Abstract
Cren7 is a crenarchaeal conserved chromatin protein discovered recently. To explore the mechanism of the DNA packaging in Crenarchaeota, the crystal structure of Cren7–GCGATCGC complex has been determined and refined at 1.6 Å resolution. Cren7 kinks the dsDNA sharply similar to Sul7d, another chromatin protein existing only in Sulfolobales, which reveals that the “bending and unwinding” compacting mechanism is conserved in Crenarchaeota. Significant structural differences are revealed by comparing both protein–dsDNA complexes. The kinked sites on the same dsDNA in the complexes with Sul7d and Cren7 show one base pair shift. For Cren7, fewer charged residues in the β-barrel structural region bind to DNA, and additionally, the flexible loop Lβ3β4 is also involved in the binding. Electrophoretic mobility shift assays indicate that loop Lβ3β4 is essential for DNA-binding of Cren7. These differences provide insight into the functional difference of both chromatin proteins, suggesting that Cren7 may be more regulative than Sul7d in the DNA-binding affinity by the methylation in the flexible loop Lβ3β4 in vivo.
Keywords: Cren7, Archaea, chromatin protein, protein–DNA complex, crystal structure
Introduction
The genomic DNAs of all cellular life are packed by various proteins, named chromatin proteins.1 Histones are the conserved major chromatin proteins in Eukaryota and Euryarchaeota, a kingdom of Archaea.2–4 The genomic DNA is wrapped on histones forming nucleosomes.2 In bacteria, the genomic DNA is packaged by a number of DNA-binding proteins, such as HU, H-NS, and so forth, forming a compact nucleoid.5,6 Crenarchaeota, another main kingdom of Archaea, lacks histones.4 A number of DNA-binding proteins, such as Sul7d, CC1, and so forth, have been identified as chromatin proteins in some crenarchaeal species. However, most of them are not conserved in Crenarchaeota.7–9 Recently, a conserved chromatin protein, Cren7, has been identified in Crenarchaeota.10 The discovery of Cren7 suggests a conserved genome packaging strategy in Crenarchaeota. Two strategies of chromatin compaction in Archaea were proposed according to these findings. The genomic DNA is packaged mainly by histones in Euryarchaeota, whereas in Crenarchaeota the genomic DNA is packaged by a “bending and unwinding” mechanism that the genomic DNA is kinked and supercoiled by minor-groove binding proteins such as Sul7d11 and Cren710 forming a compact nucleoid.
Cren7 is a small nonspecific DNA-binding protein, stabilizing dsDNA and constraining negative DNA supercoils. Besides, Cren7 is also an abundant and multiple-methylated protein associated with genomics DNA in vivo. The NMR-derived solution structure of Cren7 adopts an SH3-like fold, which shows much similarity with Sul7d, a chromatin protein existing only in Sulfolobales, although the two proteins are unrelated at the amino acid sequence level. Some biochemical and structural differences have been observed between Sul7d and Cren7. Cren7 has a large flexible loop Lβ3β4, which binds to dsDNA and an additional flexible N-terminal loop, but has no the C-terminal α-helix compared with Sul7d. The structural similarities between Cren7 and Sul7d suggested a conserved genomic packing mechanism. However, there is a question what's the functional difference if both proteins are abundant and important to the chromatin.10
Here, we report the crystal structure of Sulfolobus solfataricus P2 Cren7 and an 8-bp dsDNA complex at 1.6 Å resolution [Fig. 1(A)]. Analysis of the structure not only reveals conservation of the “bending and unwinding” compacting mechanism in Crenarchaeota but also provides insight into the functional difference between Cren7 and Sul7d.
Figure 1.
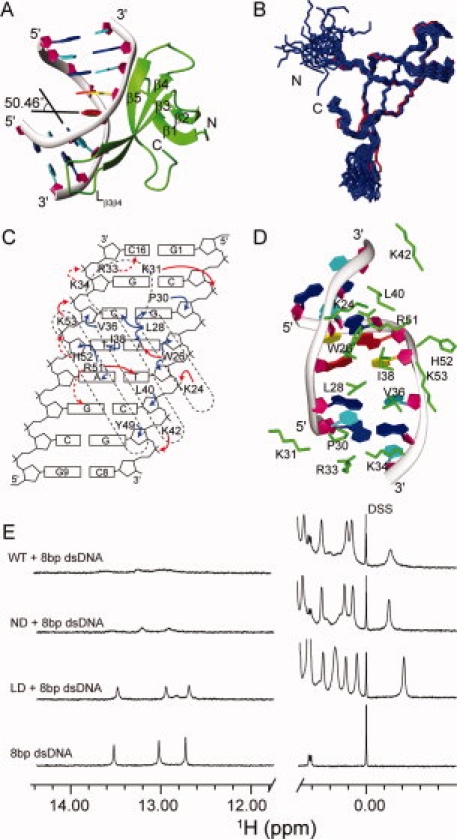
Crystal structure and protein–DNA interactions of the Cren7–dsDNA complex. A: Crystal structure of the Cren7–dsDNA complex. Cren7 is shown as green ribbon. The backbone of dsDNA is shown as gray ribbon, and the sugar ring is depicted as filled purple pentagon; A, T, C, G bases are colored in red, yellow, cyan, and blue, respectively. B: Superimposition of Cren7 in the complex (red) with the NMR solution structures of Cren7 (blue) (PDB: 2JTM). C: Schematic diagram summarizing all Cren7–DNA contacts. The filled red, filled blue, and dashed red arrows represent direct hydrogen bonds/salt bridges, van der Waals close contacts, and potential hydrogen bonds/salt bridges, respectively. D: Protein–DNA interactions in the structure. The side chains of residues involved in the interactions are shown as green sticks. E: 1D 1H NMR spectra of the Cren7–dsDNA complexes. The high field signals near 0.00 ppm are from methyl group of proteins, and the low field signals near 13.00 ppm are from dsDNA hydrogen-bonded base pairs. Full length Cren7 (WT) and Cren7ND show similar NMR spectra in the protein–dsDNA complexes. The peaks near 13.00 ppm are broadened extremely, indicating that almost all the dsDNA molecules form the complexes with Cren7 (WT). The spectrum of Cren7LD with dsDNA shows only slightly broadened peaks compared with those of the free dsDNA near 13.00 ppm, which indicates that the dsDNA in the Cren7LD-dsDNA complex is in a small fraction and fast exchange with the free dsDNA. An interactive view is available in the electronic version of the article. PRO385 Figure 1
Results and Discussion
Structure determination
To facilitate the crystallization, a construction lacking the flexible N-terminal 5 residues (Cren7ND) was used in this research. Since the N-terminal 5 residues are in the opposite side of DNA-binding surface and the deletion does not change the structures of other portions (Supporting Information Figure S1), Cren7ND and Cren7 showed similar binding affinity and patterns in the electrophoretic mobility shift assays (EMSA) and 1H NMR spectra [Supporting Information Figure S2 and Fig. 1(E)]. We shall call Cren7ND as Cren7 hereafter except where necessary. The 8-bp dsDNA (5′-GCGATCGC-3′) used for studying the Sul7d family proteins12–14 was adopted in this study. The Cren7–dsDNA complex crystallized in space group C2221 and one Cren7–dsDNA complex is in the asymmetric unit of the crystal. The complex structure was solved using the molecular replacement method and refined to 1.6 Å resolution, resulting in an Rwork of 17.59% and an Rfree of 21.70%. In the Ramachandran plot, all residues are in the most favored region (Supporting Information Table S1).
Structure of the Cren7–dsDNA complex
The X-ray structure of Cren7 in the complex is very similar to the NMR structure of Cren7 [Fig. 1(B)]. The RMSD for backbone N and C atoms of residues 8–11, 17–20, 24–28, 36–42, and 49–54 between the crystal and solution structures of Cren7 is 0.63 ± 0.06 Å. This is in agreement with previous report that the binding does not cause significant structural changes of Cren7.10
In the complex structure, the β-sheet consisting of three β-strands (β3, β4, and β5) and the loop Lβ3β4 of Cren7 are engaged in the binding with the minor groove of the dsDNA. The binding surface area is about 804.2 Å2. Residues participating in the interaction with dsDNA include the positively charged residues (K24, K31, R33, K34, K42, R51, and K53) and the hydrophobic/aromatic residues (W26, L28, P30, V36, I38, L40, and H52) [Fig. 1(C,D)]. The DNA-binding surface of Cren7 in the crystal is almost the same as that identified previously by NMR. However, the C-terminus of strand β1 (K11) does not interact with dsDNA directly in the crystal structure whereas K11 showed also the chemical shift changes on the binding of DNA in the previous NMR study. In the structure of Cren7, K11 is close to H52, which is a residue interacting with dsDNA. Thus, in the NMR study, the binding of dsDNA not only perturb the residues in the DNA-binding surface but also may perturb the resonance of K11 causing a relatively small change in its chemical shift compared with the chemical shifts of the residues binding with DNA. In the crystal, the atoms of the loop Lβ3β4 have relatively poor electronic density and high B-factor values, which characterizes the loop Lβ3β4 as a relatively flexible loop in the DNA-binding surface. However, the previous NMR relaxation experiments indicated that the loop Lβ3β4 become more rigid on the DNA-binding in solution.10
The dsDNA is bent significantly at an angle ∼60° relative to the standard B-type DNA (Supporting Information Table S2 for parameters of dsDNA) in the analysis by 3DNA.15 The bending is contributed mainly by the two kinks at the base pair steps G3pA4 and C2pG3. The roll angle of the major kinked site, namely the G3pA4p step, is 50.46° (Supporting Information Figure S3), which is resulted from the intercalating of three hydrophobic residues, L28, V36, and I38. The roll angle of the C2pG3 step is 12.50°, which is mainly caused by the interaction between P60 and G3 sugar ring. The intercalation of L28 and I38 also results in 22.27 and –19.51 buckle angles between the base pairs G3-C14 and A4-T13, respectively. W26 may play a role of stabilizing the kink by a hydrogen bond between the W26 indole NH group and the N3 of A4 base, and by the hydrophobic/van der Waals interactions with the sugar rings of A4 and T5. A hydrogen bond network is formed between the guanidinium group of R51 and the O2 atoms of T5 and C6 bases (Supporting Information Figure S4), whereas R51 also has hydrophobic interactions with the sugar rings of T13. The minor groove of dsDNA was greatly expanded in the region interacting with Cren7. The width of the minor groove is increased to ∼20 Å whereas it is 11.7 Å in the standard B-type DNA. The twist angles decrease ∼10–20° for the base pair steps from G1pC2 to A4pT5 compared with the standard B-type DNA, especially the twist angle of G3pA4 step decreases 17.04° (Supporting Information Table S2). The large decrease of twist angles would generate a significant negative supercoil as previously reported.10
Comparison of Cren7–dsDNA and Sul7d–dsDNA complexes
Both Cren7 and Sul7d bind to dsDNA at the three-stand β-sheet, and kink the dsDNA sharply by intercalating hydrophobic residues in the minor groove of dsDNA. These similarities have been proposed in our previous research based on the structural and biochemical similarity of Cren7 to Sul7d.10 The Cren7–dsDNA complex structure confirms that the “bending and unwinding” mechanism is widely conserved in Crenarchaea.
However, the structural differences were observed between the two complexes (Fig. 2). First, the binding position on dsDNA has 1-bp shift in the Cren7–dsDNA complex compared with the binding in the Sul7d–dsDNA complex, that is, the kink site in the Cren7–dsDNA complex is between G3-C14 and A4-T13 base pairs, whereas it is between C2-G15 and G3-C14 base pairs in the Sul7d–dsDNA complex although the same dsDNAs were used in both complexes [Fig. 2(A) and Supporting Information Figure S3]. Probably, Cren7 needs 1 bp more for binding of the flexible loop Lβ3β4 with dsDNA, whereas Sul7d does not have such a flexible loop as reported previously. Besides, both R51 in Cren7 and R42 in Sul7d form hydrogen bonds with the same O2 atoms of T5 and C6 of the dsDNA although 1 bp shift exists in the global binding position. These hydrogen bonds were considered to determine the sequence location of Sul7d on the dsDNA.12 It seems the different conformations of R51 and R42 might also contribute to the 1-bp shift. Second, the orientations of dsDNA relative to proteins in both complexes are different, although Cren7 and Sul7d have similar overall fold and binding region. There is a ∼17° rotation between the two dsDNAs in the complexes if Cren7 and Sul7d in the complexes are superimposed according to their structural similarity [Fig. 2(B)]. This may be due to the different interactions existing in the two complexes. Residues K7, Y8, and K9 of Sul7d are involved in the interactions with dsDNA in the complex, the corresponding interactions are absent in the Cren7–dsDNA complex. Third, the intermolecular interactions between proteins and dsDNAs, especially the electrostatic interactions, are quite different [Fig. 2(C)]. In the Sul7d–dsDNA complex, six lysine residues (K7, K9, K21, K22, K28, and K39) of Sul7d bind to the backbone of dsDNA. In the Cren7–dsDNA complex, only three (K24, K42, and K53) of the corresponding six residues bind dsDNA and three additional basic residues (K31, R33, and K34) in the loop Lβ3β4 participate in the interactions with dsDNA. Interestingly, deleting the loop (residues P30-K34), producing a mutant named Cren7LD, does not change the structure of other portions (Supporting Information Figure S1) but results in the great loss of DNA-binding affinity of Cren7 [a decrease by a factor of ∼60, see Figs. 1(E) and 2(D)]. Apparently, this loop is a key element for DNA binding of Cren7. We have found that K31 has both methylated and nonmethylated forms in vivo, whereas R33 and K34 could also be methylated.10 Since these residues are important for Cren7 in the binding with dsDNA, most probably, their methylations may be functioned as a switch in regulating the DNA-binding affinity of Cren7, like histones in Eukaryota. Such switch might be used in gene regulation, transcription, and silence. However, more experiments are needed to confirm the methylating regulation and explore the physiological roles of the switch in future.
Figure 2.
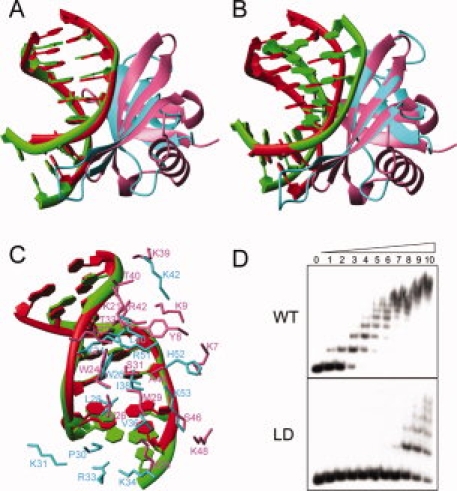
Comparison of Cren7–dsDNA and Sul7d–dsDNA (PDB: 1AZP) complexes. A: Superimposition of dsDNA in the two complexes. Structures are superimposed using the backbone phosphor atoms of G3-C8 and C10-G15 in Cren7–dsDNA complex and C2-G7 and G11-C16 in Sul7d–dsDNA complex. The dsDNA and protein in the Cren7–dsDNA complex are colored in green and cyan, respectively. The dsDNA and protein in the Sul7d–dsDNA complex are colored in red and pink, respectively. B: Superimposition of proteins in the two complexes using the backbone atoms of the interface secondary structure residues (Cre7: 24–28, 36–42, 49–53; Sul7d, 22–26, 29–35, 40–44). C: Protein–DNA interactions in the both complexes superimposed as (A). D: The electrophoretic mobility shift assays of wild-type and loop-deletion mutant Cren7LD binding with a 60 bp dsDNA. Protein concentrations were 0, 0.02, 0.04, 0.08, 0.16, 0.31, 0.63, 1.3, 2.5, 5, and 10 μM, respectively. The apparent dissociation constants are ∼0.08 μM and 5 μM for wild-type Cren7 and Cren7LD, respectively. An interactive view is available in the electronic version of the article. PRO385 Figure 2
Materials and Methods
The expression vectors of Cren7LD encoding residues 1–29 and 35–60, and Cren7ND encoding residues 6–60 were constructed from pET30a-Cren7. Cren7LD and Cren7ND were expressed and purified as described.10 The 8-bp dsDNA used in this study is 5′-GCGATCGC-3′, which was synthesized commercially, annealed and further purified by gel-filtration chromatography.
The procedures of the EMSAs are the same as that reported previously.10 Briefly, a 32P-labelled 60-bp dsDNA fragment (10 nM) was incubated with 0–10 μM proteins for 10 min at 22°C in 20 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 1 mM DTT, 100 μg/mL BSA in a total volume of 20 μL. Protein–DNA complexes were resolved by electrophoresis in nondenaturing polyacrylamide gels in 0.1× TBE buffer. After electrophoresis, the gel was dried and exposed to X-ray film.
The complex of Cren7ND and dsDNA with 1:1 ratio was crystallized using the vapor-diffusion method from 1.0 mM complex, 0.1M tri-sodium citrate dihydrate pH 5.6, 20% (v/v) iso-propanol, and 20% (w/v) polyethylene glycol 4000. Diffraction data were collected at 100 K at BL17u of Shanghai Synchrotron Radiation Facility (SSRF). The crystal of the complex is in the space group C2221 with the unit cell dimensions shown in Supporting Information Table S1. The data were processed using software HKL2000.16 The structure was solved by molecular replacement of the software PHASER17 using the NMR structure of Cren7 (PDB: 2JTM)10 and the DNA molecules in Sac7d-dsDNA complex (PDB: 1AZP)12 as searching templates. The structural model was further constructed in Coot18 and refined in Refmac519 and PHENIX20 with TLS refinement.21
NMR spectra were recorded at 298 K on a Bruker 600 MHz spectrometer equipped with a triple-resonance cryo-probe. The NMR samples contained 0.2 mM complex or 8-bp dsDNA in 50 mM potassium phosphate buffer at pH 6.0. The chemical shifts were referenced to the internal sodium 2,2-dimethylsilapentane-5-sulfonate (DSS) at 0.00 ppm.
Coordinates
Atomic coordinates and structure factors of the Cren7–dsDNA complex have been deposited at the Protein Data Bank with accession number 3KXT.
Conclusions
The high-resolution crystal structure of Cren7–dsDNA complex described here proves that Cren7 bends dsDNA with a sharp kinking angle similar to Sul7d, providing more evidences to the proposal of the conserved “bending and unwinding” packing mechanism in Crenarchaea.1,10 The Cren7–dsDNA complex structure is significantly different from the Sul7d–dsDNA complex, which correlates with the functional differences of Cren7 and Sul7d. Cren7 has an additional flexible loop Lβ3β4 which is essential to the DNA-binding affinity of Cren7, whereas Sul7d has no such loop. Cren7 may be more regulative than Sul7d in the DNA-binding affinity by the methylation of the flexible loop Lβ3β4. Both Cren7 and Sul7d play roles in the organization of the chromatin structures, whereas Cren7 might be more important in gene transcriptions or regulations.
Acknowledgments
The authors thank Dr. Jinzhong Lin, National Institute of Biological Sciences, Beijing, for the help in the structure calculation and refinement. They also thank Dr. Li Guo, Institute of Microbiology, Chinese Academy of Sciences, for the help in the EMSA experiments.
References
- 1.Luijsterburg MS, White MF, van Driel R, Dame RT. The major architects of chromatarchitectural proteins in bacteria, Archaea and eukaryotes. Crit Rev Biochem Mol Biol. 2008;43:393–418. doi: 10.1080/10409230802528488. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Sandman K, Reeve JN. Archaeal chromatin proteins: different structures but common function? Curr Opin Microbiol. 2005;8:656–661. doi: 10.1016/j.mib.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Reeve JN. Archaeal chromatin and transcription. Mol Microbiol. 2003;48:587–598. doi: 10.1046/j.1365-2958.2003.03439.x. [DOI] [PubMed] [Google Scholar]
- 5.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 6.Wu LJ. Structure and segregation of the bacterial nucleoid. Curr Opin Genet Dev. 2004;14:126–132. doi: 10.1016/j.gde.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Luo X, Schwarz-Linek U, Botting CH, Hensel R, Siebers B, White MF. CC1, a novel crenarchaeal DNA binding protein. J Bacteriol. 2007;189:403–409. doi: 10.1128/JB.01246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grote M, Dijk J, Reinhardt R. Ribosomal and DNA binding proteins of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biochem Biophys Acta. 1986;873:405–413. [Google Scholar]
- 9.Dijk J, Reinhardt R. The structure of DNA-binding proteins from Eu- and Archaebacteria. In: Gualerzi CO, Pon CL, editors. Bacterial chromatin. Berlin: Springer-Verlag; 1986. pp. 185–218. [Google Scholar]
- 10.Guo L, Feng Y, Zhang Z, Yao H, Luo Y, Wang J, Huang L. Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among crenarchaea. Nucleic Acids Res. 2008;36:1129–1137. doi: 10.1093/nar/gkm1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agback P, Baumann H, Knapp S, Ladenstein R, Hard T. Architecture of nonspecific protein-DNA interactions in the Sso7d-DNA complex. Nat Struct Biol. 1998;5:579–584. doi: 10.1038/836. [DOI] [PubMed] [Google Scholar]
- 12.Robinson H, Gao YG, McCrary BS, Edmondson SP, Shriver JW, Wang AH. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature. 1998;392:202–205. doi: 10.1038/32455. [DOI] [PubMed] [Google Scholar]
- 13.Gao YG, Su SY, Robinson H, Padmanabhan S, Lim L, McCrary BS, Edmondson SP, Shriver JW, Wang AH. The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nat Struct Biol. 1998;5:782–786. doi: 10.1038/1822. [DOI] [PubMed] [Google Scholar]
- 14.Chen CY, Ko TP, Lin TW, Chou CC, Chen CJ, Wang AH. Probing the DNA kink structure induced by the hyperthermophilic chromosomal protein Sac7d. Nucleic Acids Res. 2005;33:430–438. doi: 10.1093/nar/gki191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 17.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 19.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 20.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 21.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39:109–111. [Google Scholar]


