Abstract
Because of the high cost of commercial genotyping chip technologies, many investigations have used a two-stage design for genome-wide association studies, using part of the sample for an initial discovery of “promising” SNPs at a less stringent significance level and the remainder in a joint analysis of just these SNPs using custom genotyping. Typical cost savings of about 50% are possible with this design to obtain comparable levels of overall type I error and power by using about half the sample for stage I and carrying about 0.1% of SNPs forward to the second stage, the optimal design depending primarily upon the ratio of costs per genotype for stages I and II. However, with the rapidly declining costs of the commercial panels, the generally low observed ORs of current studies, and many studies aiming to test multiple hypotheses and multiple endpoints, many investigators are abandoning the two-stage design in favor of simply genotyping all available subjects using a standard high-density panel. Concern is sometimes raised about the absence of a “replication” panel in this approach, as required by some high-profile journals, but it must be appreciated that the two-stage design is not a discovery/replication design but simply a more efficient design for discovery using a joint analysis of the data from both stages. Once a subset of highly-significant associations has been discovered, a truly independent “exact replication” study is needed in a similar population of the same promising SNPs using similar methods. This can then be followed by (1) “generalizability” studies to assess the full scope of replicated associations across different races, different endpoints, different interactions, etc.; (2) fine-mapping or re-sequencing to try to identify the causal variant; and (3) experimental studies of the biological function of these genes. Multistage sampling designs may be more useful at this stage, say for selecting subsets of subjects for deep re-sequencing of regions identified in the GWAS.
Keywords: multistage sampling, genetic associations, replication, re-sequencing, DNA pooling, gene-environment interactions
1. INTRODUCTION
Many of the genome-wide association studies (GWAS) currently underway or already reported have used some form of multistage sampling design (Satagopan et al., 2002) because of the considerable savings in genotyping costs this approach offers. In section 2, we provide an overview of the basic approach to designing such studies, touching on such topics as the trade-offs between sample size and marker density, the selection of markers to carry forward to the second stage, methods of significance testing, the use of DNA pooling, and multistage designs for testing gene-environment (G×E) and gene-gene (G×G) interactions. Section 3 considers the general question of whether two-stage designs are still necessary in an era of declining costs and multi-purpose studies. Finally, section 4 discusses what should be done after a completed GWAS, including replication, fine mapping, generalizability and functional studies, and revisits the utility of multi-stage designs in this context.
2. BASIC PRINCIPLES OF TWO-STAGE STUDY DESIGN FOR GWAS
Two-phase case-control designs were introduced to the epidemiologic literature by White (1982) and have been extensively developed in a series of papers by Breslow and various colleagues (for a general overview of this literature, see (Breslow and Chatterjee, 1999)). The basic idea of these designs is to use information available on all subjects in the main study to draw a more informative subsample for additional, more expensive, measurements, combining the information from both phases in the analysis.
Two-stage sampling for GWAS, as introduced by Satagopan et al. (2002) is quite different, based on genotyping part of the sample using a commercial high-density panel (typically 300,000 to a million SNPs) and then genotyping the most promising SNPs using a customized panel on the remainder of the sample. A final analysis combining the information from both samples is more powerful than treating the design as a hypothesis generation followed by independent replication (Skol et al., 2006; Yu et al., 2007) because it exploits the additional information about how significant the first stage associations were, not just the fact that they exceeded some threshold. Formally, two stage designs can be conceptualized as a family of group sequential tests (one per SNP) with allowance for early stopping for “futility” (Jennison and Turnbull, 2000).
Optimization of the design is usually framed as choosing the significance levels and the allocation of samples between the two stages in such a manner as to minimize the total cost while attaining the desired genome-wide significance level and power (Kraft, 2006; Kraft and Cox, 2008; Muller et al., 2007; Saito and Kamatani, 2002; Satagopan and Elston, 2003; Satagopan et al., 2004; Service et al., 2003; Skol et al., 2007; Wang et al., 2006); alternatively, one might wish to maximize power subject to total sample size, genotype cost, and type I error, or to minimize the total sample size subject to the other factors. These optimal designs are insensitive to the genetic model (mode of inheritance, relative risk, and allele frequencies) and determined primarily by the total number of markers to be genotyped in stage I, the relative cost per genotype at stages I and II, the total available sample size, and whether (and how many) additional flanking markers will be tested around those selected from stage I. As an example of a cost minimization, the optimal design for a cost ratio of about 17.5 with 500K markers being tested in stage I and no additional SNPs being tested at stage II turns out to involve testing 30% of the sample in stage I at a significance level of 0.0037 (i.e., about 1850 markers tested in stage II), and a significance level for the joint analysis of 1.6×10−7 (Wang et al., 2006); in this case, about 87% of the total cost goes to stage I genotyping, but the total cost is only 40% that of a comparably powered one-stage design.
Several authors (Eberle et al., 2007; Gail et al., 2007; Nannya et al., 2007) have investigated the power of GWAS, either for a single-stage or the first-stage of a multistage scan, and generally concluded that sample sizes of 1000 cases and 1000 controls were sufficient to detect associations in the range of 1.7–2.0, smaller relative risks (e.g., 1.2–1.3) requiring much larger sample sizes. In general, minimum-cost two-stage designs can require considerably larger sample sizes than single-stage designs to achieve the same power. However, large costs reductions can still be achieved with a “nearly optimal” two-stage design using a sample size only slightly larger than a one-stage design (Wang et al., 2006).
2.1 Trade-off Between Sample Size and Marker Density
A crucial decision to be made is the choice of genotyping platform for stage I. At this writing, two companies—Affymetrix and Illumina—offer platforms ranging from 300K to 1M SNPs. The panels differ in the way SNPs were selected and hence their coverage (r2) of the remaining common HapMap SNPs, as well as in their laboratory performance (call rates, reproducibility, etc.). Because coverage of SNPs is highly variable across the genome and the relationship between power and r2 is nonlinear, the average power to detect an association with a random SNP is smaller than the power based on the average r2 (Jorgenson and Witte, 2006). Instead, one must average the power for a given non-centrality parameter λ at a putative causal locus across the distribution of r2s. The consequence is that one cannot simply add additional sample size to cover regions with poor coverage! For single stage studies, average power is maximized by choosing the platform with the best coverage on which it is affordable to genotype all available samples. Comparisons of recent platforms tend to show that when genotyping budget is limiting, sacrificing sample size for the higher density platform is not usually appropriate (Hao et al., 2008; Lewinger et al., 2007b; Nannya et al., 2007). A two-stage design, however, can alter the sample-size vs. coverage tradeoff in favor of higher density platforms in the first stage by allowing the use of all the available samples at a lower cost. Imputation, on the other hand, reduces the differences between SNP panels, making the lower cost, lower density platforms more attractive (Anderson et al., 2008). See also (Barrett and Cardon, 2006; de Bakker et al., 2005; Pe'er et al., 2006) for further discussion.
2.2 Design Complications
2.2.1. Additional markers
Additional markers flanking some or all hits might also be added to better characterize the full range of genetic variation in the region (Saito and Kamatani, 2002; Wang et al., 2006). With 5 additional markers being tested for each hit, the optimal design for the situation discussed above raises the first stage sample size to 49% and reduces significance levels to 0.0005 and 0.5×10−7 respectively, so that 95% of the total cost goes to stage I genotyping. In these calculations, it was assumed that the additional markers would be imputed for the first-stage sample, using methods described in §2.2.2 and more comprehensively elsewhere in this issue (Marchini, 2009), but one could instead test them directly on the stage I samples first and then decide which ones to carry forward to stage II. Further work on optimization of such designs is still needed.
While it seems intuitively appealing to also use the replication step for the purpose of fine mapping—i.e., to see whether there is another marker in the region that shows even stronger evidence for association—the yield from doing so may be minimal. Consider the three possible situations: (1) an associated marker is in perfect LD with a causal variant; (2) it is in weak LD with a causal variant; or (3) it is nowhere near the causal variant. Only in the second case would adding additional markers be of any help. Suppose the first stage sample has power 1−β1 to detect the first kind of association and 1−β2 for the second, and let πk denote the prior probability of type k. Then the prior probability that the association is of type 2 given p < α1 is
Considering the coverage of current platforms, π1 is probably larger than π2 and π3 is certainly much larger, so most detected associations are likely to be of types 1 or 3 and additional markers will not help [Peter Kraft, personal communication].
Clarke et al. (2007) have shown theoretically and by simulation that the increased penalty for multiple comparisons can defeat any possible gains in power for replication. Nevertheless, the inclusion of additional markers can be advantageous in regions of relatively high LD when the original signal is weak, such as in regions where the coverage by the original panel is poor, but then any new associations discovered in the “replication” stage would require yet further confirmation. In general, they recommend deferring fine mapping to a separate sample from that used for replication. Thus, fine mapping should be reserved for the regions that are interesting in the combined stage I and stage II data, rather than incorporated into stage II for all markers carried over from stage I. This keeps the multiple comparisons problem at a minimum whether or not a new (third) sample is used for fine mapping.
2.2.2. Haplotypes, multi-marker tests, and imputation of missing markers
The commercial panels are designed to allow for testing not just the hundreds of thousands or a million SNPs on the panel, but also all the roughly 5M common variants in the human genome they tag, including copy number variants. This entails using some form of multi-marker or haplotype-based approach to “impute” genotypes to all those variants that are not directly tested. Promising associations with imputed variants detected in the initial scan are then tested in either the original sample or the follow-up stage by direct genotyping. While at first blush it might seem that the multiple comparisons penalty for testing 5M variants would offset the advantage of using a tag-SNP approach, the correlation between tests due to LD means that the “effective number of independent tests” is only about 1M in European-descent populations or 2M in African-descent populations (Pe'er et al., 2008). Four companion papers in this issue (Chatterjee, 2009; Marchini, 2009; Visscher, 2009; Zollner, 2009) address various aspects of this topic in greater detail.
2.2.3. Family-based designs
One compelling advantage of a two-stage approach may be the opportunity to exploit different study designs, in particular family- and population-based. For example, the Cancer Family Registries for breast and colorectal cancer are currently undertaking GWASs aimed at exploiting their unique resource, combining the two sampling schemes. In the first stage, a population-based series of cases that is enriched for a positive family history or young age at onset is compared with unrelated population controls; hits from this stage are then to be tested using family-based association tests (FBAT) in the second stage using sibling or cousin controls to weed out false positives due to population stratification (see the contribution by Balding (2009) in this issue), and finally in a third stage, combining the phenotypes of all relatives from extended pedigrees with all available genotypes in a conditional segregation analysis (Hopper et al., 1999). A different two-stage design uses between-family comparisons to select a subset of SNPs with high power to detect associations in an FBAT, and tests associations with this subset using within-family comparisons in the same dataset (Van Steen et al., 2005). For further discussion of these various options, see the companion paper in this issue by Laird (2009).
If instead of a FBAT design, some form of genomic control is to be used with population-based case-control studies in a two-stage design, then problems can arise if the subjects in the two stages are derived from different populations. One approach is to estimate kinship using the available data from the different stages (a high-density chip for stage I, just the selected SNPs and perhaps some additional ancestry-informative markers in stage II).
2.2.4. More than two stages?
In principle, there is no reason why the two-stage design described above could not be extended to a multi-stage design, with successively smaller proportions of SNPs being tested in new samples at each subsequent stage. Indeed, some of the earliest studies were conducted in just that manner (Hirschhorn and Daly, 2005). Multiple stages would have the practical effect of reducing the genotyping cost ratio between the first stage and the combined later stages, perhaps by a significant factor. Inclusion of additional stages would be most cost effective when the genotyping cost ratio is 1 between the platforms used in the second and later stages (Kraft et al., 2007a). The additional complexities in both design optimization and final significance testing of results have yet to be fully explored, however.
2.3 Methods of significance testing for two-stage designs
Two-stage designs pose special challenges to significance testing in the final analysis of the combined data. The basic p-value to be computed is the probability that a given SNP would have been deemed “promising” at the first stage and that the combined data would show significance at a genome-wide level given that it was selected for testing in the second stage, under the null hypothesis that it is not associated with disease. The fact that two “hurdles” have to be crossed for each “significant” result means that the p-value of interest is actually somewhat smaller than the “nominal” p-value based on analyzing the combined data. The various two-stage design papers discussed earlier have shown how to compute this probability under simplifying assumptions and thereby optimize the design, but these approximations can often be improved upon in analyses of real data. Amongst other assumptions is that of independence across SNPs, which is necessary to derive the appropriate cutoff for genome-wide significance. An obvious way to avoid having to make such assumptions is some form of a permutation test. For a single-stage design, this is straight-forward: one could simply hold the genotypes fixed (thereby maintaining their LD structure) and randomly permute the phenotypes in a standard case-control design (or analogous methods for family-based studies, based on within-family permutation). In a two-stage design, this is not so straight-forward, however, as one must permute the entire analysis; but a random permutation of the stage-one data would yield a different set of SNPs to be tested in stage II and these genotypes are not available for permuting!
Two methods have been proposed to assess significance in two-stage studies. They both make clever use of the fully genotyped stage I subjects to mimic the effect of having two stages of genotyping. Both require large numbers of subjects in stages I and II, an assumption that would be usually met for a well powered GWAS. In Dudbridge's (2006) method, a permutation null distribution is computed by performing the full two-stage analysis on a large number of permuted datasets in which a sub-sample of the stage I subjects plays the role of the stage I sample, and the original stage I subjects play the role of the combined stage I and stage II samples. The method is valid under exchangeability of the stage I and stage II samples provided the permutation distribution “stabilizes” for large samples. The Monte Carlo method of Lin (2006) relies on the fact that the efficient scores functions have, jointly for all tests in stages I and II, a mean-zero asymptotic multivariate normal distribution under the complete null hypothesis, and that all score, Wald, or likelihood ratio test statistics commonly used to test single SNPs or haplotypes are asymptotically equivalent to simple chi-square statistics based on the efficient score functions. Assuming that the subjects are randomly chosen for stages I and II, the asymptotic variance matrix of the efficient scores can be estimated based on the observed efficient score functions for stage I only. Monte Carlo replicates can then be efficiently drawn from the estimated asymptotic multivariate normal distribution of the efficient scores, and the chi square statistics equivalent to the original tests computed for each Monte Carlo replicate. Adjusted p-values can be computed based on the Monte Carlo replicates. An advantage of Lin's method is that it does not require re-calculation for each Monte Carlo replicate of the original tests statistics that can be computationally costly, but only for the simpler equivalent chi-square tests based on the efficient scores. This can result in significant time savings. Both Lin's and Dudbridge's method can be extended to two-stage family-based GWAS but not to studies using case-control samples in stage I and families in stage II.
Methods based on Bonferroni correction using an “effective number of tests” (see §2.2.2) for a given platform in a single-stage design have typically relied on permutation tests applied to data sets where very large numbers of SNPs are genotyped in relatively small numbers of subjects (e.g., the HapMap). Just as for the methods described above, there is an implicit assumption in these calculations that the null distribution of the minimum p-value for a group of tests does not depend very strongly on the number of subjects in the analysis but only on the LD pattern between the tests considered.
The entire subject of adjustment for multiple comparisons is rapidly evolving. For a recently proposed method and a review of other methods, see Han et al. (2009).
2.4 Selection of SNPs for the next stage
Another decision entails the selection of SNPs to be carried from stage I to stage II or to be reported as “significant” at the end of the study. Of course the true causal association may not lie anywhere near the top of the distribution of p-values (Zaykin and Zhivotovsky, 2005). Furthermore, if the distribution includes some false positives due to bias (e.g., differential genotyping error), then the most significant findings are more likely to be false positives.
Most of the literature has assumed that p-values for single SNP associations will be used for selecting SNPs to carry forward, although alternatives have been suggested, including the population attributable risk (Hunter and Kraft, 2007), the False Positive Report Probability (Hunter and Kraft, 2007; Samani et al., 2007), Bayes factors or q-values (Wakefield, 2008), empirical Bayes estimates of effect size (Hunter and Kraft, 2007), or multi-marker methods like a local scan statistic (Guedj et al., 2006). But such approaches make no use of any external information that might suggest that some associations were more credible than others a priori. For example, one might wish to give greater credence to associations with SNPs located in or near genes (particularly those that may have a high prior probability of involvement in the disease) or highly conserved regions of the genome, coding SNPs, those located under a linkage peak, or those with previously reported associations. Often such information is used informally at the conclusion of a GWAS in deciding which associations to pursue with further fine mapping or functional studies.
Roeder et al. (2006; 2007) have proposed a weighted False Discovery Rate framework and Bayesian versions have been proposed by Whittemore (2007) and Wakefield (2007). All of these allow a specific variable to be used to up- or down-weight the significance assigned to each association. They showed that well chosen prior information can substantially improve the power for detecting true associations, while there was relatively little loss of power if that information is uninformative.
Each of these approaches allows only a single variable to be incorporated, with weights specified in advance. Hierarchical modeling approaches (Chen and Witte, 2007; Lewinger et al., 2007a) allow multiple sources of information to be empirically weighted in models for the probability that an association is null and the expectation of the magnitude of an association given that it is not null. Simulation studies (Lewinger et al., 2007a) showed that when there was little or no useful prior knowledge, the standard p-value ranking performed best, but when at least some of the available covariates were strongly predictive (even if one did not know which ones were truly predictive), the hierarchical Bayes ranking led to better power. See the papers by Pfeiffer et al. (2009) and Roeder (2009) in this issue for further discussion.
2.5 DNA pooling
DNA pooling offers another approach that could drastically reduce the cost of genotyping for a GWAS. While the idea has been around for some time (Bansal et al., 2002; Risch and Teng, 1998), the technical challenges in forming comparable pools and quantifying allele frequencies are formidable (Barratt et al., 2002; Feng et al., 2004; Pfeiffer et al., 2002; Sham et al., 2002; Zou and Zhao, 2004). It is only recently that it has proved feasible to apply this technique to high-density genotyping arrays (Craig et al., 2005; Docherty et al., 2007; Johnson, 2007; Meaburn et al., 2006; Sebastiani et al., 2008; Zuo et al., 2006). As currently employed, the design generally entails forming several small pools of cases and of controls in stage I and selecting SNPs on the basis of their differences in allele frequencies. These are then retested by individual genotyping in stage II, possibly on both the original and a second sample. Much remains to be done to study the best choices of design parameters (numbers of pools, sample sizes, criteria for selecting SNPs to test by individual genotyping, etc.) (Macgregor, 2007) and to estimate the statistical power and false discovery rate for this approach in practice. However, empirical applications have demonstrated that DNA pooling is capable of detecting several associations that have previously been discovered and confirmed by individual genotyping in a GWAS context (Pearson et al., 2007). Furthermore, several studies using this approach have reported novel associations (Kirov et al., 2008; Spinola et al., 2007; Steer et al., 2007), although it remains for these associations to be confirmed independently.
Several recent technological advances offer the potential to greatly improve the utility of DNA pooling. The first entails molecular “bar coding” of the individual DNA molecules contributing to each pool (Craig et al., 2008), so that the genotypes of the specific individuals contributing to the subset of pools found to contain rare variants in excess in case pools compared to control pools can be readily reconstructed without the need for further genotyping. The second development entails the use of “pools of pools” to dramatically reduce the cost, so that it now becomes feasible to obtain DNA sequence information on pools as large as 3000 (D. Duggan, TGen, personal communication). We will revisit the use of multistage designs using pooled DNA for deep-resequencing in the concluding section.
2.6 Multistage Designs for Testing Main Effects and Interactions
The NIH “Genes and Environment Initiative” has focused attention on the use of GWAS for identifying genes that modify the effects of environmental agents (Kraft et al., 2007b). Such studies pose additional methodological problems, beyond the usual challenges in assessing the main effects of genes and environmental factors, such as low power (Gauderman, 2002) (for further discussion, see the paper by Kooperberg (2009) in this issue). However, there is the opportunity to improve power by using a case-only design (Piegorsch et al., 1994) in which G×E interaction is tested by testing for association between a gene and environmental factor among cases, under the assumption that this association does not exist in the general population. Such an assumption is not likely to hold for all possible SNP × E interactions in a GWAS, but testing this assumption first in controls and deciding whether to perform a case-only or conventional case-control test accordingly can lead to substantial inflation of Type I error rates (Albert et al., 2001). Nevertheless, more appropriate methods for combining the inferences from case-control and case-only analyses of the same data have been described (Chatterjee and Carroll, 2005; Chatterjee et al., 2005; Cheng, 2006; Mukherjee et al., 2007; Mukherjee et al., 2008; Mukherjee and Chatterjee, 2008). For example, Mukherjee and Chatterjee (2008) use an empirical Bayes compromise between the case-only and case-control estimators, weighted by the estimated probability of the existence of a G−E association. Rather than limiting the analysis to an all-or-nothing choice between case-only and case-control approaches, these methods have the advantage of letting the data and a prior estimate the most appropriate weight between models. In the case of SNP×SNP interactions one may use LD information from HapMap to generate flexible priors that can greatly increase power (Li and Conti, 2008). In the context of a GWAS, various multistage designs are possible, such as using a case-only test in the combined sample of cases and controls to screen interaction effects and then confirming that subset by a standard case-control test in the same dataset (Murcray et al., 2008). This design has been shown to be substantially more efficient than a single-stage scan using a standard case-control comparison.
3. SINGLE VS. TWO-STAGE DESIGNS
As the cost of commercial chips falls relative to custom genotyping, the merits of this approach will need to be reconsidered (Hunter et al., 2007). As mentioned above, faced with a choice between density of SNPs and sample size in a single stage study, it is usually preferable to have the largest possible sample size, even if this means not being able to afford a higher density chip. A two-stage design may, however, allow a higher density chip to be used in stage I than would be affordable in a single-stage design, and hence improve power for regions of low LD and overall mean power (Lewinger et al., 2007b). The ability to combine different study designs (e.g., population-based and family-based) may also favor a two-stage design. Other considerations, however, may favor a one-stage design, such as faster study completion and simplified logistics and quality control due to use of a single genotyping platform. Additionally, multiple hypotheses can be tested using these data, say multiple phenotypes in a cohort design or various subgroup analyses or interaction tests. For example, in addition to scanning for genetic main effects, the Southern California Children's Health Study (CHS) of the health effects of air pollution aimed to identify genes that interact with two measures of air pollution, exposure to traffic, in utero and second-hand tobacco smoke, and GSTM1 (previously shown to be involved in several G×E interactions) or to differ between Hispanic and non-Hispanic children, each of these for two phenotypes, asthma and lung function development. SNPs might be selected from the initial scan for follow-up based on any of these criteria. In order to have reasonable power for detecting each of these effects, a custom panel of 12K markers or more would have been required, the cost of which begins to approach that of simply using the same high density panel as in the initial scan, so the decision was made to do a one-stage scan instead. In fact, in the NHLBI-funded STAMPEED consortium of GWAS for cardiovascular, lung, and blood disorders of which the CHS is a part (http://public.nhlbi.nih.gov/GeneticsGenomics/home/stampeed.aspx), most of the 13 participating centers are using a one-stage design. In a one-stage design, replication of SNPs attaining genome-wide levels of significance is still needed, as discussed below. However, the combination of discovery and replication phases should not be regarded as a formal two-stage design, which we define as involving the testing a large number hits in a second stage and doing a joint analysis of both. This may involve optimizing the choice of sample sizes and significance levels as discussed earlier, but these two stages combined have the same goal as a 1-stage design, namely discovery.
4. AFTER GWAS, WHAT NEXT?
Multistage sampling designs for GWAS should not be thought of as a hypothesis generation followed by independent replication approach but rather as simply a more cost-efficient way of conducting the discovery approach (Skol et al., 2006). Nevertheless, it must be appreciated that any effect estimates (e.g., odd ratios) surviving the entire discovery process will tend to be biased away from the null because attention is focused only on those that are statistically significant, a phenomenon known as the “winner's curse” (Kraft, 2008; Yu et al., 2007; Zhong and Prentice, 2008; Zollner and Pritchard, 2007). Thus, some form of truly independent replication is needed, both to confirm the existence of the reported associations and to estimate the magnitude of their effect. In the following sections, we distinguish between what we will call “exact replication” and “generalization,” the latter being aimed at determining the full extent of a replicated association across populations, phenotypes, modifiers, etc. In addition, an association initially reported may not be with the causal variant itself, but rather with some other variant it is in LD with, so further studies aimed at fine mapping or re-sequencing the region to identify the culprit may be needed. Finally, once plausible candidates for the causal variants have been identified, there is a need for further experimental studies to understand their biological function and additional in silico and epidemiologic analyses to build a comprehensive model for the causal pathway.
4.1. Replication
Failure to replicate has been a recurring problem with candidate gene association studies, hence a major concern about the new generation of GWASs (Chanock et al., 2007; Ioannidis, 2007). (The companion paper by Ioannidis in this issue (2009) explores the replication issues in greater depth.) True scientific replication must involve something more than a repetition of the study on a second random sampling from the same population using the same methods (Chanock et al., 2007; Clarke et al., 2007), since simply splitting a sample in half and requiring significance at level α in both halves is less powerful than a single analysis of the entire sample at significance level α2 (Skol et al., 2006; Thomas et al., 1985). Nevertheless, the goal at this stage should be to avoid failure to replicate because of true differences in effect between the original and follow-up populations, investigation of real heterogeneity being the subject of the next stage (“generalization”). Many granting agencies now expect investigators to discuss plans for follow-up investigations of any associations detected and some high profile journals are requiring replication studies as part of a single report of a genetic association (Anonymous, 1999; Rebbeck et al., 2004). In many cases, this might best be accomplished by collaborations with other groups with data on a genetically similar set of subjects. Failure to replicate may often be due to the use of replication data sets that were not well designed for this purpose because of heterogeneity between the original and replicate data sets or problematic study designs that were generated for different purposes originally. Replication and generalizability are often muddled together even though they are two different questions that are best addressed with different types of study populations —one selected to minimize heterogeneity and the others selected to maximize it.
One question that frequently arises is whether to restrict replication claims to the same marker detected in the initial GWA scan (“exact” replication) or to test additional markers in the region and allow association with any of them (appropriately adjusted for multiple comparisons) to be treated as evidence of replication (“local” replication) (Clarke et al., 2007). In a similar vein, associations first discovered in a GWAS by imputed SNPs should be confirmed by direct genotyping, either in the original samples, or better in independent replication samples, before a genuine association is claimed. In any event, a clear definition of replication is needed: generally this is taken to be a statistically significant association in the same direction, but now not requiring genome-wide multiple testing correction since only a subset of the top-ranking associations will be subject to replication and the magnitude of the original relative risk is likely to have been overestimated.
4.2. Generalization
Once an association has been replicated, it becomes important to investigate the full range of its effects. For example, one of the first questions to address is whether the effect differs across races. If so, this could be a sign that the association is not causal, but only a reflection of a causal effect of some other variant with which it is in LD, the patterns of LD differing across races, and would suggest that further fine mapping of the region is warranted. Furthermore, if there is heterogeneity by race/ethnicity, fine mapping within a race that exhibits the association of interest but has shorter LD blocks would help localize the signal more efficiently than in a race with longer LD blocks. Alternatively, heterogeneity by race could be a reflection of differences across races in the prevalence of some modifying factors—G×E or G×G interactions—indicating that further investigation of effect modification is warranted. Beyond the question of heterogeneity by race/ethnicity, there are other questions of generalizability worth considering. Does the variant have similar effects across different subtypes of the same disease (for example, for colorectal cancer, by location in the colon, age of onset, family history of colorectal or other cancers, presence or absence of selected molecular markers such as microsatellite instability, BRAF mutation, MLH1 methylation)? Does the variant have effects on other phenotypes — intermediate endpoints like incidence or recurrence of polyps for colorectal cancer or other cancer sites or even other diseases with which it might share a common etiologic pathway? After a result is confirmed in a properly designed replication study, we would advocate a strategic approach to the question of generalizability, guided by a careful consideration of the most important knowledge gaps about the disease, rather than the sometimes uncritical exercise of quickly testing for the reported SNP in whatever data sets are readily available (ignoring whether the result would fill an important gap in our knowledge base).
4.3. Fine-Mapping and Deep Re-sequencing
Unless there is compelling evidence that a newly discovered association with a particular SNP is indeed causal, further fine mapping of the surrounding region is generally appropriate, given that the SNPs on the discovery panel represent at most 20% of all common variants and were selected primarily for their effectiveness at tagging other variants rather than as biologically plausible candidates themselves. Furthermore, it is becoming increasingly evident that multiple rare variants may play an important role in many diseases (Fearnhead et al., 2004; Iyengar and Elston, 2007; Kryukov et al., 2007; Li and Leal, 2008; Pritchard, 2001). This search for the true culprit(s)—which could occur before, after, or concurrent with the generalization activities described above—might involve some combination of fine mapping with additional SNPs and deep re-sequencing and poses interesting challenges for study design, particularly in terms of the balance between additional efforts to fine-map a signal with additional genotyping of previously known variants versus jumping directly to deep re-sequencing for discovery.
Fine mapping might explore a relatively large region surrounding the associated SNP(s) and be informed by knowledge from HapMap of the LD structure of the region. The goal would be to genotype a denser set of tag SNPs than was possible in the initial GWAS in order to conduct haplotype or multi-marker association tests, as discussed elsewhere in this issue (Marchini, 2009; Visscher, 2009; Zollner, 2009). (For this purpose, one might wish to use a different population, such as those of African descent where LD blocks would tend to be shorter.) Deep re-sequencing would entail selection of a subset of participants from the main study for complete sequencing of the region to search for other relatively common variants (~1–5%) that may not have been characterized by HapMap. These variants would then be genotyped in the entire study sample to test for association. Because of the high cost of sequencing, it might be advisable to do the fine mapping first to narrow down the region of interest, but with the advent of next generation sequencing, DNA “bar coding,” and DNA pooling methods (Craig et al., 2008), costs are coming down so rapidly that one might want to proceed directly to sequencing. Either approach might benefit from a formal two-stage sampling design, although the cost savings are likely to be more substantial for deep re-sequencing studies.
4.3.1. Two stage designs revisited
The basic idea here would be to select a subset of subjects for additional genotyping and/or sequencing who would be most likely to carry a causal variant. This subset of subjects serves two general goals. The first may simply be to characterize the genetic variation or to discover previously unknown variants within the region to then genotype in the larger main study with a more cost-effective genotyping technology. A second goal may be to formally combine the more detailed information for the subgroup with the data from the main study on only the selected SNPs. As mentioned at the outset, this idea has been extensively developed in the literature on “two-phase sampling” in survey design and more recently in epidemiologic applications. Unlike the two-stage designs for GWAS described above, these designs typically entail using information that is readily available on the entire case-control study to select a stratified subsample. For the first goal of characterization only, a sample of only cases may be most efficient (see below). However, if one wishes to sample the subset for more detailed measurements and then combine the two datasets in a joint analysis, one may need to sample both cases and controls. Here, one needs to consider both the optimization of the informativeness of the subset for discovery as well as informativeness for the ultimate case-control analysis that takes the sampling fractions into account. In the present context, the relevant stratifying variables might include case/control status and the SNP genotypes (and possibly exposure variables if detected through a G×E interaction effect). Such an approach has been explored for candidate gene association studies (Thomas et al., 2004), where information on a dense panel of SNPs in a targeted subsample is combined with a sparser panel from the main study for the purpose of localizing the signal by LD mapping or for testing haplotype associations. Here, each region identified in a GWAS would likely target a different subsample of subjects, based on the available SNPs in that region.
A typical study might involve sequencing a sample of about 48 or 96 individuals over perhaps a 100 Kb region. Assuming that the region size has already been established based on the pattern of SNP or haplotype associations from the initial GWAS, knowledge of the LD structure of the region, and possibly additional fine-mapping, how then should this relatively small sample be selected to maximize the chances of discovering the real causal variant(s)?
Suppose first that a positive association has been found with a single SNP (Table 1, top). If not itself causal, this could theoretically reflect either a deleterious effect of another variant in positive LD with it or a protective effect of a variant in negative LD. Of the two possibilities, the former is much more likely, as negative LD with a protective minor allele is unlikely to generate a large positive association at a marker locus (see the second block in Table 1, where a perfectly protective allele in perfect negative LD yields a marker RR of only 1.067). The subjects with the highest yield of causal variants would then be cases carrying the minor allele of the associated SNP, with carrier controls somewhat lower but still much higher than either cases or controls carrying the major allele.
Table 1.
Illustrative calculation of probability of carrying a rare causal variant G among cases and controls carrying the major or minor allele at a marker locus M in LD with it: Pr(M)=0.2, Pr(G)=0.05. Bolded entries indicate the highest yield strata in each situation.
| Marker | Disease | Causal allele |
Pr(G=1|M,Y) | |
|---|---|---|---|---|
| M | Y | G=0 | G=1 | |
| Positive marker association | ||||
|
Positive LD and positive causal association δ=0.036, RRYG=2, RRYM=1.22 | ||||
| m | Controls | 0.796 | 0.004 | 0.005 |
| Cases | 0.758 | 0.008 | 0.010 | |
| M | Controls | 0.154 | 0.046 | 0.230 |
| Cases | 0.147 | 0.088 | 0.374 | |
|
Negative LD and negative causal association δ=−.010, RRYG=0, RRYM=1.067 | ||||
| m | Controls | 0.750 | 0.050 | 0.063 |
| Cases | 0.789 | 0.000 | 0.000 | |
| M | Controls | 0.200 | 0.000 | 0.000 |
| Cases | 0.211 | 0.000 | 0.000 | |
| Negative marker association | ||||
|
Negative LD and positive causal association δ=−0.010, RRYG=3, RRYM=0.889 | ||||
| m | Controls | 0.750 | 0.050 | 0.063 |
| Cases | 0.682 | 0.136 | 0.136 | |
| M | Controls | 0.200 | 0.000 | 0.000 |
| Cases | 0.186 | 0.000 | 0.000 | |
|
Positive LD and negative causal association δ=0.036, RRYG=0.5, RRYM=0.887 | ||||
| m | Controls | 0.796 | 0.004 | 0.005 |
| Cases | 0.816 | 0.002 | 0.003 | |
| M | Controls | 0.200 | 0.046 | 0.230 |
| Cases | 0.211 | 0.024 | 0.130 | |
Now suppose instead that the minor allele shows a negative association with disease. Again if not itself causal, this could reflect either a protective effect of a variant in positive LD with it or a deleterious effect of a variant in negative LD, these two scenarios now being roughly equally plausible (see bottom two blocks of Table 1, where both configurations yield similar marker RRs). In this case, the most informative subjects would be cases carrying the major allele or controls carrying the minor allele at the associated SNP, with the latter generally having a higher yield of causal variants.
To summarize the situation with a single marker, if one is purely interested in maximizing the chances of identifying a causal variant that will then be genotyped in the entire sample, then one could sample only carriers of the minor allele—cases if the marker association is positive, controls if it is negative. No weighting would be required for the analysis of the full study data for the discovered genotypes. If, on the other hand one wishes to perform a joint analysis of the main and substudy data incorporating the full sequence data on substudy subjects, then to be able to weight the analysis correctly, all four strata must be represented. The optimal sampling fractions would depend upon knowledge of the true LD and causal association parameters, but one could be guided by the general calculations illustrated in Table 1: if the association with the minor allele is positive, then sample the largest number of cases with the minor allele, then controls with the minor allele, and the smallest number of carriers of the major allele; if the association is negative, then sample most heavily controls with the minor allele, then equal numbers of cases with the minor allele and controls with the major allele, and the smallest number of controls with the minor allele.
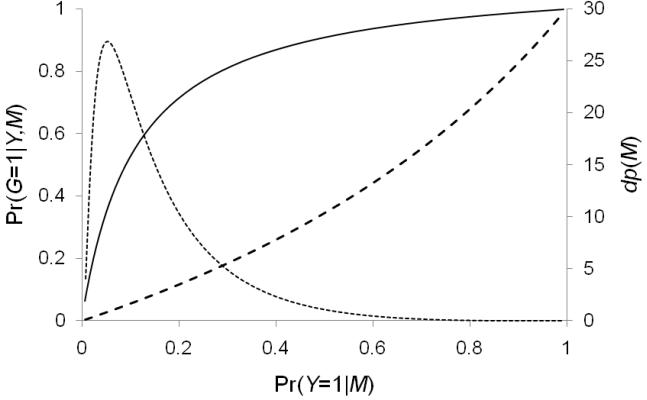
Now, suppose the association is not just with a single SNP in a region but with several. A sensible sampling design might now entail first constructing a risk index, say by logistic regression of case-control status on multiple SNPs or haplotypes and then stratifying jointly on this genetic risk index and case-control status. The concept of positive or negative association and LD is now moot and needs to be replaced by consideration of the shape of the distribution of the risk index (Figure 1). Typically, one might find a relatively small proportion of subjects with a broad range of high risk scores and a large proportion with generally low risk (Thomas et al., 2008). In this situation, it would be cases with high risk scores that are likely to be the most informative, although controls with high risk scores would also have an increased probability of carrying a causal allele. In the event that the risk distribution has a long tail of low risk, it could be worthwhile to sample controls with low risk scores, but detecting effects of rare beneficial alleles would require enormous sample sizes. (Of course, a beneficial effect of a common allele is equivalent to a deleterious effect of a rare allele.) As in the single-marker case, if the purpose is simply to discover potentially causal variants, then one could restrict the sample to high-risk cases, but if a joint analysis is planned, then a well-defined sampling scheme is required that assigns non-zero sampling probability to every individual. This could be accomplished by stratifying jointly on Y and Pr(Y=1|M) or in proportion to an estimated Pr(G=1|Y,M) for some hypothesized model.
Figure 1.
Illustration of a hypothetical distribution of an empirical risk index (M, right axis, dotted line) and yield of causal variant carriers (D=1, left axis, solid and dashed lines for cases and controls respectively) as a function of M
It must be appreciated that only very strong associations would have any power for testing associations in the subsample alone. The real purpose is simply to identify novel variants that would then be genotyped in the main study. Having completed the sequencing of this stratified sample and genotyping of selected novel variants in the main study, a joint analysis could be performed as described by Thomas et al. (2004) to test for associations with all variants discovered in the re-sequencing sample, not just those actually genotyped in the main study. This essentially involves imputation of the missing data on main study subjects using the substudy data, but requires appropriate adjustment for the sampling fractions if they depend jointly on genotypes and disease, so that that all cells of the stratification must be represented in the sample. For substudy subjects, the standard logistic model can be used by adding as an offset term the log of the ratio of genotype-specific case/control sampling probabilities. For main study subjects, the likelihood contribution becomes a more complex mixture of weighted logistic probabilities, although well approximated by a logistic function if the disease is rare. Of course, imputation of very rare variants in the main study by leveraging a substudy with only a few occurrences of such variants is of dubious value, so all potentially causal variants should be genotyped in the full sample, but imputation could be useful for exploiting LD patterns in the sequence data that might suggest regions worth closer study.
These considerations are likely to be fundamentally altered in the near future by the public availability of re-sequencing data from the “1000 Genomes Project” (http://www.1000genomes.org/), aimed at identifying variants at a frequency of 1% across the genome in approximately 1500 subjects (500 Bantu-speaking, 500 Asian, 500 Caucasian). Data at an intermediate level of detail (e.g., deep re-sequencing of 1000 genes in 1000 individuals) will be released soon. Once completed the 1000 Genomes Project will potentially reduce the need for extensive deep re-sequencing for variants in the 1–5% range, at least for studies conducted in comparable populations, but would still leave open the question of rarer variants. Methods to leverage the 1000 Genomes data for imputation purposes or joint analysis with a two-phase sampling design, allowing for possible misspecification of the G-M relationships for the specific study's target population, would be useful.
The advent of whole genome sequence using next-generation sequencing platforms (Mardis, 2008) may also resurrect interest in multistage designs, as genome-wide scans for rare variants are unlikely to be feasible on the tens of thousands of subjects that will be needed, at least until the $1000 genome becomes a reality. Whether this will ever be feasible, given the much larger multiple-testing burden, the sparseness of data on any particular variant, and the likelihood that rare variants will be less effectively tagged than common variants, remains to be seen. Nevertheless, these technologies will undoubtedly aid in following-up larger and larger regions surrounding SNPs identified in a GWAS on larger and larger samples, requiring adjustment for a much larger universe of variants, rare and common (Hoggart et al., 2008a; 2008b).
4.4. Investigating Biological Function, eQTLs, and Pathways
Once a set of highly significant and replicated SNP associations has been found, what then? The challenges posed by the study of the often broadly diverse biologic functions of hits arising from GWASs should not be underestimated. Trying to determine the functionality of even a single GWAS hit can be a daunting task. While clearly, in vitro and in vivo experimental studies would be appropriate to investigate function, the initial steps in characterization depend upon whether hits are located within genes, near known genes, or in “gene deserts.” If the hit is within a gene, various software packages and web sites could be used to assess the potential functional role of the variant, and such in silico findings could then be confirmed in vitro by molecular approaches such as quantitative RT-PCR. If the hit is near a gene, it could implicate an adjacent gene, but it could also lie in an unannotated gene, an miRNA, or an enhancer or repressor element for some gene located far away (the most likely explanation if it is in a gene desert). Unannotated genes might be identified through tiling gene expression arrays, while in silico and ChIP-chip methods might be used to identify enhancer/repressor elements.
Other types of analyses also might be undertaken that would involve more sophisticated analyses of the GWAS data, either (1) in an attempt to infer causal pathways from the pattern of associations and interactions using the kinds of network analysis tools that have been applied to gene expression and protein interaction data, or (2) to inform the search for effects in the GWAS data by incorporating external knowledge from pathway or genomic databases, literature mining, or analysis of gene expression, proteomic, metabolomic, or other –omics data, perhaps using hierarchical modeling or gene set enrichment analysis methods. See (Chasman, 2008; Gieger et al., 2008; Pan, 2005; Wang et al., 2007) for discussion of some of these approaches.
A second phase of these studies could be to examine any known biological functionality of the gene in question, and once again those applied approaches will depend upon several considerations such as the likely consequence of the SNP itself upon gene function as well as prior knowledge of the gene and understanding of its involvement, if known, in cellular pathway(s). Gene expression data (even genome-wide data) might be leveraged to identify candidate genes/pathways. A SNP that lies within the coding region of a gene may be more likely to affect the normal function of that gene either through enhancing its effect or reducing its functionality. Biochemical assays may be available that could be applied to test the effect of a coding region variant on its known gene function, such as a role in apoptosis. Alternatively where a hit lies adjacent to a gene or within a gene desert and possibly in an enhancer element or other regulatory region, the likely effect may be on the expression level of the gene—whether leading to higher or lower gene expression levels, mRNA stability, or post-translational protein levels in target tissues. The next steps of characterization would require using knowledge of that gene and related pathways to develop assays that would test the putative consequences of either elevated or reduced expression of the gene product in appropriate cells. For some genes, accumulated knowledge of its role in the cell may be extensive; however, for others that knowledge may be sparse or even non-existent. Such prior knowledge may be used to help prioritize functional biological studies. However, they could have the effect of steering us away from further characterization of potentially interesting genes that have little prior biological knowledge due to the greater challenges that they pose.
Given the potential complexity and diversity of methods that will need to be applied to follow up on any identified hit, a prioritization scheme will need to be developed that will likely involve many different considerations, such as the strength of the hit itself, whether the hit has any implications for disease subsets such as more aggressive forms of cancer, whether the hit is potentially implicated in more than one disease (such as appears to be the case for the 8q24 region, which is related to at least 4 cancers, and for several diabetes risk alleles that are found to be protective for prostate cancer) and prior biological knowledge. The hierarchical modeling approaches discussed in §2.4 may be helpful for combining the evidence from the data at hand and these external sources of knowledge to prioritize hits for follow-up functional studies. None of these kinds of studies would be likely to involve the original epidemiological study subjects, however, and further detailed investigations are likely to be gene-specific, so are beyond the scope of this article.
Two-phase sampling designs may be particularly helpful for biomarker or expression measurements to inform the analysis of pathways. Such analyses may take the form of a network of latent variables, for which the biomarkers are viewed as surrogate measurements (Thomas, 2007). In such designs, one might wish to subsample jointly on some combination of disease, exposure, and genotype(s) to select individuals for biomarker measurements. For example, in a pharmacogenetic study, one might subsample on the basis of outcomes and treatment assignment to target a GWAS or a re-sequencing study of a candidate region; or if GWAS data were already available, one might stratify by a multi-marker risk score, treatment, and outcomes for collection of biomarkers to investigate pathways (Thomas and Conti, 2007). Any study of biomarkers collected after the outcome must, however, address the problem of “reverse causation,” whereby the variable being measured (or the accuracy of its measurement) is affected by the disease or its treatment rather than the other way around.
Finally it is worth noting that an enhancement in our knowledge of the etiology of disease may have implications that transcend the merely predictive power of a specific variant. The relatively modest relative risks that have been discovered by GWASs for disease etiology could be due in part to selection against high risk variants, but this is unlikely for response to modern pharmacologic agents. For example, SNPs in HMGCR have only a small effect on low density lipoprotein levels but drugs targeting the protein encoded by HMGCR have a much larger effect (Altshuler et al., 2008). One can at least hope that solving the mystery of how variants in a gene desert such as 8q24 appear to influence the risk of a multitude of cancers would lead to methods aimed at preventing or treating the resulting diseases. Ultimately the discovery of genetic modifiers of treatment response is central to the goal of personalized medicine.
ACKNOWLEDGMENTS
Supported in part by NIH grant 5U01 ES015090. The authors are grateful to Jim Gauderman for many discussions of these issues and helpful comments on the manuscript.
REFERENCES
- ALBERT PS, RATNASINGHE D, TANGREA J, WACHOLDER S. Limitations of the case-only design for identifying gene-environment interactions. Am J Epidemiol. 2001;154:687–693. doi: 10.1093/aje/154.8.687. [DOI] [PubMed] [Google Scholar]
- ALTSHULER D, DALY MJ, LANDER ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANDERSON CA, PETTERSSON FH, BARRETT JC, ZHUANG JJ, RAGOUSSIS J, CARDON LR, et al. Evaluating the effects of imputation on the power, coverage, and cost efficiency of genome-wide SNP platforms. Am J Hum Genet. 2008;83:112–119. doi: 10.1016/j.ajhg.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANONYMOUS Freely associating. Nat Genet. 1999;22:1–2. doi: 10.1038/8702. [DOI] [PubMed] [Google Scholar]
- BANSAL A, VAN DEN BOOM D, KAMMERER S, HONISCH C, ADAM G, CANTOR CR, et al. Association testing by DNA pooling: an effective initial screen. Proc Natl Acad Sci. 2002;99:16871–16874. doi: 10.1073/pnas.262671399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARRATT BJ, PAYNE F, RANCE HE, NUTLAND S, TODD JA, CLAYTON DG. Identification of the sources of error in allele frequency estimations from pooled DNA indicates an optimal experimental design. Ann Hum Genet. 2002;66:393–405. doi: 10.1017/S0003480002001252. [DOI] [PubMed] [Google Scholar]
- BARRETT JC, CARDON LR. Evaluating coverage of genome-wide association studies. Nat Genet. 2006;38:659–662. doi: 10.1038/ng1801. [DOI] [PubMed] [Google Scholar]
- BRESLOW NE, CHATTERJEE N. Design and analysis of two-phase studies with binary outcome applied to Wilms tumor prognosis. JRSS C. 1999;48:457–468. [Google Scholar]
- CHANOCK SJ, MANOLIO T, BOEHNKE M, BOERWINKLE E, HUNTER DJ, THOMAS G, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- CHASMAN DI. On the utility of gene set methods in genomewide association studies of quantitative traits. Genet Epidemiol. 2008;32:658–668. doi: 10.1002/gepi.20334. [DOI] [PubMed] [Google Scholar]
- CHATTERJEE N, CARROLL RJ. Semiparametric maximum likelihood estimation exploiting gene-environment independence in case-control studies. Biometrika. 2005;92:399–418. [Google Scholar]
- CHATTERJEE N, KALAYLIOGLU Z, CARROLL RJ. Exploiting gene-environment independence in family-based case-control studies: increased power for detecting associations, interactions and joint effects. Genet Epidemiol. 2005;28:138–156. doi: 10.1002/gepi.20049. [DOI] [PubMed] [Google Scholar]
- CHATTERJEE N. Case-control design and haplotype analysis. Statist Sci. 2009 in press. [Google Scholar]
- CHEN GK, WITTE JS. Enriching the analysis of genomewide association studies with hierarchical modeling. Am J Hum Genet. 2007;81:397–404. doi: 10.1086/519794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHENG KF. A maximum likelihood method for studying gene-environment interactions under conditional independence of genotype and exposure. Stat Med. 2006;25:3093–3109. doi: 10.1002/sim.2506. [DOI] [PubMed] [Google Scholar]
- CLARKE GM, CARTER KW, PALMER LJ, MORRIS AP, CARDON LR. Fine mapping versus replication in whole-genome association studies. Am J Hum Genet. 2007;81:995–1005. doi: 10.1086/521952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRAIG DW, HUENTELMAN MJ, HU-LINCE D, ZISMANN VL, KRUER MC, LEE AM, et al. Identification of disease causing loci using an array-based genotyping approach on pooled DNA. BMC Genomics. 2005;6:138. doi: 10.1186/1471-2164-6-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRAIG DW, PEARSON JV, SZELINGER S, SEKAR A, REDMAN M, CORNEVEAUX JJ, et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods. 2008;5:887–893. doi: 10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE BAKKER PI, YELENSKY R, PE'ER I, GABRIEL SB, DALY MJ, ALTSHULER D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- DOCHERTY SJ, BUTCHER LM, SCHALKWYK LC, PLOMIN R. Applicability of DNA pools on 500 K SNP microarrays for cost-effective initial screens in genomewide association studies. BMC Genomics. 2007;8:214. doi: 10.1186/1471-2164-8-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUDBRIDGE F. A note on permutation tests in multistage association scans. Am J Hum Genet. 2006;78:1094–1095. doi: 10.1086/504527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EBERLE MA, NG PC, KUHN K, ZHOU L, PEIFFER DA, GALVER L, et al. Power to detect risk alleles using genome-wide tag SNP panels. PLoS Genet. 2007;3:1827–1837. doi: 10.1371/journal.pgen.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FEARNHEAD NS, WILDING JL, WINNEY B, TONKS S, BARTLETT S, BICKNELL DC, et al. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc Natl Acad Sci. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FENG Z, PRENTICE R, SRIVASTAVA S. Research issues and strategies for genomic and proteomic biomarker discovery and validation: a statistical perspective. Pharmacogenomics. 2004;5:709–719. doi: 10.1517/14622416.5.6.709. [DOI] [PubMed] [Google Scholar]
- GAIL MH, PFEIFFER RM, WHEELER W, PEE D. Probability of detecting disease-associated single nucleotide polymorphisms in case-control genome-wide association studies. Biostatistics. 2007 doi: 10.1093/biostatistics/kxm032. [DOI] [PubMed] [Google Scholar]
- GAUDERMAN WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med. 2002;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- GIEGER C, GEISTLINGER L, ALTMAIER E, HRABE DE ANGELIS M, KRONENBERG F, MEITINGER T, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUEDJ M, ROBELIN D, HOEBEKE M, LAMARINE M, WOJCIK J, NUEL G. Detecting local high-scoring segments: a first-stage approach for genome-wide association studies. Stat Appl Genet Mol Biol. 2006;5 doi: 10.2202/1544-6115.1192. Article22. [DOI] [PubMed] [Google Scholar]
- HAN B, KANG HM, ESKIN E. Rapid and accurate multiple testing correction and power estimation for millions of correlated markers. PLoS Genet. 2009;5:e1000456. doi: 10.1371/journal.pgen.1000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAO K, SCHADT EE, STOREY JD. Calibrating the performance of SNP arrays for whole-genome association studies. PLoS Genet. 2008;4:e1000109. doi: 10.1371/journal.pgen.1000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIRSCHHORN JN, DALY MJ. Genome-wide association studies for common disease and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- HOGGART CJ, CLARK TG, DE IORIO M, WHITTAKER JC, BALDING DJ. Genome-wide significance for dense SNP and resequencing data. Genet Epidemiol. 2008a;32:179–185. doi: 10.1002/gepi.20292. [DOI] [PubMed] [Google Scholar]
- HOGGART CJ, WHITTAKER JC, DE IORIO M, BALDING DJ. Simultaneous analysis of all SNPs in genome-wide and re-sequencing association studies. PLoS Genet. 2008b;4:e1000130. doi: 10.1371/journal.pgen.1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOPPER JL, SOUTHEY MC, DITE GS, JOLLEY DJ, GILES GG, MCCREDIE MRE, et al. Population-based estimate of the average age-specific cumulative risk of breast cancer for a defined set of protein-truncating mucations in BRCA1 and BRCA2. Cancer Epidemiol Biomark Prev. 1999;8:741–747. [PubMed] [Google Scholar]
- HUNTER DJ, KRAFT P. Drinking from the fire hose--statistical issues in genomewide association studies. N Engl J Med. 2007;357:436–439. doi: 10.1056/NEJMp078120. [DOI] [PubMed] [Google Scholar]
- HUNTER DJ, THOMAS G, HOOVER RN, CHANOCK SJ. Scanning the horizon: What is the future of genome-wide association studies in accelerating discoveries in cancer etiology and prevention? Cancer Causes Control. 2007;18:479–484. doi: 10.1007/s10552-007-0118-y. [DOI] [PubMed] [Google Scholar]
- IOANNIDIS J. Replication issues. Statist Sci. 2009 in press. [Google Scholar]
- IOANNIDIS JP. Non-replication and inconsistency in the genome-wide association setting. Hum Hered. 2007;64:203–213. doi: 10.1159/000103512. [DOI] [PubMed] [Google Scholar]
- IYENGAR SK, ELSTON RC. The genetic basis of complex traits: rare variants or “common gene, common disease”? Methods Mol Biol. 2007;376:71–84. doi: 10.1007/978-1-59745-389-9_6. [DOI] [PubMed] [Google Scholar]
- JENNISON C, TURNBULL BW. Group Sequential Methods with Applications to Clinical Trials. xviii. Chapman & Hall / CRC; Boca Raton: 2000. [Google Scholar]
- JOHNSON T. Bayesian method for gene detection and mapping, using a case and control design and DNA pooling. Biostatistics. 2007;8:546–565. doi: 10.1093/biostatistics/kxl028. [DOI] [PubMed] [Google Scholar]
- JORGENSON E, WITTE JS. Coverage and power in genomewide association studies. Am J Hum Genet. 2006;78:884–888. doi: 10.1086/503751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIROV G, ZAHARIEVA I, GEORGIEVA L, MOSKVINA V, NIKOLOV I, CICHON S, et al. A genome-wide association study in 574 schizophrenia trios using DNA pooling. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.33. [DOI] [PubMed] [Google Scholar]
- KOOPERBERG C. Using marginals to find interactions and interactions to find marginals in GWA analyses. Statist Sci. 2009 in press. [Google Scholar]
- KRAFT P. Efficient two-stage genome-wide association designs based on false positive report probabilities. Pac Symp Biocomputing. 2006;11:523–534. [PubMed] [Google Scholar]
- KRAFT P, CHANOCK C, HUNTER D, CHATTERJEE N, THOMAS G. Cost-efficient multi-stage designs for genome-wide association studies. In: Rao DC, editor. Genetic Dissection of Complex Traits. 2nd Ed. Elsevier; New York: 2007a. in press. [Google Scholar]
- KRAFT P, YEN YC, STRAM DO, MORRISON J, GAUDERMAN WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007b;63:111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- KRAFT P. Curses--winner's and otherwise--in genetic epidemiology. Epidemiology. 2008;19:649–651. doi: 10.1097/EDE.0b013e318181b865. discussion 657-648. [DOI] [PubMed] [Google Scholar]
- KRAFT P, COX DG. Study designs for genome-wide association studies. Adv Genet. 2008;60:465–504. doi: 10.1016/S0065-2660(07)00417-8. [DOI] [PubMed] [Google Scholar]
- KRYUKOV GV, PENNACCHIO LA, SUNYAEV SR. Most rare missense alleles are deleterious in humans: Implications for complex disease and association studies. Am J Hum Genet. 2007;80:727–739. doi: 10.1086/513473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWINGER JP, CONTI DV, BAURLEY JW, TRICHE TJ, THOMAS DC. Hierarchical Bayes prioritization of marker associations from a genome-wide association scan for further investigation. Genet Epidemiol. 2007a;31:871–882. doi: 10.1002/gepi.20248. [DOI] [PubMed] [Google Scholar]
- LEWINGER JP, DUGGAN DJ, TAVERNA DM, J GW, STRAM DO, THOMAS DC. Choosing a platform and design for genomewide association studies: cost, sample size, and power trade-offs; Paper presented at: American Society of Human Genetics; San Diego, CA. 2007b. [Google Scholar]
- LI B, LEAL SM. Methods for Detecting Associations with Rare Variants for Common Diseases: Application to Analysis of Sequence Data. Am J Hum Genet. 2008 doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI D, CONTI DV. Detecting interactions using a combined case-only and case-control approach. Am J Epidemiol. 2008 doi: 10.1093/aje/kwn339. in presss. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN DY. Evaluating statistical significance in two-stage genomewide association studies. Am J Hum Genet. 2006;78:505–509. doi: 10.1086/500812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACGREGOR S. Most pooling variation in array-based DNA pooling is attributable to array error rather than pool construction error. Eur J Hum Genet. 2007;15:501–504. doi: 10.1038/sj.ejhg.5201768. [DOI] [PubMed] [Google Scholar]
- MARCHINI J. Imputation. Statist Sci. 2009 in press. [Google Scholar]
- MARDIS ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- MEABURN E, BUTCHER LM, SCHALKWYK LC, PLOMIN R. Genotyping pooled DNA using 100K SNP microarrays: a step towards genomewide association scans. Nucleic Acids Res. 2006;34:e27. doi: 10.1093/nar/gnj027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUKHERJEE B, ZHANG L, GHOSH M, SINHA S. Semiparametric Bayesian analysis of case-control data under conditional gene-environment independence. Biometrics. 2007;63:834–844. doi: 10.1111/j.1541-0420.2007.00750.x. [DOI] [PubMed] [Google Scholar]
- MUKHERJEE B, AHN J, GRUBER SB, RENNERT G, MORENO V, CHATTERJEE N. Tests for gene-environment interaction from case-control data: a novel study of type I error, power and designs. Genet Epidemiol. 2008;32:615–626. doi: 10.1002/gepi.20337. [DOI] [PubMed] [Google Scholar]
- MUKHERJEE B, CHATTERJEE N. Exploiting gene-environment independence for analysis of case-control studies: an empirical Bayes approach to trade off between bias and efficiency. Biometrics. 2008 doi: 10.1111/j.1541-0420.2007.00953.x. in press. [DOI] [PubMed] [Google Scholar]
- MULLER HH, PAHL R, SCHAFER H. Including sampling and phenotyping costs into the optimization of two stage designs for genomewide association studies. Genet Epidemiol. 2007;31:844–852. doi: 10.1002/gepi.20245. [DOI] [PubMed] [Google Scholar]
- MURCRAY C, LEWINGER JP, GAUDERMAN WJ. Gene-environment interaction in genome-wide association studies. Am J Epidemiol. 2008 doi: 10.1093/aje/kwn353. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NANNYA Y, TAURA K, KUROKAWA M, CHIBA S, OGAWA S. Evaluation of genome-wide power of genetic association studies based on empirical data from the HapMap project. Hum Mol Genet. 2007;16:3494–3505. doi: 10.1093/hmg/ddm205. [DOI] [PubMed] [Google Scholar]
- PAN W. Incorporating biological information as a prior in an empirical Bayes approach to analyzing microarray data. Statist Appl Genet Molec Biol. 2005;4 doi: 10.2202/1544-6115.1124. Art. 12. [DOI] [PubMed] [Google Scholar]
- PE'ER I, DE BAKKER PI, MALLER J, YELENSKY R, ALTSHULER D, DALY MJ. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat Genet. 2006;38:663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- PE'ER I, YELENSKY R, ALTSHULER D, DALY MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- PEARSON JV, HUENTELMAN MJ, HALPERIN RF, TEMBE WD, MELQUIST S, HOMER N, et al. Identification of the genetic basis for complex disorders by use of pooling-based genomewide single-nucleotide-polymorphism association studies. Am J Hum Genet. 2007;80:126–139. doi: 10.1086/510686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PFEIFFER RM, RUTTER JL, GAIL MH, STRUEWING J, GASTWIRTH JL. Efficiency of DNA pooling to estimate joint allele frequencies and measure linkage disequilibrium. Genet Epidemiol. 2002;22:94–102. doi: 10.1002/gepi.1046. [DOI] [PubMed] [Google Scholar]
- PFEIFFER RM, GAIL MH, PEE D. On combining data from genome-wide association studies to discover disease-associated SNPs. Statist Sci. 2009 in press. [Google Scholar]
- PIEGORSCH W, WEINBERG C, TAYLOR J. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13:153–162. doi: 10.1002/sim.4780130206. [DOI] [PubMed] [Google Scholar]
- PRITCHARD JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REBBECK TR, MARTINEZ ME, SELLERS TA, SHIELDS PG, WILD CP, POTTER JD. Genetic variation and cancer: Improving the environment for publication of association studies. Cancer Epidemiol Biomark Prev. 2004;13:1985–1986. [PubMed] [Google Scholar]
- RISCH N, TENG J. The relative power of family-based and case-control designs for linkage disequilibrium studies of compex human diseases, I. DNA pooling. Genome Res. 1998;8:1273–1288. doi: 10.1101/gr.8.12.1273. [DOI] [PubMed] [Google Scholar]
- ROEDER K, BACANU SA, WASSERMAN L, DEVLIN B. Using linkage genome scans to improve power of association in genome scans. Am J Hum Genet. 2006;78:243–252. doi: 10.1086/500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROEDER K, DEVLIN B, WASSERMAN L. Improving power in genome-wide association studies: weights tip the scale. Genet Epidemiol. 2007;31:741–747. doi: 10.1002/gepi.20237. [DOI] [PubMed] [Google Scholar]
- SAITO A, KAMATANI N. Strategies for genome-wide association studies: optimization of study designs by the stepwise focusing method. J Hum Genet. 2002;47:360–365. doi: 10.1007/s100380200050. [DOI] [PubMed] [Google Scholar]
- SAMANI NJ, ERDMANN J, HALL AS, HENGSTENBERG C, MANGINO M, MAYER B, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATAGOPAN JM, VERBEL DA, VENKATRAMAN ES, OFFIT KE, BEGG CB. Two-stage designs for gene-disease association studies. Biometrics. 2002;58:163–170. doi: 10.1111/j.0006-341x.2002.00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATAGOPAN JM, ELSTON RC. Optimal two-stage genotyping in population-based association studies. Genet Epidemiol. 2003;25:149–157. doi: 10.1002/gepi.10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATAGOPAN JM, VENKATRAMAN ES, BEGG CB. Two-stage designs for gene-disease association studies with sample size constraints. Biometrics. 2004;60:589–597. doi: 10.1111/j.0006-341X.2004.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEBASTIANI P, ZHAO Z, ABAD-GRAU MM, RIVA A, HARTLEY SW, SEDGEWICK AE, et al. A hierarchical and modular approach to the discovery of robust associations in genome-wide association studies from pooled DNA samples. BMC Genet. 2008;9:6. doi: 10.1186/1471-2156-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SERVICE SK, SANDKUIJL LA, FREIMER NB. Cost-effective designs for linkage disequilibrium mapping of complex traits. Am J Hum Genet. 2003;72:1213–1220. doi: 10.1086/375165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHAM P, BADER JS, CRAIG I, O'DONOVAN M, OWEN M. DNA Pooling: a tool for large-scale association studies. Nat Rev Genet. 2002;3:862–871. doi: 10.1038/nrg930. [DOI] [PubMed] [Google Scholar]
- SKOL AD, SCOTT LJ, ABECASIS GR, BOEHNKE M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- SKOL AD, SCOTT LJ, ABECASIS GR, BOEHNKE M. Optimal designs for two-stage genome-wide association studies. Genet Epidemiol. 2007;31:776–788. doi: 10.1002/gepi.20240. [DOI] [PubMed] [Google Scholar]
- SPINOLA M, LEONI VP, GALVAN A, KORSCHING E, CONTI B, PASTORINO U, et al. Genome-wide single nucleotide polymorphism analysis of lung cancer risk detects the KLF6 gene. Cancer Lett. 2007;251:311–316. doi: 10.1016/j.canlet.2006.11.029. [DOI] [PubMed] [Google Scholar]
- STEER S, ABKEVICH V, GUTIN A, CORDELL HJ, GENDALL KL, MERRIMAN ME, et al. Genomic DNA pooling for whole-genome association scans in complex disease: empirical demonstration of efficacy in rheumatoid arthritis. Genes Immun. 2007;8:57–68. doi: 10.1038/sj.gene.6364359. [DOI] [PubMed] [Google Scholar]
- THOMAS D, XIE R, GEBREGZIABHER M. Two-stage sampling designs for gene association studies. Genet Epidemiol. 2004;27:401–414. doi: 10.1002/gepi.20047. [DOI] [PubMed] [Google Scholar]
- THOMAS DC, SIEMIATYCKI J, DEWAR R, ROBINS J, GOLDBERG M, ARMSTRONG BG. The problem of multiple inference in studies designed to generate hypotheses. Am J Epidemiol. 1985;122:1080–1095. doi: 10.1093/oxfordjournals.aje.a114189. [DOI] [PubMed] [Google Scholar]
- THOMAS DC. Multistage sampling for latent variable models. Lifetime Data Anal. 2007;13:565–581. doi: 10.1007/s10985-007-9061-1. [DOI] [PubMed] [Google Scholar]
- THOMAS DC, CONTI DV. Two stage genetic association studies. In: Elston RC, editor. Encycolpedia of Clinical Trials. Wiley; New York: 2007. [Google Scholar]
- THOMAS G, JACOBS KB, YEAGER M, KRAFT P, WACHOLDER S, ORR N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- VAN STEEN K, MCQUEEN MB, HERBERT A, RABY B, LYON H, DEMEO DL, et al. Genomic screening and replication using the same data set in family-based association testing. 2005;37:683–691. doi: 10.1038/ng1582. [DOI] [PubMed] [Google Scholar]
- VISSCHER PM. Statistical aspects for making predictions from GWAS using multiple markers. Statist Sci. 2009 in press. [Google Scholar]
- WAKEFIELD J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAKEFIELD J. Reporting and interpretation in genome-wide association studies. Int J Epidemiol. 2008;37:641–653. doi: 10.1093/ije/dym257. [DOI] [PubMed] [Google Scholar]
- WANG H, THOMAS DC, PE'ER I, STRAM DO. Optimal two-stage genotyping designs for genome-wide association scans. Genet Epidemiol. 2006;30:356–368. doi: 10.1002/gepi.20150. [DOI] [PubMed] [Google Scholar]
- WANG K, LI M, BUCAN M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHITE JE. A two stage design for the study of the relationship between a rare exposure and a rare disease. Am J Epidemiol. 1982;115:119–128. doi: 10.1093/oxfordjournals.aje.a113266. [DOI] [PubMed] [Google Scholar]
- WHITTEMORE AS. A Bayesian false discovery rate for multiple testing. J Appl Statist. 2007;34:1–9. [Google Scholar]
- YU K, CHATTERJEE N, WHEELER W, LI Q, WANG S, ROTHMAN N, et al. Flexible design for following up positive findings. Am J Hum Genet. 2007;81:540–551. doi: 10.1086/520678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZAYKIN DV, ZHIVOTOVSKY LA. Ranks of genuine associations in whole-genome scans. Genetics. 2005;171:813–823. doi: 10.1534/genetics.105.044206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHONG H, PRENTICE RL. Bias-reduced estimators and confidence intervals for odds ratios in genome-wide association studies. Biostatistics. 2008;9:621–634. doi: 10.1093/biostatistics/kxn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZOLLNER S, PRITCHARD JK. Overcoming the winner's curse: estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80:605–615. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZOLLNER S. Copy number variation. Statist Sci. 2009 in press. [Google Scholar]
- ZOU G, ZHAO H. The impacts of errors in individual genotyping and DNA pooling on association studies. Genet Epidemiol. 2004;26:1–10. doi: 10.1002/gepi.10277. [DOI] [PubMed] [Google Scholar]
- ZUO Y, ZOU G, ZHAO H. Two-stage designs in case-control association analysis. Genetics. 2006;173:1747–1760. doi: 10.1534/genetics.105.042648. [DOI] [PMC free article] [PubMed] [Google Scholar]