells, suggesting that p53 limits the reprogramming of differentiated cells into a self-renewing state (Hong et al. 2009; Kawamura et al. 2009; Li et al. 2009; Marion et al. 2009; Utikal et al. 2009). It remains to be determined whether these effects reflect a physiological role for p53, or are merely a consequence of stress associated with the experimental manipulation (Krizhanovsky and Lowe 2009). Nevertheless, the ability of p53 to “immortalize” cells in culture has long been linked to its role in tumorigenesis. Whether or not p53's function in stem cell biology is important for its tumor-suppressive role has not been tested directly.
Acute myeloid leukemia (AML) is a heterogeneous cancer thought to arise through an accumulation of mutations that cooperate to deregulate proliferation—typically by activating Ras signaling pathways—and impair terminal myeloid differentiation (Gilliland et al. 2004). Both HSCs and more committed myeloid progenitor cells can function as the cellular origin for the initiation of AML in some circumstances (Passegue et al. 2003). As myeloid progenitors and their differentiated progeny lack self-renewal capabilities, these cells apparently acquire this ability during the course of leukemia development. How this occurs has been a topic of much debate, although fusion proteins involving the mixed-lineage leukemia (MLL) gene and lesions enhancing Wnt/β-catenin signaling can confer this capability (Cozzio et al. 2003; Jamieson et al. 2004; Krivtsov et al. 2006).
Although p53 mutations occur in only 10%–15% of AMLs at diagnosis, they are associated with the most aggressive disease courses and drug resistance (Wattel et al. 1994; Haferlach et al. 2008; Nahi et al. 2008). Interestingly, these aggressive clinical traits can be recapitulated in mouse models. For example, our laboratory recently produced a series of mosaic mouse models of AML, and applied them to demonstrate that p53 mutations can confer resistance to the chemotherapeutic agents used to treat the human disease (Zuber et al. 2009). Furthermore, we showed that expression of MLL fusion proteins, which are also associated with poor prognosis and drug resistance (Schoch et al. 2003), produce some of these aggressive traits by disabling p53 (Zuber et al. 2009). How p53 loss influences AML development remains to be determined, but may provide insights into the most aggressive features of this disease.
In this study, we explored the genetic and biological interactions between oncogenic Kras and p53 in AML. Expressing oncogenic KrasG12D in the hematopoietic system drives hyperproliferation and differentiation of HSCs and myeloid progenitors, resulting in fatal myeloproliferative disorders (MPDs) (Braun et al. 2004; Chan et al. 2004). Unlike MLL fusion oncoproteins, oncogenic Kras does not enhance the self-renewal of myeloid progenitors. Consequently, KrasG12D-induced MPDs depend on the involvement of naturally self-renewing HSCs, and their progression to AML requires secondary lesions in progenitor populations (Sabnis et al. 2009; Zhang et al. 2009). By exploiting this well-characterized model and established assays to assess self-renewal capabilities of normal and leukemic cells, we show that p53 deficiency, together with oncogenic KrasG12D, provides one mechanism whereby committed myeloid progenitor cells acquire the capacity to self-renew indefinitely and transform into leukemia-initiating cells. Our results therefore provide new insights into AML biology and p53-mediated tumor suppression.
Results
A self-excising Cre vector that simultaneously delivers an shRNA and a GFP reporter
Studies examining the effects of oncogenic Ras in the myeloid compartment have used bitransgenic mice harboring a “lox-Stop-lox” (LSL) Kras allele (Tuveson et al. 2004) and an interferon-responsive Mx1-Cre transgene (Kuhn et al. 1995). In this system, the oncogenic Kras allele is knocked into the endogenous locus but kept silent by the LSL cassette, which can be excised by Cre-mediated recombination following injection of polyinosinic-polycytidylic acid (pIpC). As a consequence of KrasG12D expression, mice develop a fatal MPD that initiates in self-renewal-competent HSCs and cannot be transplanted into syngeneic recipient mice (Braun et al. 2004; Chan et al. 2004; Sabnis et al. 2009; Zhang et al. 2009).
To test the leukemogenic effect of p53 loss in this model, we began generating compound Mx1-Cre; LSL-KrasG12D; p53−/− mice by intercrossing the individual mutant strains. Already time-consuming and expensive, the four-allele cross did not yield sufficient offspring for studies examining leukemia onset, since mice of the desired genotype were born at sub-Mendelian ratios and frequently died shortly after birth due to uncharacterized causes (data not shown). We also attempted to control p53 deletion through production of Mx1-Cre;LSL-KrasG12D;p53loxp/loxp mice, in which both activation of Kras and inactivation of p53 were triggered simultaneously by Cre-mediated recombination. Shortly after pIpC administration, these mice developed complicated phenotypes—including myeloid hyperplasia, atypical lymphoid hyperplasia in thymus and spleen, and histiocytic sarcoma—and needed to be sacrificed (data not shown).
The difficulties we encountered producing mice harboring conditional germline Cre, KrasG12D, and p53 alleles prompted us to explore a more efficient strategy to examine the effects of p53 inactivation on leukemogenesis. Our laboratory has developed methods for mimicking tumor suppressor gene loss in mice through stable RNAi (Hemann et al. 2003, 2004; Xue et al. 2007; Zender et al. 2008). Therefore, we set out to develop a transplantation-based “mosaic” mouse model in which hematopoietic stem/progenitor cells (HSPCs) derived from LSL-KrasG12D mice could be transduced with a retrovirus harboring a shRNA targeting p53. We constructed a murine stem cell virus (MSCV)-based retroviral vector (LGmCreER) (see Fig. 1A; Supplemental Fig. S1a) containing a well-characterized p53 shRNA based on the mir30 design that efficiently triggers the RNAi machinery and can be expressed in the same transcript as green fluorescent protein (GFP) (Dickins et al. 2005; Stegmeier et al. 2005).
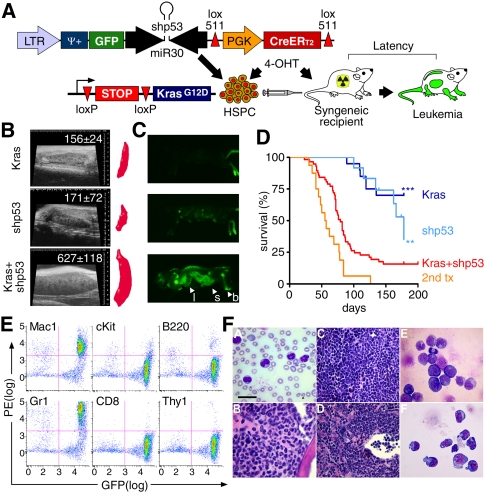
Figure 1.
Activation of endogenous KrasG12D and shRNA-mediated suppression of p53 cooperate to induce AML. (A) Schematic of the experimental strategy. Retroviral LGmCreER is used to cotransduce GFP, a miR30-based shRNA, and 4-OHT-inducible self-excising CreERT2 into HSPCs isolated from embryonic day 13.5–15.5 fetal livers of LSL-KrasG12D embryos. Cre-mediated recombination is induced by treatment with 4-OHT in vitro or in vivo. (B) Representative spleen sonogram and three-dimensional reconstruction volumetry in recipient mice of HSPCs of indicated genotypes 6 wk after transplantation. Numbers indicate average spleen volume and standard deviation in six mice for each genotype. Only KrasG12D-shp53 recipient mice show severe splenomegaly at this preleukemic stage. (C) Whole-body imaging of representative mice reconstituted with KrasG12D, shp53, and KrasG12D-shp53 HSPCs. Only KrasG12D-shp53 mice show strong accumulation of GFP-positive cells in bones (b), spleen (s), and liver (l). (D) Kaplan-Meier curve showing the survival of mice reconstituted with KrasG12D-shp53 HSPCs (n = 57) and controls. (**) P = 0.0022 and (***) P < 0.0001 (log-rank test) indicate highly significant survival reduction compared with recipients of shp53 (n = 12) and KrasG12D (n = 20) control HSPCs, respectively. Secondary transplantation (2nd tx) curve indicating survival of sublethally irradiated recipient mice transplanted with KrasG12D-shp53 leukemias (n = 16). (E) Representative immunophenotyping of KrasG12D-shp53 leukemic bone marrow showing strong infiltration of GFP-positive cells that are positive for Mac1 and Gr1. (F) Representative histopathology of KrasG12D-shp53-induced AMLs, including MPD-like leukemias (panels A–D) and acute myelomonocytic leukemia (panel E) and AML without maturation (panel F). (Panel A) Peripheral blood containing elevated numbers of neutrophilic and monocytic cells. Arrowhead denotes Howell-Jolly body. (Panel B) Sternal bone marrow filled with maturing myeloid cells. (Panel C) Spleen with vastly expanded red pulp filled with erythroid cells (top left) and maturing myeloid elements (bottom right). (Panel D) Liver with extensive perivascular and sinusoidal hematopoiesis. (Panel E) Bone marrow cytospin showing blasts and monocytic cells. (Panel F) Bone marrow cytospin showing immature blast cells. Blood smear and bone marrow cytospin: Wright-Giemsa; bar, 20 μm. Liver and spleen: hematoxylin and eosin; bar, 50 μm.
To enable coexpression of Cre recombinase from the same vector, we introduced a tamoxifen-inducible CreERT2 expressed from a constitutive PGK promoter. Since long-term Cre exposure can be toxic (Loonstra et al. 2001; Forni et al. 2006), we implemented a novel self-excising design by flanking the PGK-CreERT2 cassette with two lox511 sequences, which prevents recombination with conventional loxP sites in the targeted cells (Hoess et al. 1986) and retains an intact GFP-shRNA transgene after Cre excision. Southern blotting of DNA derived from mouse embryo fibroblasts (MEFs) infected with virus collected within 24–36 h after packaging cell transfection revealed that >70% of the integrated proviruses were full-length (Supplemental Fig. S1b). As predicted, the addition of 4-hydroxytamoxifen (4-OHT) led to the efficient excision of the PGK-CreERT2 cassette. Moreover, immunoblotting revealed that cells transduced with the LGshp53CreER vector showed potent p53 suppression (Supplemental Fig. S1c).
Next, we analyzed the effectiveness of LGmCreER at excising LSL cassettes. NIH3T3 cells containing a LSL-LacZ reporter were transduced with LGmCreER and a non-self-excising MSCV-CreERT2-IRES-GFP vector, and cell populations were examined in the absence and presence of 4-OHT (Supplemental Fig. S1d). Each vector was delivered efficiently into target cells, as assessed by GFP fluorescence. However, in the absence of 4-OHT, a high percentage of cells harboring the non-self-excising vector were β-Gal-positive, suggesting leaky activity of long terminal repeat (LTR)-driven CreERT2. Moreover, 4-OHT addition induced substantial cell death, presumably owing to Cre toxicity as reported previously (Silver and Livingston 2001). In contrast, cells transduced with the LGmCreER construct showed low basal β-Gal activity, which was induced efficiently by 4-OHT without signs of toxicity (Supplemental Fig. S1d). Thus, the LGmCreER vector could deliver a reporter (GFP) and shRNA to target cells, and, at the same time, induce Cre-mediated recombination with minimal toxicity.
Loss of p53 cooperates with KrasG12D to promote AML
To test whether p53 suppression cooperates with oncogenic KrasG12D in promoting AML, we isolated cell populations enriched for HSPCs from fetal livers of LSL-KrasG12D embryos and infected these cells with LGmCreER containing either the p53 shRNA (LGshp53CreER) or the control empty miR30 cassette (LGmCreER). Cells were then transplanted into lethally irradiated syngeneic recipient mice (Fig. 1A). To induce Cre activity, 4-OHT was either added directly to the infected HSPCs in vitro or administered to mice by intraperitoneal (i.p.) injection 4 wk after transplantation. Recipient mice were monitored periodically for signs of neoplasia, as well as overall morbidity, using sonographic spleen imaging, whole-body fluorescence imaging, and blood analysis (white blood cell counts and blood smear cytology).
As early as 6 wk post-transplantation, and well before the median onset of blood pathology, mice reconstituted with KrasG12D-shp53 HSPCs showed pronounced splenomegaly (Fig. 1B). At the same time, KrasG12D-shp53 mice had an accumulation of GFP-positive cells in the bone marrow, spleen, and liver (Fig. 1C). Eventually, mice receiving KrasG12D-shp53 HSPCs developed lethal neoplasias with a median survival of 79 d post-transplantation (n = 45) (Fig. 1D). Bone marrow isolates from animals with advanced disease were capable of transplanting leukemia to sublethally irradiated recipient mice (Fig. 1D), indicating that the neoplasias produced by cells harboring KrasG12D and p53 shRNAs were bona fide leukemias rather than an aggressive MPD (Kogan et al. 2002). Similar phenotypes were observed regardless of whether 4-OHT was administered before or after transplantation, although disease onset and progression was delayed for ∼2 wk in the animals treated post-transplantation (Supplemental Fig. S2). In contrast, mice carrying wt-shp53 or KrasG12D alone mainly developed thymic lymphomas, with much lower penetrance and longer latency compared with KrasG12D-shp53 leukemias (P < 0.0001 for KrasG12D; P = 0.0022 for wt-shp53) (Fig. 1D; data not shown).
To further characterize these leukemias, we analyzed leukemic cells and tissues from moribund mice using flow cytometry and histopathology. GFP-positive leukemic cells were present in the bone marrow, spleen, and peripheral blood, ranging between 60% and 90% of the total cell population. Immunophenotyping revealed that most leukemias were of myeloid origin, with the majority (13 out of 14) consisting of cells expressing Mac1 and Gr1 (Fig. 1E; data not shown). In ∼40% of cases (nine out of 22), leukemic mice also had enlarged thymi that were infiltrated with GFP+/Thy1+ lymphoblasts reminiscent of concomitant thymic lymphoma. Leukemic mice displayed elevated white blood cell counts and signs of severe anemia (Fig. 1F, panel A). In all cases, the spleens were massively enlarged and harbored extensive extramedullary hematopoiesis and infiltration of myelomonocytic cells (Fig. 1F, panel C), while enlarged livers showed extensive perivascular and sinusoidal leukemia infiltrates (Fig. 1F, panel D). Approximately two-thirds of these animals showed MPD-like myeloid leukemias composed of mature neutrophilic and monocytic cells and <20% blasts (Fig. 1F, panels A–D), while the other third presented with either myelomonocytic AML (Fig. 1F, panel E) or AML without maturation (Fig. 1F, panel F; Kogan et al. 2002). In contrast, recipients of HSPCs harboring shp53 or KrasG12D as a single lesion developed late-onset T-cell lymphomas, recapitulating the typical disease phenotype in p53-null mice (Donehower et al. 1992) and the predominant malignancy in transplant recipients of pIpC-treated Mx1-Cre;LSL-KrasG12D bone marrow (Sabnis et al. 2009; Zhang et al. 2009). Hence, KrasG12D and p53 loss cooperate in the hematopoietic compartment to induce aggressive AML, and thereby bias transformation toward the myeloid lineage.
We confirmed activation of KrasG12D and suppression of p53 using a variety of molecular approaches. First, using an optimized PCR strategy that enables detection of rare LSL recombination events in small subpopulations (Supplemental Fig. S3), we documented excision of the LSL cassette in peripheral blood of KrasG12D-shp53 recipient mice 7 d after 4-OHT treatment (Fig. 2A, top panel), and in leukemic cells from multiple recipient mice (Fig. 2A, bottom panel). Second, we used flow cytometry (Van Meter et al. 2007) to quantify the intracellular phosphorylation of various Ras effectors. Oncogenic Ras aberrantly activates kinase effector pathways, which can be assayed by measuring phosphorylation of Erk1/2 and S6: Accordingly, KrasG12D-shp53 leukemias consistently displayed a substantial elevation of basal p-Erk1/2 and p-S6 levels in comparison with wild-type bone marrow in both bulk and core progenitor cells (Lin−cKit+) (Fig. 2C,D; data not shown). In many cases, stimulation by GM-CSF, a growth factor involved in normal myeloid cell proliferation, did not further enhance p-Erk1/2 levels, indicating that the Raf/Mek/Erk cascade was constitutively activated at saturating levels. Finally, immunoblotting of total bone marrow lysates extracted from KrasG12D-shp53 leukemic mice revealed that p53 was virtually undetectable, even following ex vivo treatment with the DNA-damaging agent adriamycin (Fig. 2B). Taken together, these results demonstrate that the oncogenic effects of KrasG12D and RNAi-mediated suppression of p53 strongly cooperate in the myeloid lineage and promote aggressive AML.
Figure 2.
Molecular characterization of KrasG12D-shp53-induced AML. (A) PCR analysis to verify Cre-mediated recombination of the LSL cassette. (Top panel) Two-step PCR in peripheral blood of a representative KrasG12D-shp53 HSPC recipient mouse before and 5 d after 4-OHT treatment in vivo and after leukemia onset in bone marrow (bm) and spleen (spl). Recombination is indicated by the appearance of a 749-bp band after the first (I), and a 249-bp band after the second (II) PCR. (Bottom panel) LSL-KrasG12D recombination in leukemic mice (1–5) analyzed in bone marrow and spleen (left and right lane in each sample, respectively). (nc) Negative control. (B) Western blotting of p53 in wild-type bone marrow, Arf−/−, and p53−/− MLL/ENL-induced control leukemias and three independent KrasG12D-shp53 AMLs (#1, #2, and #3) without (−) or with (+) adriamycin (Adr) treatment in vitro. (C) Flow cytometry analysis of intracellular phosphorylated Mapk1/Mapk3 (pErk) and Rps6 (pS6) in bulk and core (Lin−cKit+) populations of wild-type bone marrow and KrasG12D-shp53 leukemia. (D) Elevated basal levels of intracellular phosphorylated Mapk1/Mapk3 (pErk) and Rps6 (pS6) in multiple independent KrasG12D-shp53 AMLs.
p53 suppression induces oncogenic self-renewal of KrasG12D-expressing hematopoietic progenitor cells in vitro
Loss of p53 cooperates with oncogenic Ras to promote transformation in many tissue types, although the biological basis for these genetic interactions has been controversial and may be context-dependent (Tuveson et al. 2004; Collado et al. 2005). The fact that KrasG12D expression and p53 deficiency synergize to promote AML, while neither mutation gives rise to this disease as a sole lesion, suggests unique and potent effects of Ras and p53 signaling on the myeloid lineage. Owing to the availability of well-established assays to study proliferation, differentiation, and self-renewal capabilities of hematopoietic cells, we reasoned that exploring the functional role of p53 in this model would provide unprecedented insights into its action as a tumor suppressor.
Normal hematopoiesis involves a cellular hierarchy whereby HSCs give rise to multipotential progenitors (MPPs). These cells eventually differentiate into lineage-restricted common lymphoid progenitors (CLPs) and common myeloid progenitors (CMPs), which are committed to terminal differentiation and unable to self-renew (Chao et al. 2008). Previous studies have demonstrated that oncogenic Kras drives proliferation and differentiation of HSCs and myeloid progenitors, but does not promote—and, in fact, depends on—self-renewal capabilities to initiate disease (Sabnis et al. 2009; Zhang et al. 2009). Hence, mutations that deregulate Ras signaling frequently co-occur and functionally cooperate with mutations that impair differentiation and endow myeloid progenitor cells with an aberrant and unlimited capacity to self-renew (Alcalay et al. 2003; Cozzio et al. 2003; Zuber et al. 2009).
To determine how p53 loss—alone or in cooperation with KrasG12D—influences proliferation, differentiation, and life span in the myeloid lineage, we transduced wild-type and LSL-KrasG12D HSPCs with LGshp53CreER or a control vector (LGshlucCreER, expressing a control shRNA targeting firefly luciferase), induced Cre recombination by addition of 4-OHT, and subsequently quantified stem and progenitor compartments, proliferation, and self-renewal capacity in the resulting genetic contexts (wt-shluc, wt-shp53, KrasG12D-shluc, or KrasG12D-shp53). Of note, transduced cells can be tracked easily through expression of the GFP reporter in our vector, which also enables comparisons with untransduced cells as internal controls. Cell populations that are enriched for self-renewing HSCs can be readily identified by flow cytometry using KSL markers (cKit+Sca1+Lin−) (Okada et al. 1992), or, with greater purity, by incorporating SLAM markers (CD150+CD48−KSL) (Kiel et al. 2005). Non-self-renewing myeloid progenitors can be identified as Lin−Ckit+Sca-1−IL7Rα− cells (Akashi et al. 2000). Of note, the utility of these markers for identifying HSCs in in vitro culture still remains controversial; thus, all results were validated further in vivo (see below).
We first analyzed the proliferation (BrdU [5′-bromodeoxyuridien] incorporation) and overall representation of GFP-positive HSCs and myeloid progenitors in culture (Fig. 3A,B). Consistent with previous reports (Van Meter et al. 2007; Sabnis et al. 2009), we found that acute KrasG12D activation resulted in a mild increase in BrdU incorporation in KSL cells (Fig. 3A), while the overall frequency of HSCs (CD150+CD48−KSL) among transduced cells was reduced compared with wild-type controls (Fig. 3B, top panel). Similar results were observed in cells cultured in STIFA medium (StemSpan serum-free media containing the growth factors SCF, TPO, IGF-2, FGF-1, and Angptl3), which supports the expansion of normal HSCs in vitro (Supplemental Fig. S4; CC Zhang et al. 2006; Akala et al. 2008). These results suggest that KrasG12D favors proliferation but not self-renewal of HSCs. In contrast, although consistent with two recent reports (Akala et al. 2008; Liu et al. 2009), p53 suppression increased both the proliferation and frequency of HSCs (Fig. 3A,B, top panel). p53 loss also increased the proliferation and overall frequency of myeloid progenitors in both the wild-type and KrasG12D background (Fig. 3A,B, bottom panel). Thus, in addition to its modest effects on HSCs, p53 loss dramatically promotes the expansion of the myeloid progenitor compartment independently of oncogenic Kras activation.
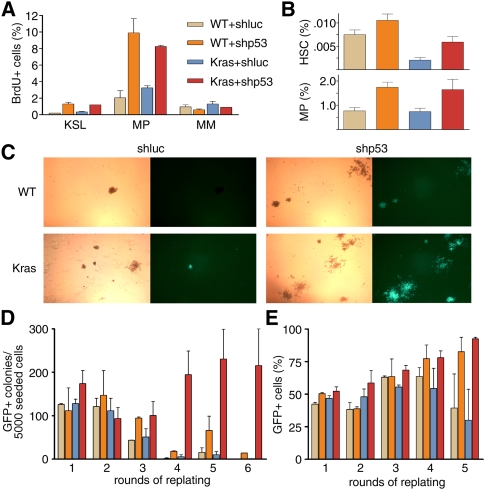
Figure 3.
p53 suppression and KrasG12D induce self-renewal of myeloid progenitor cells in vitro. (A) BrdU incorporation in transduced mature myeloid cells (MM; Gr1+), myeloid progenitors (MP; Lin−cKit+Sca1−IL7Rα−), and MPP and stem cells (KSL; Lin−cKit+Sca1+) in indicated genotypes 4 d following 4-OHT-induced activation of KrasG12D. (B) Frequency of GFP+ HSCs (Lin−cKit+Sca1+CD150+CD48−) and myeloid progenitors (MP; Lin−cKit+Sca1−IL7Rα−) in all transduced (GFP+) HSPCs of indicated genotypes (color coding as in A) 4 d after activation of KrasG12D. (C) Methylcellulose colony formation and serial replating assay of transduced 4-OHT-treated HSPCs of indicated genotypes. Representative bright-field and GFP fluorescent images showing colonies formed after the fourth round of replating. (D) Quantification of GFP+ colonies of indicated genotypes over six rounds of replating (color coding as in A). (E) Percentage of GFP+ cells of indicated genotypes at the end of each round of replating (color coding as in A).
To analyze the impact of oncogenic Kras activation and/or p53 suppression on self-renewal, we used a well-established serial replating assay, whereby the life span of progenitors is assessed by their ability to form colonies derived from single cells in methylcellulose (Lavau et al. 1997). Normal myeloid progenitors are able to form colonies in only the first two to three rounds of replating, and then exhaust their proliferative potential. We plated 5000 GFP+ wt-shluc, wt-shp53, KrasG12D-shluc, and KrasG12D-shp53 HSPCs in methylcellulose and counted the number of GFP+ colonies 7 d later. Cells were then harvested, and the same procedure was repeated.
Consistent with previous reports (Braun et al. 2004; Chan et al. 2004), wild-type and KrasG12D HSPCs lost their ability to produce colonies during the fourth round of replating, indicating that the population had exhausted its progenitor compartment (Fig. 3C,D). Conversely, wt-shp53 cells showed a steady enrichment of GFP+ cells over time (Fig. 3E), and modest enhancement in their colony-forming and replating ability (Fig. 3C,D; see also Liu et al. 2009). In marked contrast, KrasG12D-shp53 cells continued forming numerous robust GFP+ colonies through six rounds of replating, after which the experiments were terminated (Fig. 3C,D). Outgrown KrasG12D-shp53 colonies after the fourth round of replating contained myeloid progenitors and MPPs that were all positive for GFP (data not shown). Furthermore, cells recovered from KrasG12D-shp53 colonies were able to induce aggressive AML and T-cell lymphoma after transplantation into sublethally irradiated recipient mice (data not shown), indicating they were enriched for leukemia-initiating cells. Taken together, these in vitro results indicate that loss of p53 enhances proliferation of myeloid progenitor cells, and, in combination with oncogenic Kras, enables these cells to continually self-renew.
p53 loss enables KrasG12D-expressing myeloid progenitors to become leukemia-initiating cells
In contrast to KrasG12D-induced MPD, which depends on the involvement of naturally self-renewing HSCs (Sabnis et al. 2009; Zhang et al. 2009), our in vitro studies indicate that self-renewal capabilities can be acquired de novo in myeloid progenitors lacking p53. To determine how loss of p53 and activation of oncogenic Kras affect myeloid progenitors in vivo, we transduced both wild-type and LSL-KrasG12D HSPCs with LGshlucCreER or LGshp53CreER and transplanted these cells into lethally irradiated recipients. Transduction efficiencies were consistently ∼40% (data not shown), such that HSPCs of all genotypes contained a comparable fraction of nontransduced (GFP−) cells as an internal wild-type control population. In order to bypass deleterious effects of Ras activation on engraftment (Sabnis et al. 2009), KrasG12D expression was triggered upon host reconstitution by 4-OHT treatment. To determine the relative competition between cells harboring activated Kras and/or p53 knockdown (GFP+) and nontransduced internal controls, we analyzed the frequency of GFP+ cells in total bone marrow and the myeloid progenitor and HSC subcompartments 3 wk later. In parallel, we analyzed the frequency of myeloid progenitors and HSCs among all GFP+ cells, which reflects the relative growth potential of cells within the total transduced population.
Compared with bone marrow from mice transplanted with wt-shluc HSPCs, recipients of KrasG12D-shluc HSPCs showed a consistent, although not significant, decrease in GFP+ cells in bone marrow, myeloid progenitors, and HSCs (see Fig. 4A for a representative flow cytometry profile, and B for quantification), indicating that KrasG12D expression did not confer a competitive advantage to these cells in vivo. Consistent with previous findings in the Mx1-Cre;LSL-Kras MPD model (Van Meter et al. 2007) and our in vitro results, we observed a reduced frequency of myeloid progenitors (Fig. 4C) and HSCs (Fig. 4D) among all GFP+ cells. In contrast, independently of KrasG12D, shp53-expressing cells were strongly enriched in total bone marrow, HSCs, and myeloid progenitors (Fig. 4A,B), indicating that p53 suppression conferred a selective advantage in these compartments in vivo.
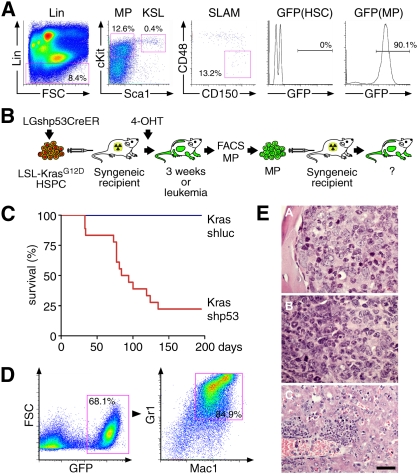
Figure 4.
p53 suppression expands myeloid progenitors in vivo and, in concert with endogenous, KrasG12D promotes leukemogenesis. (A) Representative bone marrow flow cytometry profiles of stem and myeloid progenitor compartments of mice reconstituted with HSPCs of indicated genotypes, including conventional Lin/cKit/Sca1 staining (MP; Lin−cKit+Sca1−IL7Rα−), KSL (Lin−cKit+Sca1+), SLAM markers (HSC; Lin−cKit+Sca1+CD150+CD48−), and histograms indicating the percentage of GFP+ cells in myeloid progenitors. (B) Frequency of GFP+ cells in total bone marrow (all), myeloid progenitors (MP), and HSCs of mice reconstituted with HSPCs of indicated genotypes 3 wk after in vivo 4-OHT treatment to induce KrasG12D (n = 4–5 in each group). (C) Quantification of GFP+ myeloid progenitors within GFP+ bone marrow cells of mice reconstituted with HSPCs of indicated genotypes (n = 4–5 in each group). (D) Quantification of GFP+ HSCs within GFP+ bone marrow cells of mice reconstituted with HSPCs of indicated genotypes (n = 4–5 in each group).
In the context of oncogenic KrasG12D, p53 had opposing effects on the size of HSC and myeloid progenitors compartments: While p53 suppression prevented the KrasG12D-induced depletion of myeloid progenitors to levels at or above those occurring in wt-shluc controls (Fig. 4C), the percentage of GFP+ HSCs was unchanged or even further reduced compared with KrasG12D alone (Fig. 4D). Moreover, bone marrow from mice with advanced leukemia consistently showed a strong enrichment of GFP+ cells in the myeloid progenitor compartment, while HSCs were almost exclusively GFP-negative (Fig. 5A). This suggests that HSCs might be dispensable for maintaining KrasG12D-shp53 AML in vivo. These results imply that intact p53 limits the expansion of normal and Kras mutant myeloid progenitors in vivo, while p53 loss allows oncogenic Kras to trigger aberrant proliferation of this compartment.
Figure 5.
KrasG12D-shp53 myeloid progenitor cells induce AML. (A) Representative bone marrow flow cytometry profiles of stem and progenitor compartments in KrasG12D-shp53 leukemic mice, including conventional Lin/cKit/Sca1 staining (left two panels), SLAM markers (middle panel), and a histogram indicating the percentage of GFP+ in HSCs and myeloid progenitors (right two panels). (B) Schematic overview of generation, sorting, and secondary transplantation of KrasG12D-shp53 myeloid progenitor cells. (C) Kaplan-Meier curve of lethally irradiated recipient mice transplanted with 2000–5000 doubly sorted myeloid progenitor cells isolated from six independent primary recipients of KrasG12D-shp53 HSPCs (n = 18) and KrasG12D HSPCs (n = 3). (D) Representative bone marrow flow cytometry analysis of leukemia induced by transplantation of KrasG12D-shp53 myeloid progenitors. (Left panel) The bone marrow is dominated by GFP+ cells, which are predominantly expressing Mac1 and Gr1. (E) Histopathology analysis of leukemias induced by transplantation of KrasG12D-shp53 myeloid progenitors. (Panel A) Sternal bone marrow densely packed with blasts and few maturing myeloid cells (100×). (Panel B) Spleen with areas predominated by blasts and maturing neutrophils (100×). (Panel C) liver with invasion of leukemic cells with differentiation (40×). All hematoxylin and eosin-stained. Bars: 40× images, 50 μm; 100× images, 20 μm.
To rigorously interrogate these “rescued” myeloid progenitors for aberrant self-renewal capabilities, and to assay their ability to initiate Kras-shp53-induced AML, we purified Lin−Ckit+Sca-1−IL7Rα− cells from preleukemic (3 wk post-4-OHT treatment) and leukemic mice in two consecutive rounds of fluorescence-activated cell sorting (FACS). GFP+ myeloid progenitors (2000–5000 per mouse) were injected with 200,000 wild-type syngeneic helper cells into lethally irradiated recipient mice (see Fig. 5B for schematic overview). While all mice receiving Kras-shluc control cells remained healthy, those receiving Kras-shp53 myeloid progenitors rapidly succumbed to AML (Fig. 5C). Interestingly, the resulting leukemias displayed immunophenotypes resembling the primary AMLs described above (GFP+Mac1+Gr1+) (Fig. 5D,E), while none were accompanied by concomitant thymic lymphoma (data not shown). Hence, sorted Kras-shp53 myeloid progenitors formed leukemia while maintaining their differentiation potential. Collectively, these results indicate that myeloid progenitors with oncogenic Kras activation and p53 deficiency gain self-renewal capability in vivo, and can serve as leukemia-initiating cells.
p53 gene deletion recapitulates the effects of p53 knockdown in myeloid progenitors
To rule out the possibility that technical aspects of the experimental system (such as isolation of fetal liver HSPCs; methods of Cre delivery, transplantation, and reconstitution; RNAi off-target effects; etc.) influenced our observations, we repeated key experiments in an entirely transgenic setting. To produce the precise genetic configurations described above, we intercrossed Mx1-Cre (C), LSL-KrasG12D (K), and p53loxp/loxp (P) mice. Mx1-Cre;LSL-KrasG12D;p53loxp/loxp (CKP) and control (C, CK, and CP) mice were treated with pIpC to activate KrasG12D and inactivate p53 (p53Δ2–10) at 6 wk of age. Seven days later, we harvested bone marrow from mice of each genotype to perform colony-forming and serial replating assays as well as FACS analysis for the frequency of mature myeloid cells and stem/progenitor populations. In addition, myeloid progenitors were isolated by FACS and assessed for their capacity to initiate leukemia using the same strategies described above (for overview, see Supplemental Fig. S5).
Precisely paralleling our observations using the retroviral HSPC transduction approach, p53Δ2–10 bone marrow cells displayed mildly enhanced colony-forming and replating capacity, while the combination of KrasG12D and p53Δ2–10 induced a dramatic increase in colony-forming and replating capabilities (Fig. 6A,B). Consistent with published results (Braun et al. 2004; Chan et al. 2004; Van Meter et al. 2007; Sabnis et al. 2009) and observations from our retroviral system, bone marrow analysis showed that KrasG12D expression uniformly resulted in increased frequency of mature myeloid cells (Fig. 6C, top) as well as an approximately twofold reduction in HSC frequency, which was not rescued but was aggravated further by loss of p53 (Fig. 6C, bottom). The frequency of myeloid progenitors in KrasG12D bone marrow was not reduced at this time point (10 d post-pIpC) as in fully developed MPD (∼50 d post-pIpC) (Van Meter et al. 2007). However, as in the retroviral system, combined KrasG12D;p53Δ2-10 mice showed the most dramatic expansion of myeloid progenitors in vivo (Fig. 6C, middle).
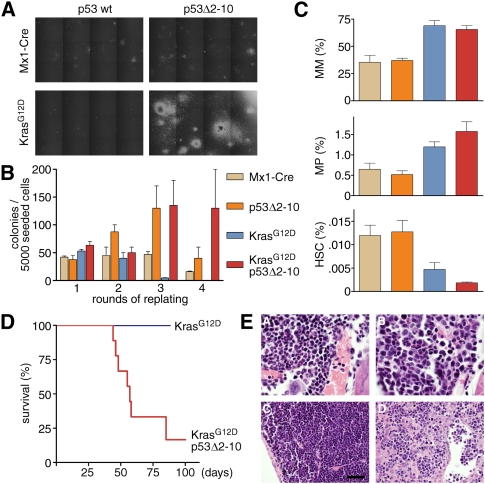
Figure 6.
Conditional p53 deletion recapitulates the effects of RNAi-mediated p53 suppression. (A) Methylcellulose cultures (bright-field images) of bone marrow cells of indicated genotypes after the third round of replating. Robust CFU-M-, CFU-G-, and CFU-GM-like colonies are only formed by KrasG12D;p53Δ2-10 bone marrow cells. (B) Quantification of colony formation in bone marrow cells of indicated genotypes over four rounds of replating. (C) Frequency of mature myeloid cells (MM; Mac1+Gr1+), myeloid progenitors (MP; Lin−cKit+Sca1−IL7Rα−), and HSCs (Lin−cKit+Sca1+CD150+CD48−) in bone marrow of mice with indicated genotypes (color coding as in B) 7–10 d after pIpC treatment (n = 3–5 for each genotype). (D) Kaplan-Meier curve of lethally irradiated recipient mice transplanted with 2000–5000 myeloid progenitor cells doubly sorted from primary Mx1-Cre;LSL-KrasG12D;p53loxp/loxp (n = 9) and Mx1-Cre;LSL-KrasG12D (n = 3) mice. (E) Histopathology analysis of donor and recipient mice of Mx1-Cre;LSL-KrasG12D;p53loxp/loxp myeloid progenitors. (Panel A) Bone marrow (100×) of a donor mouse at the time of myeloid progenitor isolation (10 d after pIpC induction of Cre) showing normal mixed hematopoeisis. (Panels B–D) Bone marrow (100×), spleen (40×), and liver sections (40×), respectively, of a myeloid progenitor recipient mouse showing leukemia infiltrates. All hematoxylin and eosin-stained. Bars: 40× images, 50 μm; 100× images, 20 μm.
As seen in the retroviral setting, combined occurrence of oncogenic KrasG12D and loss of p53 enabled myeloid progenitors to self-renew, as well as to initiate and propagate leukemia in vivo. Most recipient mice of KrasG12D;p53Δ2–10 myeloid progenitors succumbed to leukemias, with a median survival of 62 d (Fig. 6D), while all recipients of myeloid progenitors harboring KrasG12D as a single lesion remained disease-free. Histopathologic analysis showed a massive mononuclear infiltration in bone marrow, spleen, and liver, indicating the presence of leukemia (Fig. 6E; data not shown). In summary, our observations using a purely transgenic system confirm biology obtained through our rapid retroviral approach, and collectively demonstrate that loss of p53 strongly cooperates with KrasG12D to promote leukemogenesis in the myeloid lineage by rendering myeloid progenitor cells capable of indefinite self-renewal.
Discussion
A rapid and versatile approach to study the interplay of complex mutations in cancer
We used a novel approach that combines RNAi technology, Cre-lox-conditional transgenic alleles, and mosaic mouse cancer models to efficiently study the interplay between p53 and Ras signaling in AML. Using retroviral codelivery of a miR30-based shRNA, CreERT2, and GFP into HSPCs harboring a single Cre-lox-activatable KrasG12D allele, we rapidly generated compound mutations alongside the relevant control genotypes, and tagged these modified cells with a fluorescent reporter for easy tracking and isolation in vitro and in vivo. Our system minimizes the impact of Cre-mediated toxicity by using inducible CreERT2 and implementing a new strategy to excise the Cre transgene as soon as Cre is enzymatically active. Using conventional germline methods, the generation of such compound mutant conditions would have required producing mice harboring five germline alleles and, hence, many months of crossing to generate sufficient animal numbers. Moreover, unlike strictly transgenic systems, the resulting mice are “mosaics,” such that only a subset of cells in the hematopoietic system harbor the oncogenic lesions, much as would occur in human leukemia patients.
Beyond leukemia, our rapid approach can be used in other mosaic models of cancer (e.g., breast, liver, and brain). Importantly, the combination of endogenous Cre-lox conditional knock-in alleles and RNAi provides a means to model oncogenic alleles that are expressed at their endogenous loci, thereby eliminating overexpression artifacts and more accurately mimicking conditions in human cancers. As a growing number of such “endogenous” genetically engineered mice harboring Cre-lox-inducible alleles (including oncogene mutations, translocations, and deletions) become available (for review, see Frese and Tuveson 2007), our approach will provide greater flexibility in using these systems for studying the interplay of complex mutations in cancer.
Interplay between KrasG12D and loss of p53 in AML
Ras pathway mutations—including activating mutations in N- and KRAS, as well as other network components (such as FLT3, KIT, NF1, and PTPN11)—occur in the majority of human AML cases (Scholl et al. 2008). In mouse models, some of these lesions act to force both proliferation and differentiation of HSCs and myeloid progenitors, leading eventually to the development of MPD (Braun et al. 2004; Chan et al. 2004). However, Ras signaling does not override the intrinsic programs that limit the ability of myeloid cells to self-renew, and, indeed, may trigger them (Braun et al. 2004; Chan et al. 2004). Consequently, Ras-induced MPDs depend on naturally self-renewing HSCs to initiate disease (Sabnis et al. 2009; Zhang et al. 2009). Studies suggest that progression to full-blown AML requires the acquisition of one or more additional lesions that may confer aberrant self-renewal capabilities to cells within the myeloid lineage. For example, fusion proteins involving MLL promote self-renewal of myeloid progenitors (Cozzio et al. 2003; Huntly et al. 2004) and efficiently cooperate with oncogenic Ras in leukemogenesis (Zuber et al. 2009). In addition, these lesions act to potently suppress myeloid differentiation.
Unexpectedly, we found that both shRNA-mediated suppression and genomic deletion of p53 induced aberrant self-renewal in myeloid progenitor cells, which, in concert with oncogenic Kras, were able to initiate and propagate AML. Thus, in this system, p53 abrogates the requirement of HSC involvement for Ras-induced myeloid disorders, and enables myeloid progenitors to serve as leukemia-initiating cells. However, in contrast to self-renewal-promoting fusion proteins capable of inducing poorly differentiated diseases, p53 loss gives rise to predominantly differentiated AML. Thus, loss of p53 acts strictly to promote self-renewal while enabling leukemia cells to retain their differentiation program, revealing for the first time that impaired differentiation and self-renewal are separable functions during myeloid leukemogenesis. Moreover, p53 suppression may contribute to the self-renewing effects of other AML oncogenes, and, consistent with this possibility, MLL fusion proteins can promote leukemogenesis in part by disabling p53 (Zuber et al. 2009).
Our analyses confirm previous studies demonstrating that KrasG12D leads to the proliferation, differentiation, and, therefore, long-term depletion of HSCs, while loss of p53 expands this compartment (Akala et al. 2008; Liu et al. 2009; Sabnis et al. 2009). Surprisingly, p53 suppression is unable to override the Ras-induced depletion of HSCs, suggesting that p53 is not a key player protecting against Ras-induced effects in this compartment. Indeed, our ability to track Kras-p53shRNA-expressing cells using a linked GFP reporter revealed that leukemia-bearing mice harbor few, if any, Kras-shp53-expressing HSCs (see Figs. 4D, 5A, 6C). Although our data do not rule out the possibility that HSCs can serve as AML-initiating cells, they indicate that HSCs are dispensable for Kras-shp53-mediated leukemogenesis.
In contrast, we found committed myeloid progenitors to be exquisitely sensitive to p53-mediated tumor suppression in the presence of activated Ras, since the combination of both lesions strongly promoted the proliferation, subsequent expansion, and aberrant self-renewal of these cells. Interestingly, the combined activation of Kras and loss of p53 in multipotent HSPCs resulted in AML as the dominant disease phenotype, while both mutations as single lesions almost exclusively drive T-cell malignancies in the same experimental system. While molecular mechanisms underlying the hypersensitivity of myeloid progenitors to Kras-shp53-mediated transformation are not understood, it is noteworthy that p19ARF—a key inducer of p53 in response to oncogenic Ras (Sherr and McCormick 2002)—is silenced in HSCs (Park et al. 2003), yet becomes poised to fulfill its gatekeeper functions as soon as progenitor cells reach a lineage-committed stage (Williams and Sherr 2008). Interestingly, preliminary experiments applying our mosaic approach showed that combined suppression of p19Arf and p16Ink4a also promotes leukemogenesis in concert with KrasG12D (Z Zhao, J Zuber, and SW Lowe, unpubl.). Further studies will be required to dissect the role of both genes and their interplay with p53 during myeloid leukemogenesis.
Prevention of aberrant self-renewal and tumor suppression by p53
Our studies provide new insights into the tumor-suppressive functions of p53. p53 can be activated in response to various forms of cellular stress, leading to a broad spectrum of cellular responses such as apoptosis, cell cycle arrest, senescence, or autophagy in a context-dependent manner (Zilfou and Lowe 2009). The most extensively studied mechanisms underlying p53's function as a tumor suppressor involve its ability to trigger two fail-safe programs—apoptosis and senescence—in response to Ras activation and other oncogenic stimuli. In the hematopoietic system, Ras activation results in hyperproliferation of stem and progenitor cells. This proliferative burst is inseparably linked to terminal differentiation, establishing a barrier to limitless proliferation of Ras mutant cells. While phenotypic markers may vary, the ability of Ras to enforce a terminal cell fate program is conceptually similar to oncogene-induced senescence observed in fibroblasts and certain epithelial cell types (Serrano et al. 1997; Lin and Lowe 2001). Moreover, our data suggest that p53 plays an essential role in these myeloid fail-safe programs by limiting aberrant self-renewal and enforcing terminal differentiation.
p53 has been implicated in limiting self-renewal of normal HSCs and neuronal stem cells (Meletis et al. 2006; Zheng et al. 2008; Liu et al. 2009). In normal progenitor cells, p53 acts as a modulator rather than a sole barrier to self-renewal: Its inactivation delays, but does not prevent, the eventual exhaustion of the progenitor compartment. Our data demonstrate that, in the context of activated Ras, the presence or absence of p53 becomes a decisive factor determining between fail-safe elimination and limitless growth. We show that loss of p53 in myeloid progenitor cells circumvents the deleterious effects of Kras activation and establishes aberrant self-renewal, paving the way for AML development. Although we cannot rule out that additional functions of p53 contribute to tumor suppression in this context, the de novo acquisition of self-renewal in cells that normally lack this capability could be a key function of p53 mutations in AML and perhaps other tumor types where a differentiation hierarchy plays a role in disease.
Similar to activated Ras, the BCR-ABL fusion protein drives proliferation and differentiation of HSCs and myeloid progenitors, and can initiate an MPD—chronic myeloid leukemia (CML)—that requires the involvement of self-renewing HSCs (Huntly et al. 2004; Williams and Sherr 2008; Schemionek et al. 2010). If untreated, the disease inevitably progresses to fatal blast crisis (CML-BC), often resembling AML. Interestingly, the most frequent secondary alterations in myeloid CML-BC involve p53, which is mutated in 25%–30% of cases or repressed by activation of its negative regulator, MDM2 (Calabretta and Perrotti 2004). Of note, homozygous deletions encompassing ARF are a hallmark mutation in lymphoid CML-BC- and BCR-ABL-positive ALL, and are associated with poor prognosis (Williams and Sherr 2008). Given the similarity between KrasG12D and BCR-ABL in their effects on HSCs and myeloid progenitors, aberrant self-renewal caused by loss of p53 could explain the high frequency of p53 mutations in myeloid CML-BC.
Interestingly, loss of the Pten tumor suppressor, which activates the PI3K pathway downstream from Ras and can also trigger senescence, depletes the HSC compartment and can induce MPDs (Yilmaz et al. 2006; J Zhang et al. 2006). In preliminary studies, we applied our approach to introduce LGshp53CreER into Ptenfl/fl HSPCs, and saw a strong acceleration of leukemogenesis resulting from combined loss of p53 and Pten (Z Zhao, J Zuber, and SW Lowe, unpubl.). Other studies using standard germline methods noted similar effects (JY Lee and S Morrison, unpubl.), suggesting that p53 may act generally in the hematopoietic compartment to limit self-renewal in response to oncogenic stress.
p53 has long been associated with cellular immortalization in vitro (Jenkins et al. 1984), which is an acquired property that confers limitless replicative potential and is considered to be an essential property of cancer cells (Hanahan and Weinberg 2000). As such, the process of immortalization may conceptually and mechanistically overlap with the physiological process of self-renewal. Although the mechanism whereby p53 loss promotes immortalization or enhances self-renewal remains elusive, it is noteworthy that p53 can serve as a barrier to epigenetic reprogramming of somatic cells into pluripotent stem cells (Puzio-Kuter and Levine 2009). Accordingly, loss of p53 might contribute to the formation of leukemia-initiating cells, which by definition have maintained or reacquired the capacity for indefinite self-renewal through accumulated mutations and/or epigenetic changes (Passegue and Weisman 2005). Such cells have properties reminiscent of cancer stem cells, which are considered to be inherently more aggressive and refractory to chemotherapy (Guan and Hogge 2000; Jordan and Guzman 2004; Holtz et al. 2005). Consequently, our results suggest a biological explanation for the occurrence of p53 mutations in the most aggressive and drug-resistant AMLs.
Materials and methods
Retroviral constructs
LGmCreER was constructed in the MSCV backbone (Clontech) using standard cloning techniques. We sequentially inserted a GFP PCR product, a customized double-stranded oligonucleotide containing a SalI/NsiI cloning site and two lox511 sites (Hoess et al. 1986) flanking an MfeI/AvrII cloning site. The PGK promoter and CreERT2 (Feil et al. 1997) were cloned by PCR between the two lox511 sites. The empty miR30 context and previously validated miR30-embedded shRNAs targeting firefly luciferase (Silva et al. 2005) and mouse Trp53 (Dickins et al. 2005) were cloned into the SalI/NsiI site. In addition to “self-deleting” LGmCreER harboring two lox511 sites flanking PGK-CreERT2, we created a “non-self-deleting” version of LGmCreER by removing the first lox511 site. The latter vector was used in all experiments in which GFP-positive cells were traced for in vitro proliferation, colony formation, stem/progenitor cells analysis, and sorting to ensure that GFP-positive cells uniformly harbored both the shRNA and CreERT2 (typically, a minor fraction of “self-deleting” LGmCreER virus does not carry PGK-CreERT2 due to leaky Cre activity in packaging cells) (see Supplemental Fig. S1 for details). MSCV-CreERT2-IRES-GFP was cloned by PCR of CreERT2 into the MSCV-IRES-GFP backbone (Clontech). The LMP vector has been described previously (Dickins et al. 2005).
Mouse strains
All mouse strain-related experiments were performed under the approval of the Cold Spring Harbor Laboratory Animal Care and Use Committee. LSL KrasG12D and Mx1-Cre mice (kind gifts from Dr. Kevin Shannon, University of California at San Francisco) were backcrossed onto the C57BL/6 background for more than six generations. Trp53loxp/loxp mice (Jonkers et al. 2001) were backcrossed onto C57BL/6 for four generations before generating quadruple Mx1-Cre;LSL-KrasG12D;p53loxp/loxp (CPK) mice. Genotyping was performed according to standard protocols available at http://mouse.ncifcrf.gov. For induction of Cre expression, mice of ∼6 wk of age were injected i.p. with three doses of 250 μg of pIpC (Sigma-Aldrich) every other day.
HSPC isolation, retroviral transduction, and transplantation
HSPCs were isolated from fetal livers (embryonic day 13.5–15.5), cultured, and retrovirally transduced as described (Schmitt et al. 2002a), except that only early viral supernatant harvested from packaging cells 24, 30, and 36 h after transfection was used in order to minimize leaky self-deletion of PGK-CreERT2. Retroviral transduction of GFP-expressing constructs was assessed 24 h after the last infection by flow cytometry (Guava EasyCyte, Guava Technologies). To induce Cre activity in vitro, infected HSPCs were treated with 0.2 μM 4-OHT (Sigma-Aldrich; dissolved in 95% cold ethanol) for 36–48 h. Approximately 2 × 106 cells were injected into the tail vein of 8- to 10-wk-old lethally irradiated syngeneic recipient mice (9.0 Gy total in two split doses) (Gammacell 40 Exactor; MDS Nordion). To induce Cre activity in vivo, 4 mg of 4-OHT (Sigma-Aldrich; emulsified at 40 mg/mL in sunflower seed oil by sonication) was administrated i.p. for four consecutive days after reconstitution of recipient mice. For secondary leukemia transplantation, 2 × 106 leukemia cells freshly harvested from bone marrow were transplanted into sublethally irradiated syngeneic recipient mice (4.5 Gy as single dose).
HSPC proliferation and colony formation analysis and other in vitro assays
LSL-KrasG12D and wild-type HSPCs were infected with a non-self-deleting version of LGmCre vectors containing shluc or shp53 and treated with 0.2 μM 4-OHT for 48 h. For the BrdU assay, we used an APC BrdU Flow Kit (BD Pharmingen). BrdU was added 2 h before analysis at a final concentration of 1 μM; staining and analysis were carried out according to the manufacturer's protocol. For methylcellulose colony formation, 5000 GFP-positive cells were plated into Methocult GF M3534 (Stem Cell Technologies), cultured on 35-mm dishes, and imaged with GFP fluorescence and bright-field phase-contrast optics of a Zeiss Observer Z.1 inverted microscope. After 7 d of culture, images of the entire surface were collected using a 5× objective, a Zeiss MRm monochrome CCD camera, and Zeiss Axiovision 4.7.3 software, and were subsequently analyzed by ImageJ. Colonies were harvested to repeat the same procedure for serial replating. For STIFA culture, 2.5 × 106 transduced and 4-OHT-treated HSPCs were transferred into 6 mL of STIFA medium (composition exactly as described in Akala et al. 2008); cytokines were replenished every 3 d. Proliferation of stem and progenitor cells was analyzed by flow cytometry. LSL-LacZ NIH-3T3 fibroblasts (a kind gift from Ronald DePinho) were transduced with LGmCreER and MSCV-CreERT2-IRES-GFP, treated with 0.5 μM 4-OHT for 72 h or left untreated, and subsequently stained using the Stain for β-Gal Expression in Tissue kit (Millipore).
Histocytological and molecular characterization of leukemias
Peripheral blood smears (Wright-Giemsa-stained) of recipient mice were monitored starting from 4 wk after transplantation. Ultrasonic imaging was performed 7 wk after transplantation using a Vevo 770 Imaging System (Visualsonics). Fluorescence-based whole-body GFP imaging was performed using an Illumatool LT-9900 Bright Light System (LightTools, Optical Research) as described (Schmitt et al. 2002b). Animals were sacrificed by CO2 euthanasia upon severe leukocytosis and/or moribund appearance. Statistical evaluation of overall survival was based on the log-rank (Mantel-Cox) test for comparison of the Kaplan-Meier event time format. Organs were fixed in 10% neutral buffered formalin and were processed to obtain paraffin sections for histological staining. Bone marrow cells were flushed from tibias and femurs with PBS with 2% FBS. Erythrocytes were lysed in ACK (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) for 5 min.; nucleated cells were resuspended in PBS/2% FBS and filtered through a nylon screen (100 μm) to obtain a single-cell suspension. Whole bone marrow cells were stained with PE-conjugated antibodies for Sca1, cKit, CD34, Mac1, Gr1, B220, Thy1, and Ter119 (BD Biosciences) for single staining. Data were collected on an LSR-II flow cytometer (BD Biosciences), and were analyzed using FACSDiva (BD Biosciences) and FlowJo (Treestar) software.
Flow cytometry analysis and FACS
For flow cytometry analysis of phosphorylated Ras effectors, bone marrow cells were harvested from femurs and tibias into IMDM containing 1% BSA (Sigma). Erythrocytes were lysed (ACK), and mononuclear cells were incubated in harvesting medium for 45 min at 37°C. For growth factor stimulation, cells were incubated with 10 ng/mL murine GM-CSF (Invitrogen) for 15 min prior to fixation. Antibodies and experimental procedures were exactly as described previously (Van Meter et al. 2007). For stem and progenitor analysis and sorting, we used lineage markers (Mac1, Gr1, CD3, CD4, CD8, B220, and Ter119), Sca1, cKit, IL-7R, CD150, and CD48. Antibodies were conjugated directly or biotinylated and purchased from e-Bioscience, BD Biosciences, and BioLegend; cells were analyzed and sorted on an Aria-II cell sorter (BD Biosciences). HSCs, MPPs, and myeloid progenitors were identified as CD150+CD48−Sca1+Lin−cKit+, CD150−CD48−Sca1+Lin−cKit+, and cKit+Sca1−Lin−IL-7Rα− populations, respectively (Kiel et al. 2005; Kim et al. 2006; Akala et al. 2008). For mosaic transplantation recipients, only GFP-positive HSCs or myeloid progenitors were sorted. All HSCs and myeloid progenitors were doubly sorted into PBS, and were transplanted by retro-orbital venous sinus injection into B6/SJL (Ly5.1) mice lethally irradiated with 8.2 Gy (single dose), together with a radioprotective dose of 2 × 105 C57BL/6 bone marrow cells.
PCR analysis of Cre-mediated recombination
To assess Cre-mediated recombination of the LSL cassette in vivo, genomic DNA was extracted from peripheral blood prior to and after 4-OHT treatment, as well as from bone marrow and spleen of diseased mice. To detect recombination events with high sensitivity, we designed a two-step allele-specific PCR (Supplemental Fig. S3). For the first step, we used primers flanking the remaining loxP site (KLSLfw3, 5′-AAGCAAGGCAGAAGTCACAGAGG-3′; LoxedRasRv, 5′-TCCGAATTCAGTGACTACAGATGTACAGAG-3′) and amplified as follows: initially at 95°C for 10 min; 34 cycles of 95°C for 30 sec, 64°C for 45 sec, and 72°C for 60 sec; and ending with 72°C for 7 min, yielding products of 709 and 749 base pairs (bp) for wild-type and recombined alleles, respectively. To sensitively detect recombined alleles even in small cellular subfractions (e.g., peripheral blood), a 1:100 dilution of the primary PCR products was used in a secondary PCR using a nested primer set specifically amplifying alleles harboring a single remaining loxP site (KLSLfw4, 5′-GTGCAGTTTTGACACCAGCTTCG-3′; KLSLrev4, 5′-CGCATGAGCTTGTCGACATAACTTCG-3′) with the following cycling: initially at 95°C for 10 min; 28 cycles of 95°C for 30 sec, 65°C for 30 sec, and 72°C for 60 sec; and ending with 72°C for 7 min.
Immunoblotting and Southern blotting analysis
Wild-type MEFs were infected with LMPmir30, LMPshp53, LGmCreER, and LGshp53CreER. Infection efficiencies were >95%, indicated by GFP percentage analyzed 48 h post-infection. Infected cells were incubated with or without 0.5 μM 4-OHT for 48 h. Bone marrow cells were harvested as above from wild-type, Kras-shp53 leukemic, and p53−/− and p19Arf−/− control leukemic mice. Control leukemias were generated by transduction of MLL/ENL into p53−/− and p19Arf−/− HSPCs as described previously (Zuber et al. 2009). To induce p53, all samples were treated with 1 μg/mL adriamycin for 5 h and then harvested for whole-cell lysates. p53 immunoblotting was performed as described previously (Hemann et al. 2003). Genomic DNA was isolated from infected untreated or 4-OHT-treated (0.5 μM, 4 d) MEFs and packaging cells at different time points after transfection. Southern blotting of NheI-digested DNA was performed using standard techniques. Hybridization was performed in CHURCH buffer (0.25 M sodium phosphate buffer at pH 7.2, 1 mM EDTA, 1% BSA, 7% SDS) under addition of 100 μg/mL sonicated sperm DNA using a PCR-amplified GFP probe labeled with αP32-dATP (Megaprime DNA labeling system, Amersham).
Acknowledgments
We thank Kristin Diggins-Lehet, Meredith Taylor, Jacqueline Cappellani, and Janelle Simon for generating mice and HSPCs; Aigoul Nourjanova for histopathology; Lisa Bianco and her team for animal support; Pamela Moody for flow cytometry support; Stephen Hearn for microscopic imaging and data analysis; and Dr. Michelle LeBeau for SKY analysis. We gratefully acknowledge Dr. Shane Mayack for experimental suggestions and critically reviewing the manuscript. We thank Drs. Agustin Chicas, Cornelius Miething, Mona Spector, Katherine McJunkin, and Claudio Scuoppo, and other members of the Lowe laboratory for constructive discussion and advice. We are grateful to Dr. Sean Morrison for sharing unpublished data and helpful discussions. This work was supported by a Specialized Center of Research Grant from the Leukemia and Lymphoma Society, NIH grant R37 CA72614, and generous gifts from the Don Monti Memorial Research Foundation. J.Z. was supported by a research fellowship from the German Research Foundation (DFG), and is the Andrew Seligson Memorial Fellow at CSHL. S.C.K. is a Scholar of the Leukemia and Lymphoma Society; S.W.L. is a Howard Hughes Medical Institute investigator.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1940710.
Supplemental material is available at http://www.genesdev.org.
References
- Akala OO, Park IK, Qian D, Pihalja M, Becker MW, Clarke MF 2008. Long-term haematopoietic reconstitution by Trp53−/− p16Ink4a−/− p19Arf−/− multipotent progenitors. Nature 453: 228–232 [DOI] [PubMed] [Google Scholar]
- Akashi K, Traver D, Miyamoto T, Weissman IL 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404: 193–197 [DOI] [PubMed] [Google Scholar]
- Alcalay M, Meani N, Gelmetti V, Fantozzi A, Fagioli M, Orleth A, Riganelli D, Sebastiani C, Cappelli E, Casciari C, et al. 2003. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest 112: 1751–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armesilla-Diaz A, Bragado P, Del Valle I, Cuevas E, Lazaro I, Martin C, Cigudosa JC, Silva A 2009. p53 regulates the self-renewal and differentiation of neural precursors. Neuroscience 158: 1378–1389 [DOI] [PubMed] [Google Scholar]
- Bardeesy N, Bastian BC, Hezel A, Pinkel D, DePinho RA, Chin L 2001. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol Cell Biol 21: 2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 [DOI] [PubMed] [Google Scholar]
- Braun BS, Tuveson DA, Kong N, Le DT, Kogan SC, Rozmus J, Le Beau MM, Jacks TE, Shannon KM 2004. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci 101: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP 2009. Awakening guardian angels: Drugging the p53 pathway. Nat Rev Cancer 9: 862–873 [DOI] [PubMed] [Google Scholar]
- Bueso-Ramos CE, Yang Y, deLeon E, McCown P, Stass SA, Albitar M 1993. The human MDM-2 oncogene is overexpressed in leukemias. Blood 82: 2617–2623 [PubMed] [Google Scholar]
- Buttitta F, Marchetti A, Gadducci A, Pellegrini S, Morganti M, Carnicelli V, Cosio S, Gagetti O, Genazzani AR, Bevilacqua G 1997. p53 alterations are predictive of chemoresistance and aggressiveness in ovarian carcinomas: A molecular and immunohistochemical study. Br J Cancer 75: 230–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabretta B, Perrotti D 2004. The biology of CML blast crisis. Blood 103: 4010–4022 [DOI] [PubMed] [Google Scholar]
- Chan IT, Kutok JL, Williams IR, Cohen S, Kelly L, Shigematsu H, Johnson L, Akashi K, Tuveson DA, Jacks T, et al. 2004. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest 113: 528–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MP, Seita J, Weissman IL 2008. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol 73: 439–449 [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436: 725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG 2009. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138: 1083–1095 [DOI] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, et al. 2005. Tumour biology: Senescence in premalignant tumours. Nature 436: 642. [DOI] [PubMed] [Google Scholar]
- Cozzio A, Passegue E, Ayton PM, Karsunky H, Cleary ML, Weissman IL 2003. Similar MLL-associated leukemias arising from self-renewing stem cells and short-lived myeloid progenitors. Genes Dev 17: 3029–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW 2005. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 37: 1289–1295 [DOI] [PubMed] [Google Scholar]
- Donehower LA 1996. The p53-deficient mouse: A model for basic and applied cancer studies. Semin Cancer Biol 7: 269–278 [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr, Butel JS, Bradley A 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215–221 [DOI] [PubMed] [Google Scholar]
- Dumble M, Moore L, Chambers SM, Geiger H, Van Zant G, Goodell MA, Donehower LA 2007. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood 109: 1736–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL 1999. Disruption of the ARF–Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev 13: 2658–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG 2001. A gene hypermethylation profile of human cancer. Cancer Res 61: 3225–3229 [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P 1997. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 237: 752–757 [DOI] [PubMed] [Google Scholar]
- Forni PE, Scuoppo C, Imayoshi I, Taulli R, Dastru W, Sala V, Betz UA, Muzzi P, Martinuzzi D, Vercelli AE, et al. 2006. High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. J Neurosci 26: 9593–9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese KK, Tuveson DA 2007. Maximizing mouse cancer models. Nat Rev Cancer 7: 645–658 [DOI] [PubMed] [Google Scholar]
- Gilliland DG, Jordan CT, Felix CA 2004. The molecular basis of leukemia. Hematology Am Soc Hematol Educ Program 2004: 80–97 [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Klatt P, Delgado S, Conde E, Lopez-Rios F, Sanchez-Cespedes M, Mendez J, Antequera F, Serrano M 2006. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature 440: 702–706 [DOI] [PubMed] [Google Scholar]
- Guan Y, Hogge DE 2000. Proliferative status of primitive hematopoietic progenitors from patients with acute myelogenous leukemia (AML). Leukemia 14: 2135–2141 [DOI] [PubMed] [Google Scholar]
- Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T 2008. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia 22: 1539–1541 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000. The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C, Hannon GJ, Lowe SW 2003. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet 33: 396–400 [DOI] [PubMed] [Google Scholar]
- Hemann MT, Zilfou JT, Zhao Z, Burgess DJ, Hannon GJ, Lowe SW 2004. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci 101: 9333–9338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H, Eick D 1994. Mediation of c-Myc-induced apoptosis by p53. Science 265: 2091–2093 [DOI] [PubMed] [Google Scholar]
- Hoess RH, Wierzbicki A, Abremski K 1986. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res 14: 2287–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz MS, Forman SJ, Bhatia R 2005. Nonproliferating CML CD34+ progenitors are resistant to apoptosis induced by a wide range of proapoptotic stimuli. Leukemia 19: 1034–1041 [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S 2009. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature 460: 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntly BJ, Shigematsu H, Deguchi K, Lee BH, Mizuno S, Duclos N, Rowan R, Amaral S, Curley D, Williams IR, et al. 2004. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell 6: 587–596 [DOI] [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, et al. 2004. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med 351: 657–667 [DOI] [PubMed] [Google Scholar]
- Jenkins JR, Rudge K, Currie GA 1984. Cellular immortalization by a cDNA clone encoding the transformation-associated phosphoprotein p53. Nature 312: 651–654 [DOI] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T 2001. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410: 1111–1116 [DOI] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet 29: 418–425 [DOI] [PubMed] [Google Scholar]
- Jordan CT, Guzman ML 2004. Mechanisms controlling pathogenesis and survival of leukemic stem cells. Oncogene 23: 7178–7187 [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC 2009. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ 2005. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Kim I, He S, Yilmaz OH, Kiel MJ, Morrison SJ 2006. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood 108: 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, Cardiff RD, Carter JS, de Coronado S, Downing JR, Fredrickson TN, et al. 2002. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood 100: 238–245 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. 2006. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442: 818–822 [DOI] [PubMed] [Google Scholar]
- Krizhanovsky V, Lowe SW 2009. Stem cells: The promises and perils of p53. Nature 460: 1085–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K 1995. Inducible gene targeting in mice. Science 269: 1427–1429 [DOI] [PubMed] [Google Scholar]
- Lavau C, Szilvassy SJ, Slany R, Cleary ML 1997. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J 16: 4226–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Oren M 2009. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer 9: 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M 2009. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AW, Lowe SW 2001. Oncogenic ras activates the ARF–p53 pathway to suppress epithelial cell transformation. Proc Natl Acad Sci 98: 5025–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Elf SE, Miyata Y, Sashida G, Liu Y, Huang G, Di Giandomenico S, Lee JM, Deblasio A, Menendez S, et al. 2009. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J 2001. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci 98: 9209–9214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ 2003. Tumor suppression by Ink4a–Arf: Progress and puzzles. Curr Opin Genet Dev 13: 77–83 [DOI] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF Jr, Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. 1990. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250: 1233–1238 [DOI] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA 2009. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW 2009. Tumour suppression by p53: A role for the DNA damage response? Nat Rev Cancer 9: 714–723 [DOI] [PubMed] [Google Scholar]
- Meletis K, Wirta V, Hede SM, Nister M, Lundeberg J, Frisen J 2006. p53 suppresses the self-renewal of adult neural stem cells. Development 133: 363–369 [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Morrison SJ 2004. Diverse mechanisms regulate stem cell self-renewal. Curr Opin Cell Biol 16: 700–707 [DOI] [PubMed] [Google Scholar]
- Nahi H, Lehmann S, Bengtzen S, Jansson M, Mollgard L, Paul C, Merup M 2008. Chromosomal aberrations in 17p predict in vitro drug resistance and short overall survival in acute myeloid leukemia. Leuk Lymphoma 49: 508–516 [DOI] [PubMed] [Google Scholar]
- Okada S, Nakauchi H, Nagayoshi K, Nishikawa S, Miura Y, Suda T 1992. In vivo and in vitro stem cell function of c-kit- and Sca-1-positive murine hematopoietic cells. Blood 80: 3044–3050 [PubMed] [Google Scholar]
- Orsulic S, Li Y, Soslow RA, Vitale-Cross LA, Gutkind JS, Varmus HE 2002. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell 1: 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302–305 [DOI] [PubMed] [Google Scholar]
- Passegue E, Weisman IL 2005. Leukemic stem cells: Where do they come from? Stem Cell Rev 1: 181–188 [DOI] [PubMed] [Google Scholar]
- Passegue E, Jamieson CH, Ailles LE, Weissman IL 2003. Normal and leukemic hematopoiesis: Are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sci 100: 11842–11849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzio-Kuter AM, Levine AJ 2009. Stem cell biology meets p53. Nat Biotechnol 27: 914–915 [DOI] [PubMed] [Google Scholar]
- Rambhatla L, Bohn SA, Stadler PB, Boyd JT, Coss RA, Sherley JL 2001. Cellular senescence: Ex vivo p53-dependent asymmetric cell kinetics. J Biomed Biotechnol 1: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabnis AJ, Cheung LS, Dail M, Kang HC, Santaguida M, Hermiston ML, Passegue E, Shannon K, Braun BS 2009. Oncogenic Kras initiates leukemia in hematopoietic stem cells. PLoS Biol 7: e59 doi: 10.1371/journal.pbio.1000059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schemionek M, Elling C, Steidl U, Baumer N, Hamilton A, Spieker T, Gothert JR, Stehling M, Wagers A, Huettner CS, et al. 2010. BCR-ABL enhances differentiation of long-term repopulating hematopoietic stem cells. Blood 115: 3185–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW 2002a. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell 1: 289–298 [DOI] [PubMed] [Google Scholar]
- Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW 2002b. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 109: 335–346 [DOI] [PubMed] [Google Scholar]
- Schoch C, Schnittger S, Klaus M, Kern W, Hiddemann W, Haferlach T 2003. AML with 11q23/MLL abnormalities as defined by the WHO classification: Incidence, partner chromosomes, FAB subtype, age distribution, and prognostic impact in an unselected series of 1897 cytogenetically analyzed AML cases. Blood 102: 2395–2402 [DOI] [PubMed] [Google Scholar]
- Scholl C, Gilliland DG, Frohling S 2008. Deregulation of signaling pathways in acute myeloid leukemia. Semin Oncol 35: 336–345 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Sevignani C, Wlodarski P, Kirillova J, Mercer WE, Danielson KG, Iozzo RV, Calabretta B 1998. Tumorigenic conversion of p53-deficient colon epithelial cells by an activated Ki-ras gene. J Clin Invest 101: 1572–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, McCormick F 2002. The RB and p53 pathways in cancer. Cancer Cell 2: 103–112 [DOI] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. 2005. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet 37: 1281–1288 [DOI] [PubMed] [Google Scholar]
- Silver DP, Livingston DM 2001. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell 8: 233–243 [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ 2005. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci 102: 13212–13217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- TeKippe M, Harrison DE, Chen J 2003. Expansion of hematopoietic stem cell phenotype and activity in Trp53-null mice. Exp Hematol 31: 521–527 [DOI] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, et al. 2004. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 5: 375–387 [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K 2009. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter ME, Diaz-Flores E, Archard JA, Passegue E, Irish JM, Kotecha N, Nolan GP, Shannon K, Braun BS 2007. K-RasG12D expression induces hyperproliferation and aberrant signaling in primary hematopoietic stem/progenitor cells. Blood 109: 3945–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattel E, Preudhomme C, Hecquet B, Vanrumbeke M, Quesnel B, Dervite I, Morel P, Fenaux P 1994. p53 mutations are associated with resistance to chemotherapy and short survival in hematologic malignancies. Blood 84: 3148–3157 [PubMed] [Google Scholar]
- Williams RT, Sherr CJ 2008. The INK4–ARF (CDKN2A/B) locus in hematopoiesis and BCR-ABL-induced leukemias. Cold Spring Harb Symp Quant Biol 73: 461–467 [DOI] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445: 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441: 475–482 [DOI] [PubMed] [Google Scholar]
- Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. 2008. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CC, Kaba M, Ge G, Xie K, Tong W, Hug C, Lodish HF 2006. Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells. Nat Med 12: 240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. 2006. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441: 518–522 [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang J, Liu Y, Sidik H, Young KH, Lodish HF, Fleming MD 2009. Oncogenic Kras-induced leukemogeneis: Hematopoietic stem cells as the initial target and lineage-specific progenitors as the potential targets for final leukemic transformation. Blood 113: 1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, et al. 2008. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature 455: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilfou JT, Lowe SW 2009. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol 1: a001883 doi: 10.1101/cshperspect.a001883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Radtke I, Pardee TS, Zhao Z, Rappaport AR, Luo W, McCurrach ME, Yang MM, Dolan ME, Kogan SC, et al. 2009. Mouse models of human AML accurately predict chemotherapy response. Genes Dev 23: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]