Abstract
Protein export mediated by the general secretory Sec system in Escherichia coli proceeds by a dynamic transfer of a precursor polypeptide from the chaperone SecB to the SecA ATPase motor of the translocon and subsequently into and through the channel of the membrane-embedded SecYEG heterotrimer. The complex between SecA and SecB is stabilized by several separate sites of contact. Here we have demonstrated directly an interaction between the N-terminal residues 2 through 11 of SecA and the C-terminal 13 residues of SecB by isothermal titration calorimetry and analytical sedimentation velocity centrifugation. We discuss the unusual binding properties of SecA and SecB in context of a model for transfer of the precursor along the pathway of export.
Keywords: chaperone, SecA, SecB, protein export, protein–protein interaction, titration calorimetry, analytical sedimentation velocity centrifugation
Introduction
The general secretory, or Sec, system in Escherichia coli transfers precursor polypeptides through the cytoplasmic membrane into the periplasmic space. This translocation must occur before the polypeptides have acquired stable tertiary structure. To accomplish this, the export system comprises not only a pathway through the membrane, the SecYEG translocon, but also cytosolic proteins that bind the precursor polypeptides before they fold. Although SecA, the ATPase motor of the secretion machine, can interact directly with non-native precursor polypeptides, often SecB, a small cytosolic chaperone, first interacts with the ligand. Subsequent binding of SecA forms a ternary complex, which engages the translocon through the affinity of SecY for SecA. Binding of the complex to the translocon results in stimulation of the ATPase activity of SecA, and a cycle of ATP binding, hydrolysis and release is coupled to the movement of the precursor through the translocon. To achieve translocation, the system must transfer the ligand from the chaperone SecB to SecA and through the channel of SecYEG.
Unfolded ligands bind to SecB with high affinity through extensive contacts over most of the surface of the chaperone. For most efficient translocation, two protomers of SecA (protomer mass, 102 kDa) must be present in complex with the ligand-bound SecB.1 The binding between SecB and SecA involves multiple areas of contact. The side of the SecB tetramer, which is a flat eight-stranded β-sheet, and the extreme C-terminal 13 aminoacyl residues of SecB have been shown to serve as contact sites for SecA.2–4 The surface on SecA that interacts with SecB overlaps with the surface that binds precursors as well as that which binds the SecYEG translocon.5–7 Transfer of the ligand among these binding partners is a dynamic process in which the contacts between given proteins are likely to be replaced by contacts with other partners.
To further elucidate this process, we have used isothermal titration calorimetry and analytical centrifugation to define the cognate binding site on SecB for the amino terminus of SecA.
Results and Discussion
The first identified site of interaction between SecA and SecB was that between the flat β-sheet of SecB and the zinc-containing C-terminal 21 residues of SecA.8,9 The existence of additional sites of contact became apparent when it was demonstrated that a truncated form of SecA (SecAN880) missing the 21 carboxyl-terminal residues retained the ability to form stable complexes with SecB.10 It was concluded that these complexes were stabilized by interactions with the C terminus of SecB because a truncated species of SecB, SecB142, which lacks the C-terminal 13 aminoacyl residues was unable to form a complex with SecAN880.3 The cognate binding site on SecA for the C-terminal tail of SecB was shown indirectly to be the amino terminus of SecA. This conclusion was based on the demonstration that if the proteins could not form the contact between the zinc site and the side of SecB, then additional removal of aminoacyl residues 2 through 11 rendered SecA incapable of binding SecB.4
To further investigate this interaction, we synthesized a peptide mimic of residues 2 through 11 of SecA. We herein show that this decapeptide competes with SecA for binding to SecB using analytical ultracentrifugation and further determine the thermodynamic parameters of the interaction using titration calorimetry.
We begin with a description of the calorimetric studies. Interaction of the peptide was examined with both wild-type full-length SecB and a series of SecB proteins truncated from the C terminus (Fig. 1). Binding studies were performed at 8°C in Tris, imidazole, and Hepes buffers at pH 7.6. Figure 2 displays representative data for the titration in Tris buffer of full-length, wild-type SecB with the peptide. The injection heats measured in the presence of Hepes are significantly larger than those measured in Tris, indicating that association of the peptide with SecB is accompanied by deprotonation. The slope of a plot of the observed ΔH as a function of the enthalpy of ionization of the buffer (Fig. 3) suggests that the binding event is accompanied by the release of ∼0.2 proton at pH 7.6. The intercept of the plot provides an estimate of the intrinsic binding enthalpy, independent of buffer ionization, ∼5 kcal/mol.
Figure 1.

Position of C-terminal truncations of SecB. SecB comprises 155 aminoacyl residues. The sequence is given from residue 140 to the end with positions of truncations indicated by arrows.
Figure 2.
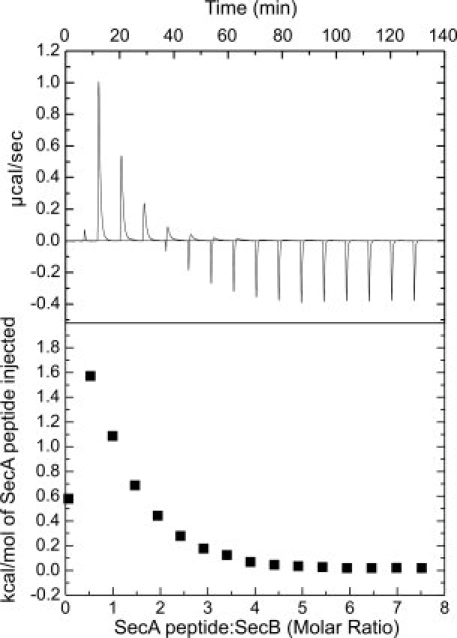
Binding of SecA peptide to SecB. Wild-type SecB was loaded into the cell at a concentration of 17 μM tetramer in 20 mM Tris-Cl, 50 mM KOAc, 5 mM Mg(OAc)2, pH 7.6. The peptide (held in the syringe at 3.3 mM) in the same buffer was added in a sequence of 15 injections of 15 μL after a first injection of 2 μL, which is included to expel any air that might be in the tip of the syringe. The heat of the 2 μL injection is not included in the analysis. (Upper) Raw data. (Lower) Integrated area of heat as a function of the molar ratio of the reactants.
Figure 3.
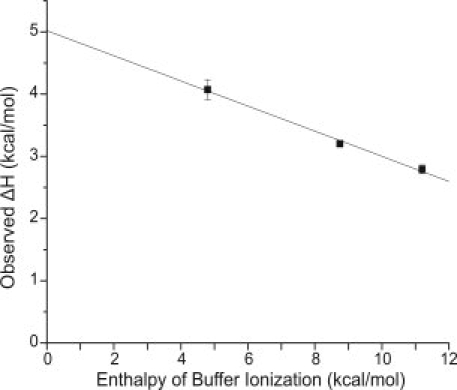
Dependence of observed ΔH on heat of ionization of buffers. The buffers used and their corresponding enthalpy of ionization at 8°C were Hepes, 4.8 kcal/mol; imidazole, 8.75 kcal/mol; and Tris, 11.5 kcal/mol. The solid line is the linear regression analysis that gives a slope of −0.2 (the number of protons released to solvent per mole of complex formed) and an intercept of ∼5 kcal/mol (the corrected ΔH of binding). The titration in Tris was carried out four times, in imidazole once, and in Hepes twice. The standard deviation is shown. The standard deviation is contained within the data point for Tris.
Because ΔG for the reaction is negative, this positive, energetically unfavorable, value for enthalpy indicates that the ΔS term is favorable (i.e., positive) because ΔG = ΔH − TΔS. Clearly, the driving force for association of the peptide with SecB is entropic. Although the predicted net charge of the SecA peptide is +3 at pH 7.6, the sequence is predominantly hydrophobic. Thus, the observed binding energetics may include a large contribution from peptide desolvation and the accompanying decomposition of clathrate structures: enthalpically unfavorable and entropically favorable.
The interaction of SecB and the peptide was also examined using SecB variants harboring deletions from the C terminus of 10–13 aminoacyl residues (Fig. 1). The corresponding binding parameters are summarized in Table I. Although removal of the C-terminal 10 residues from SecB, to produce SecB145, had little impact on the association with the SecA peptide, further truncation of SecB by removal of one amino acid at a time resulted in stepwise decreases in affinity. The removal of the entire 13 amino acid C-terminal tail resulted in a total loss of 1.1 kcal/mol in free energy of stabilization (ΔΔG) of the complex. Half of that loss (0.55 kcal/mol) occurred upon removal of amino acid Gln143 (Fig. 4).
Table I.
Thermodynamic Parameters for Interaction of Peptide with SecB
| Protein species | Kd (μM) | ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS (kcal/mol) |
|---|---|---|---|---|
| SecB | 32 ± 1 | −5.78 | 2.79 ± 0.07 | 8.57 |
| SecB145 | 35 ± 3 | −5.73 | 2.06 ± 0.08 | 7.79 |
| SecB144 | 52 ± 2 | −5.50 | 2.17 ± 0.07 | 7.67 |
| SecB143 | 85 ± 7 | −5.23 | 1.99 ± 0.18 | 7.22 |
| SecB142 | 224 ± 23 | −4.69 | 2.51 ± 0.20 | 7.20 |
The relationships used to obtain Kd and TΔS are: Kd = 1/Ka, ΔG = −RTlnKa, and ΔG = ΔH − TΔS.
Figure 4.
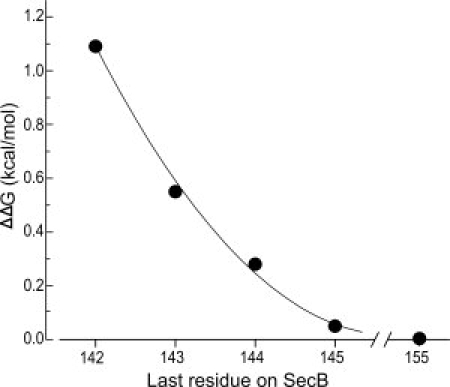
Effect on the free energy of stabilization by truncation of SecB. The values of ΔΔG were calculated from data given in Table I. The line is drawn to aid the eye.
The fact that binding to the tail of SecB is enthalpically unfavorable is consistent with the observation that the truncated SecB, SecB142, binds SecA with a threefold higher affinity than does the full-length SecB.11 This result supports the idea that the energetically unfavorable contact is present in the native SecA:SecB complex. Furthermore, it is of interest that the addition of one amino acid, Gln143, had the greatest effect on binding by SecB to both full-length SecA and to the N-terminal peptide, even though the effects were in opposite directions.
To demonstrate directly that the interaction between the N-terminal region of SecA and the C-terminal tail of SecB is present within a complex between full-length proteins, we carried out sedimentation velocity centrifugation of equimolar mixtures of SecA and SecB in the presence and absence of the SecA peptide.
When protein is loaded into a sample cell and subjected to centrifugation, the boundary of protein, which moves away from the meniscus, not only sediments with time but also spreads because of diffusion [Fig. 5(A)]. Analysis of SecA-SecB interaction is complicated because the system contains multiple species which are in equilibrium. Therefore, it is advantageous to extract the s value independently of the diffusional spreading by using the method of van Holde and Weischet12 (for a detailed description see Demeler et al.13). In this analysis, the sedimenting boundaries are divided into 50 equally spaced segments along the concentration axis, and the apparent sedimentation coefficient, s*, is calculated for each segment. Plots of s* as a function of the inverse of the square root of time allow extrapolation to infinite time [Fig. 5(B)]. Because diffusion is proportional to the square root of time, whereas sedimentation is proportional to the time of centrifugation, the effect of diffusion is eliminated.
Figure 5.
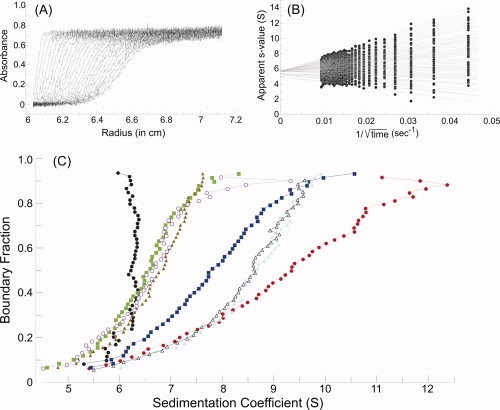
Sedimentation velocity centrifugation. (A) Raw data. The analytical ultracentrifuge cell contained SecA at 4 μM dimer in 10 mM Hepes-KOH, 300 mM KOAc, 5 mM Mg(OAc)2, 1 mM TCEP, pH 7.6. Thirty-five successive scans are shown to display the sedimenting boundary. Scans three through 35 were subjected to analysis as shown in B. (B) van Holde and Weischet analysis.12 Each point is the apparent sedimentation coefficient (s*) of each fraction of the boundary, and each vertical array of 50 points represents one boundary. Each line is the best fit through the points for a particular boundary fraction. (C) Distribution plot of extrapolated s values. Protein samples were subjected to centrifugation, and the raw data were analyzed as described above. The intercepts of each line in the van Holde and Weischet analysis were plotted versus the boundary fraction to which the line pertained. The samples subjected to analysis were SecA (black circles), SecA:SecB (red circles); SecA:SecB and N-terminal SecA peptide (blue squares); SecA:SecB and both the N-terminal SecA peptide and zinc-containing peptide (green squares); SecA:SecB142 (light blue triangles); SecA:SecB142 and N-terminal SecA peptide (gray open triangles); SecA:SecB142 and both the N-terminal SecA peptide and zinc-containing peptide (brown triangles); SecAdN10:SecB142 (purple open circles). The proteins were present at 4 μM SecA dimer:4 μM SecB tetramer, 120 μM N-terminal peptide and 40 μM zinc-containing peptide.
Figure 5 shows the raw data [Fig. 5(A)] and the analysis [Fig. 5(B)] of the sedimentation of SecA loaded into the cell at 4 μM dimer. The distribution plot for SecA [Fig. 5(C), black circles] shows that the majority of the population has a sedimentation coefficient of 6.3 S with slightly lower s values at low concentrations. This behavior is consistent with the known tendency for SecA to dimerize with an equilibrium constant of ∼1 μM.14
An equimolar mixture of SecA and SecB comprises multiple species. The proportion of the population that is in complex is a function of the equilibrium constant for the complex (Kd 1.5 μM)11 and the concentration of the proteins loaded in the cell (4 μM SecA dimer:4 μM SecB tetramer). Thus, the sample exhibits a distribution in which the s value smoothly increases as a function of concentration [Fig. 5(C), red circles].
Addition of the SecA peptide to the SecA:SecB mixture causes a change in the sedimentation behavior so that the population sediments more slowly [Fig. 5(C), blue squares]. Because addition of the peptide to either SecA or SecB alone caused no change in s value (data not shown), we conclude that the peptide competes with the N terminus of SecA for binding. Further evidence that the peptide acts by breaking the contact between SecA and SecB is provided by analysis of the complex between SecA and the truncated SecB, SecB142, which binds the peptide only weakly (Table I). Addition of the peptide to this complex caused no change in sedimentation behavior [Fig. 5(C), compare light blue triangles with gray open triangles]. It is remarkable that the free SecA decapeptide competes with the C-terminal tails on SecB, which is held in complex with SecA through other interactions. The peptide was added at a concentration fourfold over the dissociation constant (120 μM vs. 32 μM) and at only a 7.5-fold molar excess over the concentration of the tails on the SecB in complex (120 μM vs. 16 μM), which have the advantage of being tethered to SecA.
To demonstrate that the more slowly sedimenting complex has the same stoichiometry as that of the wild-type complex with all contacts in place (SecA dimer:SecB tetramer) a peptide mimic of the C-terminal zinc-containing site on SecA was added. This peptide has been demonstrated previously to release one protomer of SecA by disrupting contact between the SecA C terminus and its binding site on SecB.4 As expected, after addition of the zinc-containing peptide to mixtures containing SecA, SecB (either full-length SecB or the truncated SecB142), and the N-terminal SecA peptide the populations display distributions of s values [Fig. 5(C), green squares and brown triangles] characteristic of an equilibrium mixture in which the complex comprises one protomer of SecA bound to tetrameric SecB. The sedimentation profile of a mixture of SecAdN10 (SecA having amino acids 2 through 11 deleted) and SecB142, which were shown previously to form a complex of stoichiometry SecA protomer:SecB tetramer,4 is included for reference [Fig. 5(C), purple open circles].
Concluding Remarks
We have shown previously that SecA interacts with its diverse partners using one surface comprising multiple overlapping binding sites.5 The residues involved in contacts lie on one face of SecA along the mouth of a wide cleft (Fig. 6). They are distributed along the long helix of the helix scaffold domain (HSD), on a short linker helix, which connects the HSD to nucleotide binding fold 2 (NBD2), and near the amino terminus. Additional sites of interaction extend from this face into the cleft and are located on the precursor binding domain (PBD). The fact that the multiple ligands, which include lipids, the translocon SecYEG, SecB and precursor polypeptides, share a common surface with overlapping sites for binding on SecA may be crucial to function in export. To ensure that polypeptides arrive at the translocon in an unfolded state that is compatible with transfer through the channel, the chaperone SecB binds them before they acquire stable tertiary structure. SecA then joins the complex. In the formation of this ternary complex, SecB binds the amino terminus of SecA. These same residues on SecA make interfacial contacts in the antiparallel dimer in which the PBD adopts the closed form.15 Opening of the dimer interface by SecB might allow the SecA protomers to undergo conformational changes to acquire the open state seen in the structure of the SecA monomer.16 The formation of the cleft that results from rotation of the PBD would poise SecA to receive the precursor polypeptide, which is wrapped around the surface of SecB. In subsequent steps, the amino terminus of SecA would release SecB so that it could interact with its other demonstrated binding partners: lipids and the SecYEG translocon. This dynamic interplay is likely to involve the making and breaking of subsites of interaction to accomplish transfer without complete dissociation of the complexes. In this context, it is noteworthy that, even though both proteins display twofold symmetry, the binding between SecA and SecB is asymmetric. When the interaction between the zinc site on SecA and the side of SecB is disrupted, only one protomer of SecA is released.4 If release from SecB were accompanied by the transfer of a portion of the precursor to the free protomer, the protomer might bind SecY using the surface formerly occupied by SecB. The protomer could then transfer a stretch of polypeptide, whereas the second protomer would remain bound to SecB and the remainder of the precursor. This idea is supported by our previously published observation that when SecB is involved in export the efficient transfer of the polypeptide requires the participation of two protomers of SecA.1,4
Figure 6.
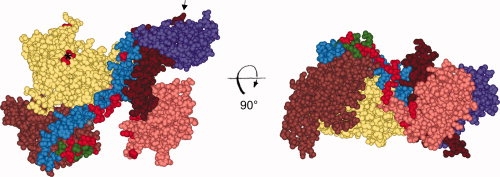
The surface on SecA that binds ligands. The contact sites for the binding partners of SecA including precursors, SecB, lipids and SecYEG are shown in red. The CPK model of the protomer of SecA (PDB ID 2FSF with the PBD modeled in by A. Economou) is colored by domain: NBD1 (yellow), NBD2 (light brown), linker helix (green), HSD (blue), PBD (pink), two-helix finger (dark brown), the helical wing domain, HWD, (purple). The first residue at the N terminus that is resolved (Val9) is the red CPK model indicated by the asterisk. The final 65 residues from the C terminus were not resolved but would emerge from residue Glu836 indicated by the arrow. The identification of the individual residues which contact each of the ligands can be found in our previous publication.5
It is of interest that, depending on the context within the SecA:SecB complex, interaction with the C-terminal 13 aminoacyl residues of SecB can be stabilizing or destabilizing. When all possible interactions between SecA and SecB are present in the complex, the SecB tails make an unfavorable contribution to the binding energy. If interaction between the SecA zinc-site and the side of SecB is disrupted, one protomer of SecA completely dissociates and, in that case, contact with the SecB tails stabilizes the binding of the other protomer. In fact, as a result of enthalpic/entropic compensation, the affinity is the same for both types of complex.
In conclusion, the complexes between SecA and SecB display properties that may be vital to efficient transfer of the unfolded precursor polypeptide along the pathway of export. The asymmetric nature of the binding and the enthalpic/entropic compensation allows one protomer of SecA to dissociate without changing the binding of the other. Additionally sites of binding are common to several partners. The SecA amino terminus has been shown to interact with precursors,5 lipids,5,17,18 and SecYEG.5,7 Here we have characterized the specific interaction between the SecA amino terminus and the C-terminal tails of SecB. It seems likely that the contact between SecA and the SecB tails must be broken so that the amino terminus of SecA is free to bind other partners along the pathway of transfer of polypeptides through the membrane.
Materials and Methods
Protein purification
Wild-type SecB and the truncated variants were purified using the published procedure as described.4 Concentrations of SecB species were determined spectrophotometrically at 280 nm, using a molar absorptivity of 47,600 M−1 cm−1 for the SecB tetramer.
Peptide synthesis
A peptide with the sequence LIKLLTKVFG with the terminal carboxylic acid blocked by amidation was synthesized with the multiple peptide synthesizer 396 Omega from AAPPTec (Louisville, KY), using standard Fmoc chemistry and solid-phase synthesis. The peptide was purified by preparative reverse-phase HPLC (Beckman Coulter, Brea, CA). The final product was subjected to analytical HPLC and ESI-MS (Finnigan, Thermo Scientific, Waltham, MA) and found to be greater than 98% pure with a molar mass of 1129.7 (calculated mass, 1130.8).
Titration calorimetry
SecB (17 μM tetramer) was titrated with the N-terminal SecA peptide (3.3 mM) at 8°C in a VP-ITC (MicroCal LLD, Northampton, MA). Injections of peptide (15 μL each) were made at 500 s intervals. The titration protocol also included a 2 μL preinjection, the heat from which was neglected during analysis. For all titrations summarized in Table I, sample and titrant were both prepared in 20 mM Tris, 50 mM KOAc, and 5 mM Mg(OAc)2, pH 7.6. The heat effects associated with injection of the concentrated SecA peptide into buffer alone were constant (data not shown), indicating that the peptide does not undergo concentration-dependent self-association under these experimental conditions. Data from two (SecB142, SecB145), three (SecB, SecB143), or four (SecB144) experiments were subjected to simultaneous (global) nonlinear least-squares minimization, using Origin v. 7.5 (OriginLab, Northampton, MA).
The injection heat for the ith titrant addition was fit to the following equation:
| (1) |
where Qi and Qi−1 are the cumulative heats following the ith and (i − 1)th injections, dV is the injection volume, Vo is the calorimeter cell volume, and bl is the heat of titrant dilution. The second term in Eq. (1) corrects for the heat associated with the solution displaced from the sample cell by the titrant addition. The cumulative heat following the ith injection is equal to
| (2) |
where n is the reaction stoichiometry, ΔH is the apparent enthalpy of binding, and [MX]i is the concentration of the complex between SecB and the N-terminal SecA peptide. The latter value is given by
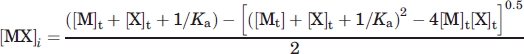 |
where [M]t is the total SecB concentration, [X]t is the total peptide concentration, and Ka is the peptide association constant. Ka and ΔH are global parameters, varied to maximize agreement between the observed and calculated values for each of the replicate experiments. The global least-squares treatment permits the stoichiometry and heat of dilution, bl, to be treated as titration-specific local parameters. The stoichiometry for all titrations was near one per monomeric subunit. The average value for the 14 titrations was 1.03 ± 0.08 (the error is the standard deviation).
In experiments to determine heat of protonation, the solutions contained the same salts buffered at pH 7.6 with Hepes, imidazole, or Tris. The values used for the change in enthalpy of buffer ionization at 8°C were calculated using values for changes in enthalpy and heat capacity for deprotonation of the buffers taken from the literature.19,20
Analytical centrifugation
Solutions containing mixtures of proteins at the concentrations indicated in 10 mM Hepes-KOH, 300 mM KOAc, 5 mM Mg(OAc)2, 1 mM tris(2-carboxyethyl) phosphine (TCEP), pH 7.6 were subjected to centrifugation by using the XL-I ultracentrifuge (Beckman Coulter, Brea, CA). Samples (415 μL) were loaded into cells with two-sector centerpieces in the An-60Ti rotor, and after equilibration to 6°C, were centrifuged at 48,000 rpm for 3 h. Absorbance was measured as a function of radial position at 280 nm, at 5 min intervals. The data were analyzed by the method of van Holde and Weischet,12 using the UltraScan Data Analysis Program, version 9.9, from Borries Demeler (University of Texas Health Science Center, San Antonio, TX). The values used for density and viscosity of the buffer relative to water were 1.014 and 1.063, respectively. The s values reported are corrected to water at 20°C.
Acknowledgments
The authors thank Fabio Gallazzi of the Structural Biology Core, University of Missouri, for the synthesis of the peptide and Jennine M. Crane for valuable discussions. We are grateful to Anastassios Economou for providing the coordinates of E. coli SecA with the PBD.
References
- 1.Mao C, Hardy SJS, Randall LL. Maximal efficiency of coupling between ATP hydrolysis and translocation of polypeptides mediated by SecB requires two protomers of SecA. J Bacteriol. 2009;191:978–984. doi: 10.1128/JB.01321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fekkes P, de Wit JG, Boorsma A, Friesen RH, Driessen AJ. Zinc stabilizes the SecB binding site of SecA. Biochemistry. 1999;38:5111–5116. doi: 10.1021/bi982818r. [DOI] [PubMed] [Google Scholar]
- 3.Randall LL, Crane JM, Liu G, Hardy SJS. Sites of interaction between SecA and the chaperone SecB, two proteins involved in export. Protein Sci. 2004;13:1124–1133. doi: 10.1110/ps.03410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Randall LL, Crane JM, Lilly AA, Liu G, Mao C, Patel CN, Hardy SJS. Asymmetric binding between SecA and SecB two symmetric proteins: implications for function in export. J Mol Biol. 2005;348:479–489. doi: 10.1016/j.jmb.2005.02.036. [DOI] [PubMed] [Google Scholar]
- 5.Cooper DB, Smith VF, Crane JM, Roth HC, Lilly AA, Randall LL. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol. 2008;382:74–87. doi: 10.1016/j.jmb.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer BW, Rapoport TA. Mapping polypeptide interactions of the SecA ATPase during translocation. Proc Natl Acad Sci USA. 2009;106:20800–20805. doi: 10.1073/pnas.0910550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimsey HH, Dagarag MD, Kumamoto CA. Diverse effects of mutation on the activity of the Escherichia coli export chaperone SecB. J Biol Chem. 1995;270:22831–22835. doi: 10.1074/jbc.270.39.22831. [DOI] [PubMed] [Google Scholar]
- 9.Fekkes P, de Wit JG, van der Wolk JP, Kimsey HH, Kumamoto CA, Driessen AJ. Preprotein transfer to the Escherichia coli translocase requires the co-operative binding of SecB and the signal sequence to SecA. Mol Microbiol. 1998;29:1179–1190. doi: 10.1046/j.1365-2958.1998.00997.x. [DOI] [PubMed] [Google Scholar]
- 10.Woodbury RL, Topping TB, Diamond DL, Suciu D, Kumamoto CA, Hardy SJS, Randall LL. Complexes between protein export chaperone SecB and SecA. Evidence for separate sites on SecA providing binding energy and regulatory interactions. J Biol Chem. 2000;275:24191–24198. doi: 10.1074/jbc.M002885200. [DOI] [PubMed] [Google Scholar]
- 11.Patel CN, Smith VF, Randall LL. Characterization of three areas of interactions stabilizing complexes between SecA and SecB, two proteins involved in protein export. Protein Sci. 2006;15:1379–1386. doi: 10.1110/ps.062141006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Holde KE, Weischet WO. Boundary analysis of sedimentation velocity experiments with monodisperse and paucidisperse solutes. Biopolymers. 1978;17:1387–1403. [Google Scholar]
- 13.Demeler B, Saber H, Hansen JC. Identification and interpretation of complexity in sedimentation velocity boundaries. Biophys J. 1997;72:397–407. doi: 10.1016/S0006-3495(97)78680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodbury RL, Hardy SJS, Randall LL. Complex behavior in solution of homodimeric SecA. Protein Sci. 2002;11:875–882. doi: 10.1110/ps.4090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 16.Osborne AR, Clemons WM, Jr, Rapoport TA. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci USA. 2004;101:10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breukink E, Keller RC, de Kruijff B. Nucleotide and negatively charged lipid-dependent vesicle aggregation caused by SecA. Evidence that SecA contains two lipid-binding sites. FEBS Lett. 1993;331:19–24. doi: 10.1016/0014-5793(93)80289-7. [DOI] [PubMed] [Google Scholar]
- 18.Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B. The C terminus of SecA is involved in both lipid binding and SecB binding. J Biol Chem. 1995;270:7902–7907. doi: 10.1074/jbc.270.14.7902. [DOI] [PubMed] [Google Scholar]
- 19.Fukada H, Takahashi K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins. 1998;33:159–166. [PubMed] [Google Scholar]
- 20.Goldberg RN, Kishore N, Lennen RM. Thermodynamic quantities for the ionization reactions of buffers. J Phys Chem Ref Data. 2002;31:231–370. [Google Scholar]


