Abstract
The Lac repressor has been used as a tool to understand protein–DNA recognition for many years. Recent experiments have demonstrated the ability of the Lac repressor to control gene expression in various eukaryotic systems, making the quest for an arsenal of protein–DNA binding partners desirable for potential therapeutic applications. Here, we present the results of the most exhaustive screen of Lac repressor-DNA binding partners to date, resulting in the elucidation of functional rules for Lac–DNA binding. Even within the confines of a single protein–DNA scaffold, modes of binding of different protein–DNA partners are sufficiently diverse so as to prevent elucidation of generalized rules for recognition for a single protein, much less an entire protein family.
Keywords: specificity, lac operon, protein engineering, LacI-GalR
Introduction
DNA binding proteins play an essential role in genetic and epigenetic functions in all organisms. The molecular basis of DNA recognition has been a topic of interest for biologists since the discovery of the genetic code. In 1976, Seeman et al.1 were the first to discuss the role of hydrogen bonding in specific recognition. Subsequently, Pabo and Sauer2 suggested the existence of a “Recognition Code,” or a simple set of rules that predict amino acid–nucleotide binding partners. Shortly thereafter, Matthews3 argued that while there are general rules for protein–DNA recognition, the complexity and individuality of each complex argues against the possibility of a simple Recognition Code. This did not deter the field from attempting to identify rules for recognition. Over the next 20 years, analyses of protein–DNA interactions on different molecular levels have contributed to our knowledge of protein–DNA specificity.
In the pioneering work of Seeman et al.,1 it was proposed that sequence-specific DNA recognition is accomplished by amino acid side chains that make two hydrogen bonds to basepairs. As protein–DNA structures became available, more quantitative analyses were performed by various groups. Pabo and Sauer2 examined three complexes and identified a list of amino acid base contacts, most notably bidentate and/or bifurcated bonds. They argued that amino acids might differentiate between nucleotides based on the local environment or the ability to form bifurcated bonds. From analysis of 20 complexes, Suzuki4 proposed that a protein–DNA recognition code exists and is explained by chemical rules (favorable interactions between side chains and bases) and stereochemical rules (accessible contact positions between amino acids and bases, depending on geometry and residue size).
The theories of recognition suggested by Seeman et al., Suzuki, and Mandel-Gutfreund et al. have been supported by more recent work. Luscombe et al.5 examined hydrogen bonds, van der Waals contacts, and water-mediated bonds in 129 protein–DNA complexes. Consistent with previous results, they found that arginine and lysine account for the majority of interactions with guanine and that asparagine and glutamine prefer adenine. They discovered that van der Waals contacts make up two-thirds of all protein–DNA interactions, whereas hydrogen bonds and water-mediated bonds account for one-sixth each. They concluded that van der Waals contacts do not generally contribute to specificity, with the exception of threonine and aromatic residues which make favorable interactions. The majority of nonhydrogen bonded pairings can be explained by random but neutral dockings between protein and DNA. They suggested that water-mediated bonds are mostly used as space fillers for stability, but in certain contexts, can be used for specificity, as in the Trp repressor.6 A year later, Luscombe and Thornton used the same set of proteins to perform a family-level analysis of the effect of mutations on DNA-sequence recognition.7 They argued that certain rules apply to all families of DNA-binding proteins (see also Choo and Klug, 19978) but specific recognition of DNA can only be understood by studying protein families individually.
Experiments and analyses of zinc-finger proteins (ZFPs) have supported the concept that DNA recognition should be examined at the protein-family level. The phage display technique9 has identified thousands of ZFPs that recognize various DNA sequences. Through this and other methods, functional rules for ZFP-DNA binding have been determined mostly by experimental techniques (for a review, see Sera, 200910).
It is obvious that a recognition code for ZFPs is not valid for other protein domains. However, it is not necessarily valid for tandem zinc-fingers or in vivo functions either.11 Zinc-fingers proteins have provided a vehicle to study amino acid specificity, docking arrangements, and other properties of protein–DNA binding. Crystallographic and solution structures of ZFPs and other DNA-binding proteins provide a means to understand how a particular protein binds a particular DNA sequence, but cumulative knowledge to date is insufficient to allow for the prediction of a protein sequence that will bind a particular DNA sequence or vice versa. To fully understand protein–DNA interactions, studies of other DNA binding proteins are required and re-examination of current theory may be required.
In contrast to ZFP's, the Lac repressor contains a helix-turn-helix DNA binding motif. The second helix in the helix-turn-helix domain is generally responsible for specific nucleotide sequence recognition. The Lac repressor has been the model system for studying prokaryotic transcriptional regulation since the pioneering work of Jacob and Monod in 1961.12 Recently, applications of the Lac operon to nonbacterial systems have highlighted its use as a regulator of desired genes in vivo. It has been used to regulate expression of target genes in mammals,13–15 stem cells,16 plants,17 and breast cancer lines.18 Most promising for potential gene therapy applications is the integration of the Lac molecular switch into mice.13 The largest obstacle for using the Lac operon as a therapeutic genetic regulator is the requirement that the DNA binding site is pseudosymmetric. Recent experiments in our lab have produced a heterodimeric Lac repressor mutant, which can recognize an asymmetric operator sequence.19 Combined with the heterodimeric Lac, a functional recognition code could be a powerful tool for potential therapeutics.
A variety of experiments have been performed to identify the amino acid–nucleotide pairs that are most important to complex formation between the Lac repressor and DNA; they are residues 17, 18, and 22 of the repressor and operator positions 4, 5, and 620–23 (Fig. 1). These and other studies have identified mutant Lac repressors that bind to various operator sequences that are functional in vivo.24–26 In 1990, Lehming et al. argued that rules of recognition had been elucidated for Lac repressor–operator interactions and they likely apply to other bacterial repressors.21
Figure 1.
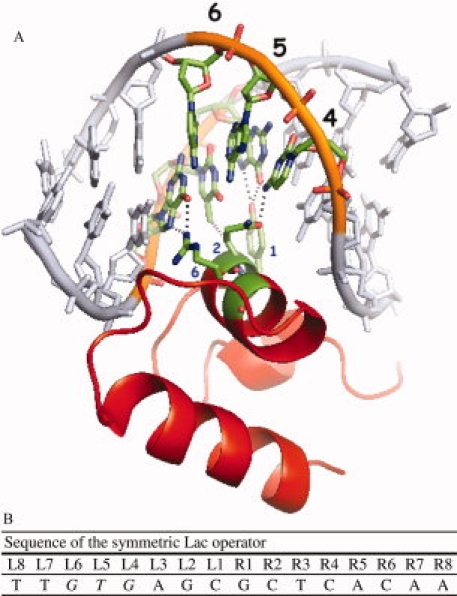
(A) The structure of the Lac repressor bound to the symmetric operator. The recognition helix fits into the major groove of the DNA and residues 1, 2 and 6 of the recognition helix (Y17, Q18, R22) make specific interactions with operator positions 4, 5 and 6 of the operator (Bell and Lewis, 2000). Reprinted with permission from Oxford University Press, Daber and Lewis, Towards Evolving a Better Repressor, Protein Engineering Design and Selection, 2009, vol. 22, no. 11, p 673–683. (B) The symmetric Lac operator as identified by Sadler et al. (1983) and typically referred to as ‘symL (−1)’. “L” and “R” denote the left and right half-sites of the operator. An is available in the electronic version of the article. PRO389 Figure 1
Here, we describe a set of experiments that constitute the most exhaustive screen of Lac repressor–operator binding pairs to date. A library of Lac repressor mutants consisting of fully randomized amino acids at residues 17, 18, and 22 was screened against 64 symmetric Lac operator variants. The results suggest that rules for recognition are not simple even for the single, well defined Lac repressor–operator system, much less for the LacI-GalR protein family in general. While trends can often be identified for some pairs, such as conservation of particular amino acids, no single set of rules can be applied to all repressor–operator pairs. Ultimately, the principles that underlie protein–DNA binding events cannot be generalized and recognition must be distilled down to the chemical rules determining geometry and electrostatics for each protein and DNA surface.
The results presented here provide the largest set of protein–DNA binding partners to date. They are useful for theoretical and computational analyses of protein–DNA interactions and contribute to methods for developing therapeutic genetic regulators. Combining the functional rules presented here with the recent work in our lab demonstrating that a heterodimeric Lac repressor can bind to an asymmetric DNA sequence,19 we have begun to address the one of the greatest challenges for using the Lac repressor as a therapeutic agent.
Results and Discussion
The ability of mutant repressors to prevent transcription of various operators was measured using an in vivo two plasmid system where one plasmid contains a Lac repressor gene and the other contains the GFPmut3.1 gene controlled by the Lac promoter and a given operator.24 If the repressor mutant binds the operator variant, GFP expression is decreased; the stronger the binding interaction, the lower the expression. A repressor plasmid library was transformed into competent cells containing a given, single, operator. Fluorescence Activated Cell Sorting (FACS) was used to screen individual cells of the transformation mixture to separate low and high fluorescing populations. Functional repressors were sequenced, purified, and assayed again with their corresponding operators to obtain a quantitative measure of how well they repress and derepress expression of GFP (fractional expression in the repressed and induced state).
Sixty-four different operators were cloned into the reporter plasmid, all taking the form: 5′-A ATT XXX AGC GCT YYY AAT T-3′ where “X” is any nucleotide and “Y” represents the nucleotides required to make the operator fully symmetric (the symmetric operator 5′-AATTGTGAGCGCTCACAATT-3′, also called SymL(−1), was previously identified by Sadler et al.27). Operator variants are named by the nucleotides that exist at positions 6, 5, and 4 in the left half-site of the operator [Fig. 1(B)]. For example, an operator with the sequence 5′-A ATT CTG AGC GCT CAG AAT T-3′ would be named “CTG.” In addition, nine reporter plasmids were constructed which contain the natural operators for the Galactose, Maltose, Ribitol, Fructose, Purine, Ribose, Cytosine, Raffinose, and Sucrose repressors (Supporting Information Table sI). All reporters were analyzed in the absence of repressor to ensure that the alterations did not affect GFP expression. Repressor mutants are named by the amino acids that exist at residues 17, 18, and 22; for example, a mutant with alanine, threonine, and arginine at residues 17, 18, and 22 would be named “ATR.”
General statistics
In total, 332 unique repressor–operator combinations were identified (Fig. 2, Supporting Information Table sII). Out of the 64 different operator sequences analyzed here, 26 were bound by at least one repressor mutant. The most frequently found nucleotide was guanine, followed by thymine, then adenine and cytosine. Out of the 8000 possible repressor mutants, 195 were found to bind one or more operators. Of the 20 amino acids (AA), all 20 were found at residue 17, 15 were found at residue 18, and 13 at residue 22 (Fig. 2). Arginine is the most common AA, followed by alanine, serine, asparagine, and threonine. Predictably, aspartic acid and glutamic acid are the least common residues, followed by tryptophan and phenylalanine. Hydrophobicity, molecular weight, and isoelectric point do not correlate with amino acid frequency at any of the residues, whether considering all the mutants together, or grouping them according to which operators they bind (data not shown).
Figure 2.

Matrix comparison of the frequency of amino acid — base pair combinations for the Lac repressor — operator binding partners. Possible nucleotides are listed down the left side and amino acids are listed across the top. Residue 22 interacts with operator position 6 (A) and residues 18 (B) and 17 (C) interact with positions 5 and 4, respectively. High frequency interactions are boxed.
There are 117 repressor mutants that bind a single operator sequence (unique repressors). Notably, one of these is the wild-type repressor, YQR, which binds only to the operator sequence GTG. The number of unique repressors does not correlate with the total number of repressor mutants for any one operator. For example, none of the 16 repressor mutants that bind GCA are unique, whereas all nine that bind GAC are. This fact suggests that the chemical properties of each operator variant make it more or less likely to attract promiscuous repressors. Similarly, the properties of each repressor mutant make it more or less likely to interact uniquely with a particular operator. For the 36 repressor mutants that bind three or more operator sequences, they are likely interacting with regions of DNA that are not unique to the particular operator. This demonstrates that the uniqueness of a particular repressor–operator interaction is not simply a matter of chance, but rather a fine balance of chemical properties from both the DNA and the protein.
Interaction between operator position 6 and residue 22
Only one universal rule was observed: when there is a guanine at position 6 of the operator, there is always an arginine at residue 22. The repressor–operator pairs that have a guanine at operator position 6 account for 69% of the total number of functional pairs and this rule is true for every one of them. Similarly, an asparagine at residue 22 is observed 70% of the time when operator position 6 is a thymine. An alanine at residue 22 is observed 45% of the time when operator position 6 is a cytosine (Fig. 2). This trend is strengthened by comparing the mutants which bind operators differing only at position 6 (Table I). Very often, there are mutants that have the same AA composition at residues 17 and 18, but different AA's at residue 22. This suggests that residues 17 and 18 are acting to recognize operator positions 4 and 5 and residue 22 is recognizing operator position 6, in agreement with previous studies.22,23
Table I.
Selected Repressor–Operator Pairs and Corresponding Repression Values
| GGT | Eo | TGT | Eo | CGT | Eo |
| ASR | 0.008 | ASN | 0.048 | ASA | 0.216 |
| KSR | 0.024 | KSN | 0.053 | KSA | 0.149 |
| TSR | 0.019 | TSN | 0.105 | TSA | 0.301 |
| PSN | 0.036 | PSA | 0.170 | ||
| GTA | Eo | TTA | Eo | CTA | Eo |
| IAR | 0.030 | IAN | 0.066 | IAA | 0.161 |
| TAR | 0.031 | TAN | 0.107 | TAA | 0.204 |
| AAR | 0.042 | AAN | 0.165 | ||
| GAR | 0.036 | GAN | 0.209 | ||
| STR | 0.041 | STN | 0.289 | ||
| TGR | 0.047 | TGN | 0.325 | ||
| VAR | 0.039 | VAN | 0.383 | ||
| GAC | Eo | TAC | Eo | ||
| AKR | 0.053 | AKN | 0.241 | ||
| PKR | 0.038 | PKN | 0.189 | ||
| GAG | Eo | TAG | Eo | ||
| HGR | 0.067 | HGN | 0.112 | ||
| PAR | 0.023 | PAN | 0.116 | ||
| RSR | 0.075 | RSL | 0.156 | ||
| GGA | Eo | TGA | Eo | ||
| AAR | 0.016 | AAN | 0.032 | ||
| AGR | 0.050 | AGN | 0.055 | ||
| ATR | 0.018 | ATN | 0.040 | ||
| GGG | Eo | TGG | Eo | ||
| ASR | 0.036 | ASN | 0.038 | ||
| HGR | 0.033 | HGN | 0.048 | ||
| KAR | 0.037 | KAN | 0.109 | ||
| RSR | 0.027 | RSN | 0.034 | ||
| TTT | Eo | CTT | Eo | ||
| HTN | 0.021 | HTA | 0.096 |
The operator name is displayed in italics and the letters represent the nucleotides at positions L6, L5, and L4 of the symmetric Lac operator, respectively. The mutants that bind the operator are listed beneath. The three letters in each repressor name represent the amino acids present at residues 17, 18, and 22, respectively. Eo is the ratio of RFU in the repressed state to RFU of the reporter only. Partial list of repressor–operator pairs for which the operators differ only at position 6 and the repressors differ only at residue 22. The full list is available in Supporting Information Table s2.
The number of repressor–operator pairs decreases from operators with a guanine at position 6, to those with a thymine at position 6 to those with a cytosine at position 6. This may be explained through further exploration of the trends in repression values for the interactions listed in Table I. In almost all cases, repression is strongest for the guanine–arginine interaction, followed by thymine–asparagine and weakest for the cytosine–alanine interaction. Since the only thing changing between these repressor–operator pairs is the position 6-residue 22 interaction, the decrease in repression can be attributed to a decrease in the interaction strength. In a situation where the affinity decreases enough to prevent functional binding, a repressor must have an alternative binding scheme. For instance, Table I shows the repressor mutants with identical AA's at residues 17 and 18 for the operators GTA, TTA, and CTA. Only two of the mutants that bind operator CTA fit on this table, whereas there are 10 between GTA and TTA. The eight other mutants that bind CTA have alternative binding schemes, or amino acid patterns not observed for any other operator (all CTA-binding mutants described below). In these other CTA mutants, residue 22 is much more variable while there are a small number of functional combinations for residues 17 and 18. This trend differs significantly from those of the GTA and TTA operators, which almost universally require an arginine and asparagine, respectively, at position 22.
In cases where alternative schemes are not possible, there are simply fewer functional repressor–operator pairs. This explanation associates the decrease in repression strength with a decrease in number of repressor–operator pairs and an increase of repressors with apparent alternative binding schemes for the repressor–operator pairs that were identified.
The arginine–guanine interaction is one of the most common interactions observed in protein–DNA complexes and asparagine is commonly found interacting with an adenine.5,28 It is probable that the strong correlation observed between asparagine and thymine is more likely the result of the asparagine reaching across the DNA to the opposite strand to interact with the adenine. Interestingly, repressor–operator pairs that have an adenine at operator position 6 are only observed in one case (repressor mutant KSL with operator AGG), and the AA at residue 22 is not an asparagine. This suggests that the orientation of the protein relative to the DNA is rigid enough to prohibit favorable interactions between asparagine at residue 22 and adenine at position 6, but the geometry permits asparagine to interact with an adenine on the opposite strand. A similar explanation can be used to describe why arginine is so rarely observed at any residue other than 22—favorable geometry is satisfied only when arginine occupies that position.
Trend analysis
While universal rules for protein–DNA recognition are difficult or impossible to define even for this single, well-controlled system, there are trends present in the data that deserve recognition. In some instances, the trends exist for numerous operators. For others, the trend is restricted to a single operator. These trends become obvious upon examination of the repressor–operator pairs listed in Figure 2 and Supporting Information Table sII.
Comparison with the ZFP functional rules
The Barbas group conducted a series of studies where they identified various ZFP's that recognize 51 of the 64 possible three-nucleotide sequences.29–32 In each experiment, they randomized a region of the recognition helix of one of the three fingers of the ZFP Zif268 and used phage display to identify ZFP-DNA binding partners. As six residues were always randomized in these studies, the residues of particular interest are −1, 3, and 6, as they are the ones involved in specific contacts with nucleotides. Here, Zif268 mutants are named by the amino-acid at those positions, equivalent to the naming scheme for Lac mutants. Similar to the Lac system, the amino acid residues interact with the nucleotides in opposite sequential order: residue 6 of Zif268 interacts with the 5′ nucleotide and 3 and −1 interact with the middle and 3′ nucleotides.
The compiled list of ZFP-DNA binding partners can be seen in Supporting Information Table sIV. The ZFP experiments produced mutants that exhibit better coverage of sequence space than the Lac experiments (51 vs. 26 of the 64 possible nucleotide sequences for ZFP's vs. Lac). One reason for this is because the libraries used in the ZFP experiments randomized six residues of the recognition helix, when compared with three residues in Lac. While the other residues may not be interacting specifically with the nucleotides, it is likely that the added flexibility often allows residues to adopt favorable positions that would otherwise be prohibited. The Lac experiments, on the other hand, often produced significantly more mutants for the sequences that worked.
The predominance of the guanine–arginine interaction is preserved between the two sets of protein–DNA binding pairs. In contrast to Lac, amino acids other than arginine are found at residue 6 of Zif268 when the first nucleotide is a guanine, though it is rare. The interaction between adenine and asparagine at the same positions is also preserved, though similar to the results with Lac, variations that diverge from this trend are more frequent than those that vary from the guanine–arginine trend. There are two nucleotide sequences that return the same mutant: GGT – TSR and GTG – RAR. In the Zif268 dataset, there is a strong correlation between glutamic acid at residue 6 and cytosine in the first position. This interaction is not observed in the Lac dataset. In fact, the proportion of negatively charged residues is significantly higher with the Zif268 mutants than with Lac.
It appears that, like Lac, there is no simple set of rules to determine protein–DNA recognition for this ZFP.32 There are clear trends for both Lac and for Zif268 and these trends are distinct from each other. Proteins clearly have specific requirements for DNA recognition, but they are not generalizable to protein families, or even individual proteins.
Comparison with Muller-Hill
Muller-Hill and colleagues were the first to develop a two plasmid in vivo bacterial repressor and reporter system that could be used to measure the repression capability of a Lac repressor mutant for an operator variant.22 They measured the repression values for various Lac repressor mutants with exchanges at residues 17, 18, and 22 and symmetric operator variants with exchanges at positions 4, 5, and 6.22,23 With a few exceptions, the mutants that were tested are in good agreement with the data reported here. The variation in relative repression capabilities may be accounted for by the different origin of replication used for the repressor and reporter plasmids between our two labs and also by the differing cell strain.24
In 1990, Muller-Hill and colleagues performed a set of experiments where they constructed 86 out of the 400 possible combinations of residues at position 17 and 18 of the Lac repressor. They measured repression values against the 16 possible variants of the symmetric Lac operator which varied at positions 4 and 5 (position 6 remained guanine). They deduced that residues 17 and 18 interact with the operator independently in an additive manner and they calculated predicted repression values for all the other combinations of residues 17 and 18 with the 16 operator variants.21
Out of the 61 mutants that Muller-Hill predicted to be functional with various operators, 34 were observed experimentally here. While the functional repressors for some operators were predicted reasonably well, such as GAA, GAT, and GGT, Muller-Hill's values drastically under predict the number and diversity of functional mutant repressors. This is likely because residues 17 and 18 do not always act independently of each other, as proposed by Mossing and Record based on their studies of the Lac repressor33 and even by Muller-Hill in previous work.22 This is illustrated by a couple of mutants that bind the operator GAA and their corresponding repression values (Table II). For mutants with the sequence IXR, repression is strongest when X is an alanine, followed by threonine and then serine. For mutants with the sequence TXR, repression is strongest when X is an alanine, followed by serine and then threonine. Threonine is a more favorable interaction when there is an isoleucine at residue 17 but serine is more favorable if there is a threonine at residue 17. This occurs regularly throughout the dataset, demonstrating that residues 17 and 18 do not act independently in all cases and theoretical factors cannot accurately describe the probability of a favorable amino acid–base pair interaction, even for a single operator.
Table II.
Partial List of Repressor Mutants that Bind the Operator Sequence GAA
| GAA | Eo |
|---|---|
| IAR | 0.0118 |
| ISR | 0.0302 |
| ITR | 0.0165 |
| TAR | 0.0185 |
| TSR | 0.0438 |
| TTR | 0.0531 |
Comparison with studies of lac family members
The Lac repressor is a member of the LacI-GalR family of bacterial transcriptional regulators.34 The members exhibit very high sequence homology, 60% across 21 members,35 and the three-dimensional structure of the Lac and Purine repressors exhibit extraordinary similarity.20,36 The corresponding operators of the LacI-GalR family members are similar to each other. It has been hypothesized that the similar regions of the operator are molecular attractors for all family members and divergent regions of the operator are important for operator discrimination.35 Weickert and Adhya argue that alteration of divergent residues changes the specificity of one repressor to another in the LacI-GalR family. As evidence, they use an example from Lehming et al. where residues 17 and 18 of the Lac repressor were swapped with the respective residues in the Gal repressor, valine and alanine. This Lac repressor mutant, VAR, was observed to bind to a variant of the symmetric Lac operator that is similar to the Gal operator.22
Although not explicitly stated, it appeared that altering residues 17, 18, and 22 of the Lac repressor to those of a family member was sufficient to alter specificity of the repressor to that of the family members' operator. However, the rest of the operator is similar but not identical and the binding capacity of VAR with the natural Gal operator was not reported by either group. The results of that experiment are reported here [Fig. 3(A)]. There is not a significant difference between expression in the repressed state versus expression in the induced state for the VAR mutant with the natural Gal operator. Repression is not strong enough to classify VAR as a repressor of the Gal operator. Therefore, while mutations in 17 and 18 are sufficient to alter operator specificity at two nucleotides, it is not sufficient to alter specificity to the DNA sequence of another family member.
Figure 3.
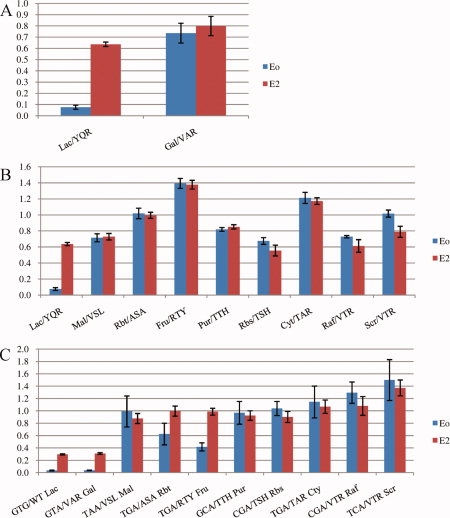
Fractional expression values for Lac repressor mutants with various reporters in the repressed (Eo) and induced (E2) state. Eo is defined as normalized GFP signal with repressor/normalized GFP signal in the absence of repressor for the given operator. E2 is defined as normalized GFP signal of the repressor plus operator in the presence of 2.5mM IPTG/normalized GFP signal of the reporter plasmid alone. Fractional expression for the Lac repressor mutant VAR with the natural Galactose operator compared to the wild-type Lac repressor and operator (A), fractional expression for Lac repressor mutants predicted to bind to LacI-GalR family operators compared to the wild-type Lac repressor and operator (B) and Lac repressor mutants predicted to bind to symmetric Lac operator variants (C). All labels read ‘operator sequence’/‘Lac repressor mutant’. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Consistent with this finding, not a single functional mutant was identified when screens were performed of the recognition helix library and the natural operators of nine different family members (listed in Supporting Information Table sI). As a follow-up experiment, the nine family member operators were directly tested with Lac repressor mutants containing alterations at residues 17, 18, and 22 corresponding to the AAs that exist in the wild-type family member at those sites [Fig. 3(B)]. Again, none were functional.
Further, familial analysis is not sufficient for predicting which mutants will bind symmetric Lac operators swapped at positions 4, 5, and 6 for the corresponding positions in the nine family members (LacSym family operators). If the example of VAR binding to the operator GTA were applicable to all LacI-GalR family members, then swapping the amino acids of the Lac repressor at residues 17, 18, and 22 for those amino acids in the wild-type family members should confer binding to the corresponding LacSym family operators. These LacSym family operators were contained in the 64 operators screened against the recognition helix library. The only case where the predicted mutant was identified is the Gal repressor (the mutant VAR and operator GTA, as above). When the predicted mutants were tested directly with their corresponding LacSym family operators, the only pair that was functional by our standards was, again, VAR with the LacSym Gal operator and RTY with the LacSym Fru operator by a slim margin [Fig. 3(C)].
Examination of the crystal structure of the Purine repressor was performed to gain insight into this apparent discrepancy.36 The amino acids that exist in Purine at residues 17, 18, and 22 are threonine, threonine, and histidine (TTH). The histidine does not appear to be making any direct contact with nucleotides of the operator. Upon further inspection, an arginine at residue 26 (equivalent to residue 28 in Lac) in the loop downstream of the recognition helix reaches back toward the operator to make hydrogen bonding interactions with position 6 of the operator. The histidine appears to base stack with the guanidinium group of the arginine, acting to align it properly to make contacts with the nucleotide. In the Lac repressor, residue 28 is a serine; this interaction is not possible and explains why the Lac repressor mutant TTH is not functional. Interestingly, the mutant TTR was identified as a functional repressor to the LacSym Pur operator in the screen. In the case of the Purine repressor, the crystal structure allows insight into why the TTH Lac repressor mutant does not bind the LacSymPur operator. There are likely clear reasons why the other predicted mutants did not work with the LacSym family operators. Simply identifying the recognition residues in Lac and extrapolating to the other family members is not sufficient to understand protein–DNA specificity for the whole LacI-GalR family.
The example of VAR and the LacSym Gal operator suggests that simple rules for recognition exist at the family level but it is the exception rather than the rule. While the secondary structures of the LacI-GalR family members are quite similar, the details of specificity distill down to individual molecular and chemical interactions that differ even between proteins of remarkable similarity.
Agreement with other DNA binding studies
Direct comparison of amino acid–nucleotide interactions between this work and previously published studies is difficult without explicit structural information about all the repressor–operator binding pairs. However, trends can certainly be compared, as well as conclusions about protein–DNA specificity. The correlation between an arginine at residue 22 of the repressor and a guanine at operator position 6 is consistent with a wealth of data suggesting that multiple hydrogen bonds are occurring between these residues. The second most prominent correlation is an asparagine at residue 22 and a thymine at operator position 6. This interaction is also consistent with previously published results if the assumption is made that the asparagine is interacting with the adenine on the opposite DNA strand. All of the mutants that bind the operator CGG have a lysine at residue 17. This is consistent with published results of a strong interaction between lysine and guanine. Additionally, lysine is very often found at residue 17 for mutants that bind the operators TGG and GGG; every mutant that binds the operator GAC has a lysine at residue 18. Consistent with literature, serine and threonine are often found at one of the recognition residues.
While the importance of Van der Waals interactions has been discussed by numerous groups,2,5 only threonine and aromatic residues have been proposed to make favorable hydrophobic interactions with DNA.2,5 Here, tyrosine, phenylalanine, and tryptophan are not commonly found as recognition residues but histidine is. Alanine was not mentioned as a potential favorable interaction partner for protein–DNA complexes. However, from the sheer number of times alanine is found in DNA binding mutants here, it has to be contributing favorably to binding or its small size is especially advantageous for conferring binding to DNA for some mutants. In the case where it is selected for because of its small size, it is making fewer or less significant disfavorable contacts than any other small amino acid.
On the level of inter-residue interactions, the results presented are consistent with previously published literature. Regarding general rules for protein–DNA interactions, however, our results diverge. There is consensus within the field that protein–DNA interactions cannot be understood on a general level but should be examined at the family-level. Results of the semiexhaustive studies performed here suggest that rules for recognition cannot be determined at even the protein-level, much less the family-level. While there are some trends that transcend family-level analysis, they are simply interactions that are more likely than others. The chemical environment dictated by each altered complex restricts particular interactions and favors others. In some cases, the chemical environment allows for many favorable interactions, in others it allows for none. This body of work suggests that even within a well-defined and well-restricted system, the chemical environment of each altered complex is potentially different enough that rules for protein recognition cannot be generalized.
Materials and Methods
Recognition helix library and reporter construction and phenotypic screening
A GFP reporter plasmid and a repressor plasmid library were created and described previously.24 The reporter plasmid was designed so that the GFPmut3.1 gene is under control of the Lac promoter which contains the natural Lac operator. Quickchange (Stratagene) mutagenesis was performed as described to create the 73 altered operator sequences listed in the results section. The repressor plasmid library contains 203 or 8000 mutant repressors and was created by introducing all 20 amino acids to the repressor at residues 17, 18, and 22.
The bacterial strain DH5α was used for all phenotypic screening experiments. The repressor library was transformed into cells containing one of the operator variants (reporter plasmid). FACS was used to separate repressing and nonrepressing populations based on their fluorescent phenotype. The low fluorescing population was plated on LB plus 100 μM Ampicillin and 50 μM Chloramphenicol (A+C), and colonies were subsequently selected for in vivo induction analysis. Details of phenotypic screening have been previously described.24
In vivo induction analysis
Induction analysis was performed to verify that the isolated repressor mutants were capable of repression (repressing transcription) in the absence of Isopropyl-β-d-thiogalactopyranoside (IPTG) and induction (derepression) upon addition of 2.5 mM IPTG. The morning after plating, 96–384 individual colonies were chosen with sterile applicator sticks and inoculated into a wells containing 0.5–1 mL LB plus A+C in a 96-well block. The same sterile sticks were used to inoculate wells containing LB plus A+C plus 2.5 mM IPTG. The blocks were grown to saturation at 37°C and 0.2 mL of each sample was aliquoted to a 96-well flat bottom plate. The samples were analyzed for fluorescence (495 nm excitation, 510 nm emission) and optical density (OD; A590) on a Perkin Elmer Victor3 plate reader. Fluorescence values were normalized by subtracting each measurement from a sample of LB plus A+C only and dividing by the OD. Induction is defined as the ratio of reporter signal in the induced state to reporter signal in the repressed state. It indirectly measures gene expression when repression is relieved. Mutants that exhibited a twofold induction were designated “hits” and sent for sequencing at the University of Pennsylvania Sequencing Facility. Subsequently, repressor mutants were isolated and fractional expression values determined.
Repressor mutant isolation and measurement of fractional expression
Mutant repressor plasmids were separated from the reporter plasmids by agarose gel electrophoresis, excised and purified with a Nucleospin Gel Extraction Kit (Macherey-Nagel, Duren, Germany). To properly depict the phenotype of each repressor–reporter combination, fractional expression in the repressed (E0) and induced (E2) state were measured. Fractional expression in the repressed state is defined as normalized relative fluorescence units (RFU), or normalized GFP signal, in the repressed state over normalized RFU of the reporter plasmid alone; it provides an indirect measure of binding affinity.24 Fractional expression in the induced state is defined as normalized RFU of the repressor plus reporter in the presence of 2.5 mM IPTG over normalized RFU of the reporter plasmid alone. Final E0 and E2 values were measured by the same method described for induction analysis above with the following differences: the purified mutant repressor was cotransformed with the reporter plasmid into chemically competent DH5α cells, colonies were chosen in triplicate so as to calculate standard deviation values for each repressor–reporter pair and a reporter-only sample was included so as to enable calculation of fractional expression values.
Conclusions
The list of Lac repressor–operator pairs described here is, in essence, a functional code, not a recognition code. It can be used for the purpose of evolving the Lac repressor for use in therapeutic applications. Analysis of this code provides insights into protein–DNA interactions, some previously identified, some novel:
While there are specific amino acid–nucleotide combinations that are statistically more probable than others, generalized rules for specificity in protein–DNA recognition do not exist for protein families, nor do they exist for a single system.
When comparing repressor mutants that bind to operators that differ only at position 6, as binding affinity decreases, the number of alternative binding schemes increases and/or the number of functional repressors decreases.
Individual amino acids that confer specificity for a protein to a particular nucleotide sequence sometimes contribute to binding independently; sometimes they function as a single unit.
While the three-dimensional structures are strikingly similar and simple genetic experiments suggest the LacI-GalR family members recognize their targets in similar ways, the Lac repressor cannot be altered to bind any operator in the LacI-GalR family by altering the recognition residues only.
Ultimately, the most profound conclusion is that specificity in protein–DNA interactions is more complicated than expected, even for this well-controlled system. While general and unique trends exist for a given system, and functional rules may be obtainable for many systems, the rules that regulate protein–DNA interactions are no more simple or general than the fundamental principles of geometry and electrostatics. To fully understand protein–DNA interactions well enough to predict them, computational analysis of the molecular details of protein surfaces must be compared with that of DNA surfaces and their compatibility evaluated.
Acknowledgments
Special thanks to Jesse Cohen for help with induction assays.
References
- 1.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pabo CO, Sauer RT. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 3.Matthews BW. Protein-DNA interaction. No code for recognition. Nature. 1988;335:294–295. doi: 10.1038/335294a0. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki M. A framework for the DNA-protein recognition code of the probe helix in transcription factors: the chemical and stereochemical rules. Structure. 1994;2:317–326. doi: 10.1016/s0969-2126(00)00033-2. [DOI] [PubMed] [Google Scholar]
- 5.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawson CL, Carey J. Tandem binding in crystals of a trp repressor/operator half-site complex. Nature. 1993;366:178–182. doi: 10.1038/366178a0. [DOI] [PubMed] [Google Scholar]
- 7.Luscombe NM, Thornton JM. Protein-DNA interactions: amino acid conservation and the effects of mutations on binding specificity. J Mol Biol. 2002;320:991–1009. doi: 10.1016/s0022-2836(02)00571-5. [DOI] [PubMed] [Google Scholar]
- 8.Choo Y, Klug A. Physical basis of a protein-DNA recognition code. Curr Opin Struct Biol. 1997;7:117–125. doi: 10.1016/s0959-440x(97)80015-2. [DOI] [PubMed] [Google Scholar]
- 9.Smith GP. Surface presentation of protein epitopes using bacteriophage expression systems. Curr Opin Biotechnol. 1991;2:668–673. doi: 10.1016/0958-1669(91)90032-z. [DOI] [PubMed] [Google Scholar]
- 10.Sera T. Zinc-finger-based artificial transcription factors and their applications. Adv Drug Deliv Rev. 2009;61:513–26. doi: 10.1016/j.addr.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, Cathomen T, Voytas DF, Joung JK. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 13.Cronin CA, Gluba W, Scrable H. The lac operator-repressor system is functional in the mouse. Genes Dev. 2001;15:1506–1517. doi: 10.1101/gad.892001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu MC, Davidson N. The inducible lac operator-repressor system is functional in mammalian cells. Cell. 1987;48:555–566. doi: 10.1016/0092-8674(87)90234-0. [DOI] [PubMed] [Google Scholar]
- 15.Mills AA. Changing colors in mice: an inducible system that delivers. Genes Dev. 2001;15:1461–1467. doi: 10.1101/gad.909301. [DOI] [PubMed] [Google Scholar]
- 16.Caron L, Prot M, Rouleau M, Rolando M, Bost F, Binetruy B. The lac repressor provides a reversible gene expression system in undifferentiated and differentiated embryonic stem cell. Cell Mol Life Sci. 2005;62:1605–1612. doi: 10.1007/s00018-005-5123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muhlbauer SK, Koop HU. External control of transgene expression in tobacco plastids using the bacterial lac repressor. Plant J. 2005;43:941–946. doi: 10.1111/j.1365-313X.2005.02495.x. [DOI] [PubMed] [Google Scholar]
- 18.Lee AV, Weng CN, McGuire SE, Wolf DM, Yee D. Lac repressor inducible gene expression in human breast cancer cells in vitro and in a xenograft tumor. Biotechniques. 1997;23:1062–1068. doi: 10.2144/97236st02. [DOI] [PubMed] [Google Scholar]
- 19.Daber R, Lewis M. A novel molecular switch. J Mol Biol. 2009;391:661–670. doi: 10.1016/j.jmb.2009.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell CE, Lewis M. A closer view of the conformation of the lac repressor bound to operator. Nature Struct Biol. 2000;7:209–214. doi: 10.1038/73317. [DOI] [PubMed] [Google Scholar]
- 21.Lehming N, Sartorius J, Kisters-Woike B, von Wilcken-Bergmann B, Muller-Hill B. Mutant lac repressors with new specificities hint at rules for protein-DNA recognition. EMBO J. 1990;9:615–621. doi: 10.1002/j.1460-2075.1990.tb08153.x. erratum in EMBO J 1990; 9:1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehming N, Sartorius J, Niemoeller M, Genenger G, Von WBB, Mueller HB. The interaction of the recognition helix of lac repressor with lac operator. EMBO J. 1987;6:3145–3153. doi: 10.1002/j.1460-2075.1987.tb02625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sartorius J, Lehming N, Kisters B, von Wilcken-Bergmann B, Muller-Hill B. lac repressor mutants with double or triple exchanges in the recognition helix bind specifically to lac operator variants with multiple exchanges. EMBO J. 1989;8:1265–1270. doi: 10.1002/j.1460-2075.1989.tb03500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daber R, Lewis M. Towards evolving a better repressor. Protein Eng Des Sel. 2009;22:673–683. doi: 10.1093/protein/gzp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolkhof P. Specificities of three tight-binding lac repressors. Nucleic Acids Res. 1992;20:5035–5039. doi: 10.1093/nar/20.19.5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopke Salinas R, Folkers GE, Bonvin AM, Das D, Boelens R, Kaptein R. Altered specificity in DNA binding by the lac repressor: a mutant lac headpiece that mimics the gal repressor. Chembiochem. 2005;6:1628–1637. doi: 10.1002/cbic.200500049. [DOI] [PubMed] [Google Scholar]
- 27.Sadler JR, Sasmor H, Betz JL. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc Natl Acad Sci USA. 1983;80:6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandel-Gutfreund Y, Schueler O, Margalit H. Comprehensive analysis of hydrogen bonds in regulatory protein DNA-complexes: in search of common principles. J Mol Biol. 1995;253:370–382. doi: 10.1006/jmbi.1995.0559. [DOI] [PubMed] [Google Scholar]
- 29.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., III Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 30.Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas CF., III Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 31.Segal DJ, Dreier B, Beerli RR, Barbas CF., III Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc Natl Acad Sci USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, Yang WP, Barbas CF., III Building zinc fingers by selection: toward a therapeutic application. Proc Natl Acad Sci USA. 1995;92:344–348. doi: 10.1073/pnas.92.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mossing MC, Record MT., Jr Thermodynamic origins of specificity in the lac repressor-operator interaction. Adaptability in the recognition of mutant operator sites. J Mol Biol. 1985;186:295–305. doi: 10.1016/0022-2836(85)90106-8. [DOI] [PubMed] [Google Scholar]
- 34.von Wilcken-Bergmann B, Muller-Hill B. Sequence of galR gene indicates a common evolutionary origin of lac and gal repressor in Escherichia coli. Proc Natl Acad Sci USA. 1982;79:2427–2431. doi: 10.1073/pnas.79.8.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weickert MJ, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 36.Schumacher MA, Choi KY, Zalkin H, Brennan RG. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices. Science. 1994;266:763–772. doi: 10.1126/science.7973627. [DOI] [PubMed] [Google Scholar]


