Abstract
Drosophila endogenous small RNAs are categorized according to their mechanisms of biogenesis and the Argonaute protein to which they bind. MicroRNAs are a class of ubiquitously expressed RNAs of ~22 nucleotides in length, which arise from structured precursors through the action of Drosha–Pasha and Dicer-1–Loquacious complexes1–7. These join Argonaute-1 to regulate gene expression8,9. A second endogenous small RNA class, the Piwi-interacting RNAs, bind Piwi proteins and suppress transposons10,11. Piwi-interacting RNAs are restricted to the gonad, and at least a subset of these arises by Piwi-catalysed cleavage of single-stranded RNAs12,13. Here we show that Drosophila generates a third small RNA class, endogenous small interfering RNAs, in both gonadal and somatic tissues. Production of these RNAs requires Dicer-2, but a subset depends preferentially on Loquacious1,4,5 rather than the canonical Dicer-2 partner, R2D2 (ref. 14). Endogenous small interfering RNAs arise both from convergent transcription units and from structured genomic loci in a tissue-specific fashion. They predominantly join Argonaute-2 and have the capacity, as a class, to target both protein-coding genes and mobile elements. These observations expand the repertoire of small RNAs in Drosophila, adding a class that blurs distinctions based on known biogenesis mechanisms and functional roles.
Drosophila melanogaster expresses five Argonaute proteins, which segregate into two classes. The Piwi proteins (Piwi, Aubergine and AGO3) are expressed in gonadal tissues and act with Piwi-interacting RNAs (piRNAs) to suppress mobile genetic elements10,11. The Argonaute class contains AGO1 and AGO2. AGO1 binds microRNAs (miRNAs) and regulates gene expression8,9. The endogenous binding partners of AGO2 have remained enigmatic.
We generated transgenic flies expressing epitope-tagged AGO2 under the control of its endogenous promoter. Tagged AGO2 localized to the cytoplasm of germline and somatic cells of the ovary (Supplementary Fig. 1). Immunoprecipitated AGO2-associated RNAs differed in their mobility from those bound to AGO1 (Fig. 1a). Deep sequencing of small RNAs from AGO1 and AGO2 complexes yielded 2,094,408 AGO1-associated RNAs and 916,834 AGO2-associated RNAs from Schneider (S2) cells, and 455,227 AGO2-associated RNAs from ovaries that matched perfectly to the Drosophila genome. We also sequenced three libraries derived from 18–29-nucleotide RNAs (936,833 sequences from wild-type ovaries, 1,042,617 sequences from Dicer-2 (Dcr-2) mutant ovaries, and 1,946,339 sequences from loquacious (loqs) mutant ovaries) and an 18–24-nucleotide library from wild-type testes (522,848 sequences). Finally, we added to our analysis 92,363 published sequences derived from 19–26-nucleotide RNAs from S2 cells15.
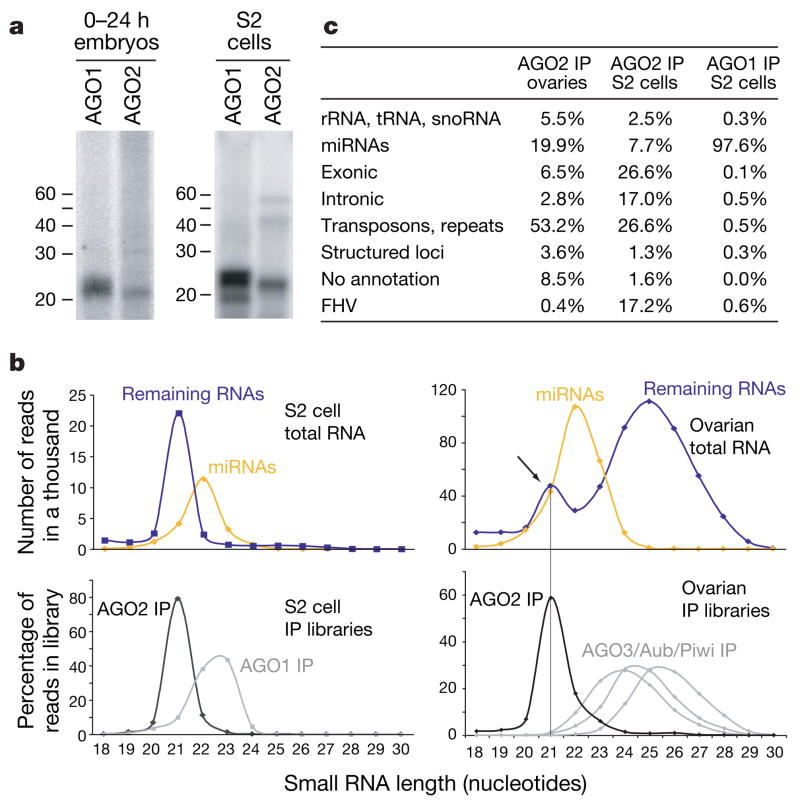
Figure 1. AGO2 binds endogenous small RNAs.
a, RNA was isolated from AGO1 and AGO2 immunoprecipitates (IP) from embryos and S2 cells. b, Length profiles for small RNAs isolated from S2 cells and ovaries are shown. Species are split into miRNAs and remainder, and are compared to those obtained from individual Argonaute complexes (as indicated). c, Annotation of AGO1- and AGO2-associated small RNAs from ovaries and S2 cells. rRNA, ribosomal RNA; snoRNA, small nucleolar RNA; tRNA, transfer RNA.
We noted that among the ~50% of AGO2-associated RNAs from S2 cells that did not match the genome, ~17% matched the flock house virus (FHV), a pathogenic RNA virus and reported target for RNAi in flies16,17. These probably arose because of persistent infection of our S2 cultures.
After excluding presumed degradation products of abundant cellular RNAs, we divided each of the total RNA libraries into two categories: annotated miRNAs and the remainder (Fig. 1b). For the S2 cell library, the size distribution of these populations formed two peaks, with non-miRNAs lying at 21 nucleotides and miRNAs exhibiting a broader peak from 21 to 23 nucleotides. Libraries derived from AGO1 and AGO2 complexes almost precisely mirrored these two size classes. In the ovary library, this approach revealed three size classes. Whereas two reflected those seen in S2 cells, a third class comprised piRNAs. Again, RNA size profiles from AGO2 or Piwi family immunoprecipitates12 mirrored those within the total ovary library. These data demonstrate that AGO2 is complexed with a previously uncharacterized population of small RNAs.
Whereas known miRNAs comprised more than 97% of AGO1-associated RNAs in S2 cells, they made up only 8% or 20% of the AGO2-bound species in S2 cells or ovaries, respectively. The remaining small RNAs in AGO2 complexes formed a complex mixture of endogenous siRNAs (endo-siRNAs; Fig. 1c). Among these, transposons and satellite repeats contributed substantially to AGO2-associated small RNAs in S2 cells (27%) and ovaries (53%). The nature of the transposons giving rise to abundant siRNAs in ovaries and S2 cells differed substantially (Fig. 2a), probably reflecting differential expression of specific transposons in these tissues. Unlike piRNAs12,13,18,19, neither somatic nor germline siRNAs exhibited a pronounced enrichment for sense or antisense species (Supplementary Fig. 2a).
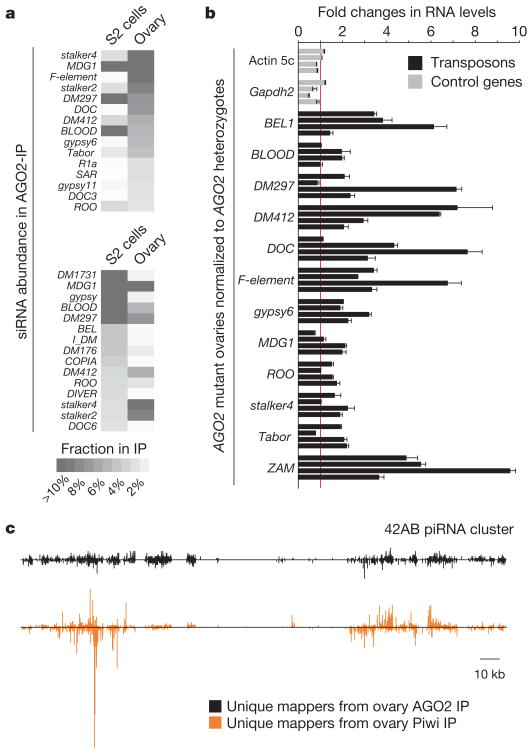
Figure 2. A subset of endo-siRNAs originates from transposons.
a, Indicated are the cloning frequencies of AGO2-bound siRNAs in ovaries and S2 cells that match individual transposons. b, RNA levels of twelve transposons and two control genes in ovaries mutant for AGO2 as compared to AGO2 heterozygotes (four biological replicates; error bars indicate technical variation). c, Distributions of AGO2-bound siRNAs (black) and Piwi-bound piRNAs (orange) from ovaries on the piRNA cluster at cytological position 42AB (ref. 12; relative abundances of both populations can be estimated from Supplementary Fig. 2d).
In accord with these findings, knockdown of AGO2 in S2 cells leads to increased expression of several mobile elements20. In the germ line, the Piwi–piRNA system has been reported as the dominant transposon-silencing pathway19. Nevertheless, we found that several transposons, with a potential to be targeted by siRNAs, were substantially derepressed in AGO2 mutant or Dcr-2 mutant ovaries (Fig. 2b and Supplementary Fig. 2c). Although comparisons of relative abundance were difficult, both piRNAs and siRNAs mapped to piRNA clusters, with the regions that generate uniquely mapping species generally overlapping (Fig. 2c and Supplementary Fig. 2d). Thus, piRNA loci are a possible source for antisense RNAs matching transposons and might serve a dual function in small RNA generation. Considered together, these data suggest that endo-siRNAs repress the expression of mobile elements, in some tissues acting alongside piRNA pathways.
To probe the nature of the remaining endo-siRNAs, we computationally extracted genomic sites, which give rise to multiple uniquely mapping RNAs that do not fall into heterochromatic regions. These generally segregated into two categories, which we term structured loci and convergently transcribed loci.
Transcripts from structured loci can fold to form extensive double-stranded RNA directly. The two major loci, termed esi-1 and esi-2 (Fig. 3a and Supplementary Fig. 3), gave rise to half of the 20 most abundant endo-siRNAs in ovaries and also generated siRNAs in embryos, larvae and adults (not shown). esi-1, annotated as CG18854, can produce an ~400-base pair (bp) dsRNA through interaction of its 5′ and 3′ untranslated regions (UTRs; Supplementary Fig. 3). esi-2 overlaps with CG4068 and consists of 20 palindromic ~260-nucleotide repeats (Fig. 3a). All siRNAs derived from these two loci arise from one genomic strand. In some previously characterized instances (for example, Arabidopsis trans-acting-siRNAs21) Dicer generates ‘phased’ siRNAs with 5′ ends showing a 21-nucleotide periodicity. In all tissues examined, esi-1 and esi-2 produced phased siRNAs, consistent with a defined initiation site for Dicer processing (Fig. 3a and Supplementary Fig. 3). Phasing was not observed for viral or repeat-derived siRNAs. Finally, siRNAs from both loci also joined AGO1 in proportions greater than siRNAs produced from transposons and repeats, perhaps owing to the imperfect nature of the dsRNA that they produce22,23 (Fig. 1c).
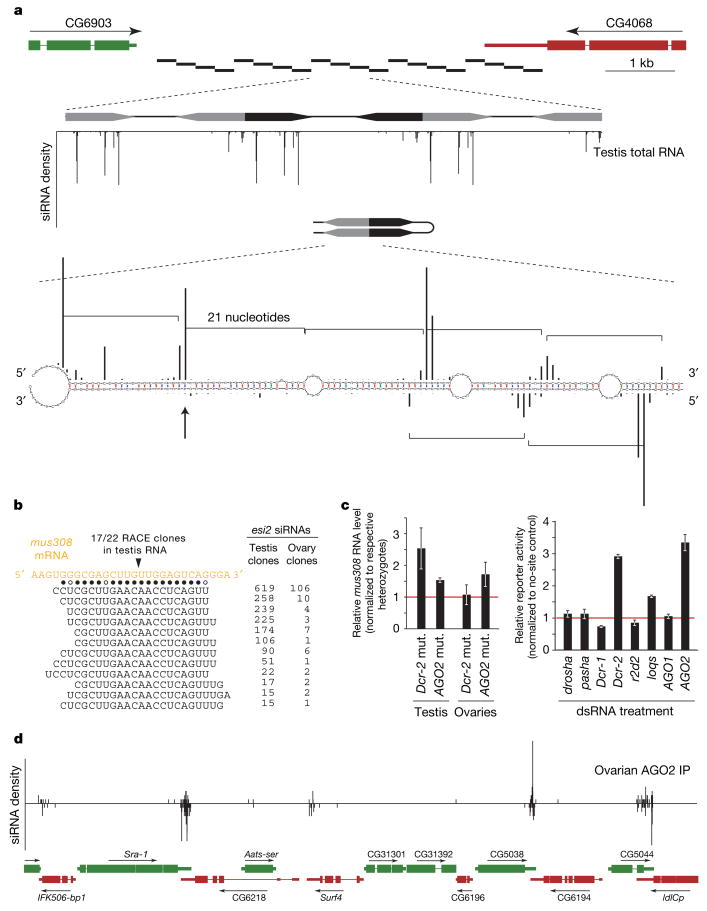
Figure 3. Two types of genic endo-siRNA loci in Drosophila.
a, FlyBase gene structure at the esi-2 locus indicating the twenty ~260-bp repeats of esi-2 (black bars). Below, three repeats are magnified and the small RNA density is depicted. Most siRNAs match up to 20 times and cannot be unambiguously assigned to a chromosomal location. Each repeat is a palindromic sequence, offering a multitude of possible structures. One structure is shown with the 5′ ends of cloned siRNAs as black bars (the height correlates with cloning frequency; phasing is indicated by brackets). b, An abundant siRNA from esi-2 (indicated by an arrow in a) is highly complementary to the mus308 mRNA. Shown is the mus308 target site and cloned siRNAs (solid dots indicate canonical base pairs; open dots indicate GU base pairs). The only detected cleavage site within this duplex is indicated above. c, Shown are mus308 transcript levels from AGO2 and Dcr-2 mutant (mut.) flies compared to their respective heterozygotes (error bars indicate standard deviation). To the right, average reporter levels (error bars indicate standard deviation) of a construct containing two mus308 target sites in S2 cells depleted of the indicated genes by RNAi are shown. d, An example of siRNAs arising from convergent transcription units. A 30-kb region containing multiple instances of convergently transcribed genes is displayed, with the density of AGO2-associated RNAs in ovaries shown above.
AGO2 regulates gene expression by cleavage of complementary sites rather than by recognition of seed sites typical of AGO1–miRNA-mediated regulation23. We searched for possible targets of endo-siRNAs by identifying transcripts with substantial complementarity. A highly abundant siRNA from esi-2 is highly complementary to the coding sequence of the DNA-damage-response gene mutagen-sensitive 308 (mus308). Using a modified rapid amplification of cDNA ends (RACE) protocol, we detected mus308 fragments with 5′ ends corresponding precisely to predicted endo-siRNA cleavage sites (Fig. 3b). Moreover, AGO2 and Dcr-2 loss consistently increased mus308 expression in testis and to a lesser extent in ovaries, consistent with the relative abundance of esi-2 siRNAs in these tissues (Fig. 3b, c). Finally, a reporter gene containing two mus308 target sites was significantly derepressed in S2 cells on depletion of Dcr-2 or AGO2 but not of Dcr-1 or AGO1 (Fig. 3c). Although extensive complementarity between other endo-siRNAs and messenger RNAs was rare, we found several esi-1-derived siRNAs complementary to CG8289 (Supplementary Fig. 3), suggesting a potential regulatory interaction in vivo.
A second group of siRNA-generating loci contained regions in which dsRNAs can arise from convergent transcription. If sorted for siRNA density, most of the top 50 ovarian and S2 cell siRNA loci lay in regions where annotated 3′ UTRs or expressed-sequence-tags corresponding to convergently transcribed protein-coding genes overlap (Supplementary Tables 1 and 2). Typically, siRNAs arise on both genomic strands but only from overlapping portions of convergent transcripts (Fig. 3d). Examining all 998 convergently transcribed gene pairs in the Drosophila genome with annotated overlapping transcripts, we found the peak abundance of ovarian siRNAs to be at the centre of the overlap, with sharp declines away from this region (Supplementary Fig. 4). In an alternative arrangement, Pgant35A produces sense and antisense siRNAs across its entire annotated transcript, consistent with expressed-sequence-tag support for antisense transcription traversing this locus (Supplementary Fig. 5).
Thus, a large number of Drosophila genes generate endogenous siRNAs, with most having perfect complementarity to the 3′ UTRs of neighbouring genes. Relative levels of endo-siRNAs generated from each convergent transcription unit were low (not shown), and we found no or little change (up to a ~1.3-fold increase) in the expression of such genes in AGO2 mutant ovaries. Possibly, the level of small RNAs produced by this genomic arrangement is inconsequential, amounting to noise within silencing pathways. However, there are probably circumstances wherein regulation by such arrangements might substantially impact expression.
In S2 cells, two neighbouring loci encoded nearly 16% of AGO2-associated RNAs (Supplementary Table 2). These reside within a large intron of klarsicht (Supplementary Fig. 6) and did not generate siRNAs in any other tissue. A similar locus, corresponding to CG14033, was found within an intron of thickveins (Supplementary Fig. 7) and gave rise to testis-specific siRNAs. Although the function of both siRNA clusters is unclear, the thickveins cluster shares considerable complementarity to CG9203, and loss of AGO2 and Dcr-2 mildly increased CG9203 mRNA levels in testis but not in ovaries (Supplementary Fig. 7).
Dcr-2 has been implicated in the production of siRNAs from viral replication intermediates or exogenously introduced dsRNAs, whereas Dcr-1 has been linked to miRNA biogenesis6,16,17. In agreement with these observations, all endo-siRNA classes were lost in Dcr-2 mutant ovaries (Fig. 4a). To obtain more insight into the genetic requirements for endo-siRNA biogenesis and stability, we depleted components of siRNA and miRNA pathways in S2 cells and analysed levels of abundant siRNAs derived from structured loci (Fig. 4b and Supplementary Fig. 8). Although depletion of Dcr-2 and AGO2 resulted in substantial reductions in siRNA levels, little or no changes were observed on Drosha, Pasha, Dcr-1 or AGO1 depletion. Unexpectedly, we found virtually no requirement for the Dcr-2 partner R2D2 (ref. 14) but a strong requirement for the Dcr-1 partner Loquacious1,4,5. Only one analysed siRNA exhibited partial dependence on R2D2, potentially correlating with the extensive dsRNA character of its precursor duplex (Supplementary Fig. 9). Artificial sensors for endo-siRNAs from esi-1 and esi-2 in S2 cells gave patterns of de-repression that matched our analysis of endo-siRNA levels (Figs 3c and 4c).
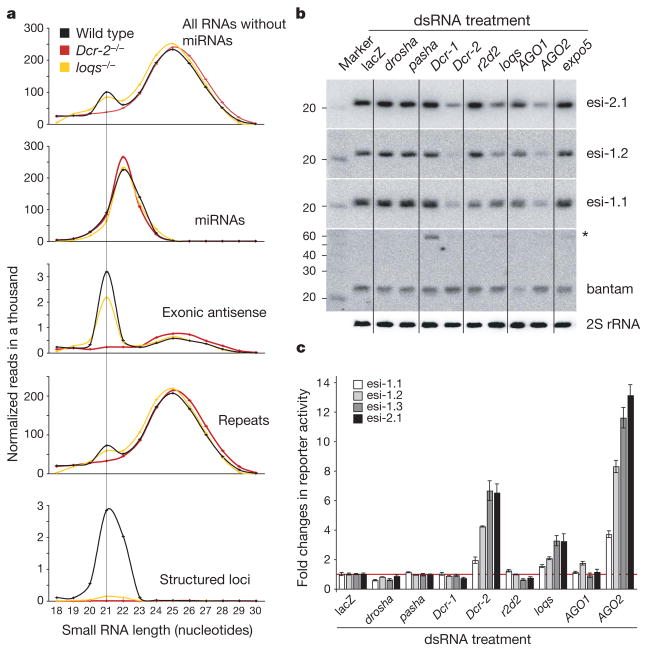
Figure 4. Genetic requirements for siRNA biogenesis.
a, Length distributions of small RNAs from total RNA libraries obtained from wild-type (black), Dcr-2 mutant (red) and loqs mutant (yellow) ovaries. Excluding miRNAs and piRNAs (23–29 nucleotides), the population of 21-nucleotide siRNAs mapping to structured loci, genes and repeats is lost from Dcr-2 mutants and those mapping to structured loci are strongly reduced in loqs mutants (all libraries are adjusted to the same total small RNA count). b, Northern blots showing levels of three siRNAs encoded from structured loci esi-1 and esi-2 (esi-2.1, esi-1.1 and esi-1.2) in S2 cells treated with dsRNA against the genes indicated. As controls, northern blots of bantam (pre-miRNA indicated by the asterisk) and 2S rRNA are shown below. c, Renilla luciferase reporter assays are shown for the siRNAs examined in b and an additional esi-1-derived species (esi-1.3) in S2 cells treated with dsRNAs against the indicated genes (error bars indicate standard deviation; n = 3).
Analysis of the most abundant siRNA from esi-2 in flies mutant for Dcr-2, AGO2, r2d2 or loqs extended our findings from cell culture (Supplementary Fig. 10). To examine the unexpected requirement for loqs more broadly, we sequenced small RNAs from loqs-mutant ovaries and observed a near complete loss of endo-siRNAs from structured loci (Fig. 4a). A much smaller impact of loqs was seen on endo-siRNAs derived from repeats and convergent transcription units. However, an involvement of Loqs and not R2D2 in the function of siRNAs derived from perfect dsRNA precursors was supported by analysing the impact of depleting siRNA/miRNA pathway components on the ability to suppress FHV replication in our infected S2 cell cultures (Supplementary Fig. 11).
Our results uncover an unanticipated role for Loqs in siRNA biogenesis and suggest that R2D2 has a lesser impact on at least two types of endogenous siRNAs. It is well established that Loqs partners with Dcr-1 for miRNA processing. To probe a molecular interaction with Dcr-2, we catalogued Loqs binding partners using quantitative proteomics. Dcr-1 and Dcr-2 were both abundant in Loqs immunoprecipitates from cultured cells and flies (Supplementary Fig. 12), supporting a physical interaction between Dcr-2 and Loqs.
Among animals, endo-siRNA pathways have so far been restricted to Caenorhabditis elegans24–27. Our results extend the prevalence of such systems to Drosophila and parallel recent discoveries of an endo-siRNA pathway in mouse oocytes28,29. These systems have many common features but also key differences. In both, siRNAs collaborate with piRNAs to repress transposons. Also, mouse and Drosophila both generate endo-siRNAs from structured loci. In mouse, dsRNAs can form by pairing of sense protein-coding transcripts with antisense transcripts from pseudogenes. Whether or not transcripts from unlinked sites lead to siRNA production in Drosophila is unclear. However, transposon sense transcripts may hybridize to antisense sequences transcribed from piRNA clusters to form endo-siRNA precursors. In flies, a much larger number of genic loci enter the pathway as compared to mice because convergent transcription of neighbouring genes frequently creates overlapping transcripts. Overall, annotation of the Drosophila genome indicates that a significant proportion is transcribed in both orientations, providing widespread potential for dsRNA formation. This property is shared by many other annotated genomes, raising the possibility that the RNAi pathway has broad impacts on gene regulation. Viewed in combination, our studies suggest an evolutionarily widespread adoption of dsRNAs as regulatory molecules, a property previously ascribed only to miRNAs.
METHODS SUMMARY
The fly stocks used were Dcr-2L811Fsx (ref. 6), AGO2414 (ref. 30), loqsf00791 (ref. 1) and r2d21 (ref. 14). Recombineering was used to insert a Flag–haemagglutinin (HA) tag at the amino terminus of the AGO2 coding sequence in the context of the genomic AGO2 locus including flanking regulatory regions (for details, see Methods). Polyclonal anti-AGO1 antibody was obtained from Abcam (lot number 113754). Small RNAs for library production were isolated from ovarian total RNA or from Argonaute immunoprecipitates. Libraries were produced as described12 and sequenced using the Illumina platform (protocol available on request). A description of the bioinformatics methods can be found online. For quantitative real-time PCR (qRT–PCR) analyses, we used total RNA preparations and random hexamer primers. Details and all primer sequences are given in the Supplementary Information. S2 cell knockdown treatments were for eight -days with two sequential dsRNA soakings. For the reporter experiments, inducible expression plasmids for Renilla and firefly were transfected into S2 cells together with dsRNA for the desired knockdown target. Renilla constructs contained two target sites for endogenous siRNAs, whereas the firefly construct was used for normalization. For details on plasmids, dsRNAs and target sites, see Supplementary Information.
Supplementary Material
Acknowledgments
We thank R. Carthew, H. Siomi, P. Zamore and D. Smith for reagents. We are grateful to M. Rooks, E. Hodges and D. McCombie for help with deep sequencing. B.C. was supported by the German Academic Exchange Service. C.D.M. is a Beckman fellow of the Watson School of Biological Sciences and is supported by an NSF Graduate Research Fellowship. R.Z. is a Special Fellow of the Leukemia and Lymphoma Society. M.D. is an Engelhorn fellow of the Watson School of Biological Sciences. J.B. is supported by the Ernst Schering foundation. A.S. is supported by an HFSP fellowship. This work was supported in part from grants from the NIH to G.J.H. and N.P. and a gift from K. W. Davis (G.J.H.).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Small RNA sequences were deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE11086. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Forstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 4.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1–Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang F, et al. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 8.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 9.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 10.Aravin AA, Hannon GJ, Brennecke J. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 11.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 12.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 13.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 15.Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila. Nature Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 17.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 20.Rehwinkel J, et al. Genome-wide analysis of mRNAs regulated by Drosha and Argonaute proteins in Drosophila melanogaster. Mol Cell Biol. 2006;26:2965–2975. doi: 10.1128/MCB.26.8.2965-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruby JG, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 25.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 26.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 27.Yigit E, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 28.Tam OH, et al. Pseudogene-derived siRNAs regulate gene expression in mouse oocytes. Nature. 2008 10 April; doi: 10.1038/nature06904. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008 10 April; doi: 10.1038/nature06908. advance online publication. [DOI] [PubMed] [Google Scholar]
- 30.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stark A, et al. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.