Abstract
N-glycosylation of the I-like domain of β1 integrin plays an essential role in integrin structure and function, and the altered sialylation of β1 integrin regulates β1 integrin binding to fibronectin. However, the structural basis underlying the effect of altered sialylation of the β1 I-like domain on β1 integrin binding to fibronectin remains largely unknown. In this study, we used a combination of molecular dynamics simulations and binding free energy analyses to investigate changes in binding thermodynamics and in conformation of the glycosylated β1 I-like domain-FN-III9-10 complex caused by altered sialylation of the β1 I-like domain. Binding free energy analyses showed that desialylation of β1 I-like domain increased β1 integrin binding to fibronectin, consistent with experimental results. Interaction analyses showed that altered sialylation of the β1 I-like domain resulted in significant changes in the interaction of the N-glycans of the I-like domain with both the I-like domain and fibronectin, and these changes could directly affect the allosteric regulation of the interaction between the I-like domain and fibronectin. Altered sialylation of the β1 I-like domain caused significant conformational changes in key functional sites of both the β1 I-like domain and fibronectin. In addition, altered sialylation of the β1 I-like domain resulted in changes in the degree of correlated motions between residues in the I-like domain and residues in fibronectin, and in the degree of motion changes in fibronectin, which could affect β1 integrin binding to fibronectin. We believe results from this study provide thermodynamic and structural evidence for a role of altered sialylation of β1 integrin in regulating β1 integrin binding to fibronectin and it's induced cellular activities.
Introduction
Cell adhesion, regulated by interactions between the cell and the extracellular matrix (ECM), plays an important role in cellular functions such as cell growth, migration, differentiation, and proliferation (1). Studies have shown that interactions between the transmembrane glycoprotein α5β1 integrin and the extracellular matrix protein fibronectin are fundamental for vertebrate development and have been suggested to be involved in cardiovascular events and tumor invasion (2–6). N-glycosylation of α5β1 integrin is required for αβ heterodimer formation and for the interaction between α5β1 integrin and fibronectin (7–12). Desialylation of α5β1 integrin leads to increased adhesion to fibronectin, affecting cell adhesion and differentiation in myeloid cells (11,13). The I-like domain of β integrin, a region crucial for ligand binding (14), carries N-glycans attached to Asn-192, Asn-249, and Asn-343 (13). A recent study has shown the essential role of N-glycosylation of the I-like domain of β1 integrin for α5β1 integrin expression, heterodimer formation, and fibronectin-mediated biological functions, including cell adhesion and cell spreading (7). The I-like domain of β1 integrin contains the critical functional sites required for ligand binding, including the specificity-determining loop (SDL) (15), the metal ion-dependent adhesion site containing a DLSYS motif (16,17), the α1 and α7 helices (18), and other critical residues (Asn-224, Asp-226, Glu-229, Asp-233, Asp-267, and Asp-295) (19). Our recent molecular dynamics (MD) study regarding the effects of altered sialylation on the structure of the I-like domain of β1 integrin shows that altered sialylation of the β1 I-like domain changes the interactions between glycans and the I-like domain, affects accessibility of the SDL region for ligand binding, and results in significant conformational changes in most of the key functional regions of the β1 I-like domain involved in ligand binding (20).
The extracellular matrix protein fibronectin (FN) is a ligand for a large number of different integrins, including α5β1 integrin (21,22). For the integrin-FN complex, fibronectin not only provides a substrate for cell anchorage to the ECM, but also serves as a regulatory factor in many cellular processes including cell adhesion, growth, migration, differentiation, and proliferation (1). Structurally, fibronectin is a multimodular extracellular protein composed of >20 modules per monomer. The structure of fibronectin is rod-like, and is composed of three different types of homologous, repeating modules, type I (FN-I), type II (FN-II), and type III (FN-III) (21). FN-III module 10 (FN-III10) contains the RGD (Arg-Gly-Asp) sequence required for specific recognition by integrins (21). In addition to the RGD sequence, fibronectin also contains a synergy site that selectively enhances binding of certain integrins to the neighboring RGD loop (23,24). Some specific integrins, including α5β1, can recognize both the synergy site and RGD-loop of FN. One apparent function of the synergy site is to allow integrin α5β1 to bind to FN preferentially over other RGD-containing matrix proteins (25–27). Previous studies have shown that the binding of α5β1 to FN is affected by altered N-glycosylation of β1 integrin (7,11,13). The structure of the FN-III repeats, including the seventh through the synergy region-containing ninth and the RGD-containing tenth repeats, has been crystallized successfully (28), providing a basis for structural studies of fibronectin interactions with glycosylated α5β1integrin.
The molecular and structural basis for the effect of the altered sialylation of the β1 I-like domain on α5β1 integrin binding to fibronectin remains unclear. In this study, we used a combination of explicit solvent MD simulations and implicit solvent binding free energy analyses to investigate changes in binding thermodynamics, conformation and motion of the glycosylated β1 I-like domain-FN-III9-10 complex caused by altered sialylation of β1 I-like domain. Results from this study provide thermodynamic and structural evidence for the role of altered sialylation of β1 integrin in regulating the interaction of β1 integrin and fibronectin and its induced cellular activities.
Results and Discussion
Materials and methods for model of the glycosylated βI-like domain-fibronectin complex, molecular dynamics simulations, binding free energy analyses, interaction analyses, conformational and motion analyses, and statistical methods are detailed in the Supporting Material.
Constructed model of the glycosylated β1 I-like domain-fibronectin complex
With the choice of a relatively simple biantennary complex carbohydrate with the same composition used in our previous study (20), along with a well developed and validated force field for carbohydrates (29), using the enhanced sampling techniques of a simulated annealing and replica exchange MD simulation (REMD) (Supporting Material), we obtained an adequately sampled conformation of the oligosaccharide (Fig. 1 A). The convergence of REMD simulation for oligosaccharide was evaluated through the cluster population analysis method (30), by comparing the populations of the clusters between the two independent REMD simulations with different initial conformations. The populations of the clusters in each ensemble were highly correlated (a correlated coefficient of R2 = 0.956 and a slope of 1.08) (Fig. S1). The highly correlated populations of the clusters between the two independent REMD simulations indicated that the relative population of the conformation of oligosaccharide is highly converged. Using the crystal structure of the extracellular segment of integrin αvβ3 in complex with an RGD ligand (PDB ID: 1L5G) as a template, we constructed a model for the β1 I-like domain in complex with an RGD ligand using the MODELER 9v2 homology modeling package (31) (Supporting Material) (Fig. 1 B). We evaluated the constructed model of the β1 I-like domain in complex with the RGD ligand with PROCHECK (32), ERRAT (33), and WHATCHECK (34). The evaluation results of the constructed model of the β1 I-like domain in complex with the RGD ligand were very similar to the template crystal structure (Table S1), suggesting a valid structure for the constructed model. The complex of the β1 I-like domain with FN-III9-10 was then constructed by replacing the RGD ligand in the constructed complex with the RGD region of the crystal structure of FN-III10 with geometrical fitting (Supporting Material). The orientation of FN-III9-10 was adjusted with the RGD region fixed to avoid any steric clashes between the β1 I-like domain and FN-III9-10, as carried out in a previous study (35). The fully sampled glycan was covalently linked to the amide nitrogens of Asn-192, Asn-249, and Asn-343 of the β1 I-like domain-fibronectin complex using VMD software (36) to obtain the glycosylated I-like domain-fibronectin complex (Fig. 1 C). Due to the uncertainty of the exact orientations of N-glycans relative to the β1 I-like domain and the highly flexible nature of the glycans (37), we constructed five pairs of glycosylated I-like domain-fibronectin complexes with sialic acid and without sialic acid with different starting orientations of the glycans relative to the I-like domain to obtain statistically meaningful results. Equilibration of the simulated systems is detailed in the Supporting Material.
Figure 1.
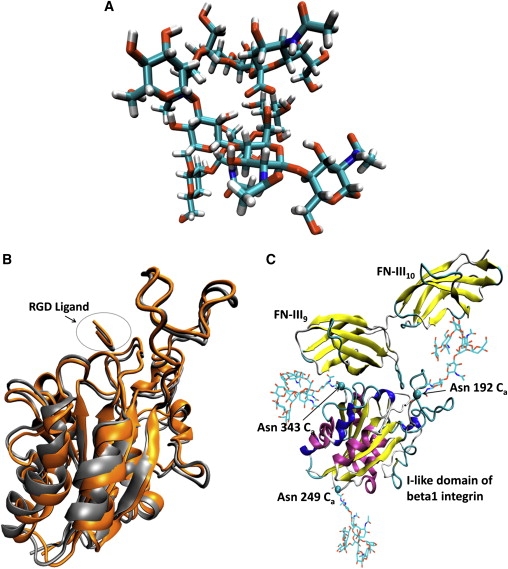
(A) Conformation of the N-linked oligosaccharide with α2-6 sialic acid (ASO). (B) Superposition of the homology built I-like domain β1 integrin in complex with the RGD ligand and crystal structure of I-like domain of β3 integrin in complex with RGD ligand. (C) Model of the glycosylated I-like domain of β1 integrin in complex with the crystal structure of FN-III9-10. The images were made with VMD software support.
Binding thermodynamics analysis
Experiments show that desialylation of β1 integrin enhances the interaction between β1 integrin and fibronectin (11,13). To validate the constructed glycosylated β1 I-like domain-fibronectin complex and simulation results, we calculated the binding free energy changes of the glycosylated β1 I-like domain-fibronectin complex by the desialylation of β1 I-like domain and qualitatively compared these results to experimental results about the tendency for the effect of hyposialylation of α5β1 integrins on its binding to fibronectin. The calculated binding free energy results, including the energy components contributing to the binding free energy, were calculated using the last 35 ns of simulation trajectories of the five sets of MD simulations that had different initial N-glycan orientations (see Table 1 for the results averaged over the five sets of MD simulations, and for the results of each individual set of MD simulations). From the 15 ns–50 ns simulation trajectories of the five sets of MD simulations, the mean of the binding free energy was obtained by averaging over trajectories, and the standard deviation was calculated with the independent periods of the MD simulation trajectories, using the bootstrap statistics method described in the Materials and Methods section of the Supporting Material. The autocorrelation time used to resample the MD simulation trajectories was calculated based on the binding free energy complexes of the β1 I-like domain and fibronectin as a function of cumulative time over the 50 ns MD simulations. The independent conformational region of the glycosylated I-like domain and fibronectin complexes were further displayed in the pairwise backbone RMSD matrix for the entire glycosylated I-like domain and fibronectin complex structure (Fig. S5). The RMSD matrix showed four blue squares along diagonal of the RMSD matrix for the system without sialic acid (Fig. S5 A), and also showed four blue squares along diagonal of the RMSD matrix for the system with sialic acid (Fig. S5 B). The blue represented low RMSD, whereas red represented high RMSD, and each blue square along the diagonal represented an independent conformational region. The calculated results showed that desialylation of the β1 I-like domain resulted in a decreased binding free energy of 12.4 ± 5.2 kcal/mol, indicating that desialylation of the β1 I-like domain has a positive effect on the binding between β1 integrin and fibronectin. The experimental studies showed that hyposialylation of α5β1 integrins stimulates the activity of myeloid fibronectin receptors (11) and the enzymatic removal of sialic acids from purified α5β1 integrins increases α5β1 integrins binding to fibronectin (13). The calculated results of the positive effect of desialylation of the β1 I-like domain on the binding between β1 integrin and fibronectin were qualitatively consistent with the experimental observations about the positive effect of hyposialylation of α5β1 integrins on its binding to fibronectin (11,13). Based on changes in the energy components contributing to the binding free energy, the change in binding free energy of the complex caused by desialylation could result from one or more possibilities. These possibilities include the change in charge caused by desialylation (sialic acid has a −1e charge), changed interactions of glycans with the I-like domain and fibronectin and the conformational changes in the I-like domain and fibronectin by the changed allosteric regulation by the glycans with different degree of sialylation.
Table 1.
Calculated binding free energy for glycosylated β1 I-like domain-fibronectin complex (kcal/mol)
| With α2-6 sialic acid (3ASO) | Without α2-6 sialic acid (3ANS) | |
|---|---|---|
| ΔEelectrostatic | −383.5 ± 23.9 | −408.7 ± 28.8 |
| ΔEvdW | −98.2 ± 3.5 | −104.0 ± 5.4 |
| ΔGsolvation-polar | 447.5 ± 23.8 | 463.2 ± 28.4 |
| ΔGsolvation-nopolar | −16.4 ± 0.8 | −18.1 ± 0.7 |
| −ΔTS | 37.2 ± 3.6 | 41.8 ± 3.3 |
| ΔGbinding | −13.4 ± 4.5 | −25.8 ± 5.8 |
| ΔΔGbinding | −12.4 ± 5.2∗ | |
| Experimental result (11) | Weaker | Strong |
All the mean values in this table were averaged over the five sets of MD simulation trajectories and the standard deviations were obtained based on the averages of all resamples datasets from five sets of MD simulation trajectories using bootstrap method. ΔEvdW is the van der Waals energy, ΔEelectrostatic is the electrostatic energy, ΔGsolvation-polaris the polar solvation energy, ΔGsolvation-nopolar is the nonpolar solvation energy, ΔTS is the energy contributed from solute entropy, ΔGbinding is the binding free energy for the complex, and ΔΔGbinding is the relative binding free energies with respect to the complex without sialic acid. ∗Differences are statistically significant (Student's t-test, p < 0.05).
After the qualitative validation of the calculated results regarding the positive effect of desialylation of the β1 I-like domain on the β1 integrin-fibronectin binding with the experimentally observed positive effect of hyposialylation of α5β1 integrins on its binding to fibronectin, we further analyzed the effects of altered sialylation on changes in interaction of glycans with the I-like domain and fibronectin and the conformational and motion changes of the I-like domain and fibronectin in the glycosylated β1 I-like domain-fibronectin complexes.
Interaction changes of N-glycans of the β1 I-like domain with the β1 I-like domain-fibronectin complex
The glycans of the glycoprotein are highly flexible (37), and the glycans of the I-like domain appeared to interact with the β1 I-like domain and fibronectin during the dynamics simulations, shown by the formation of hydrogen bonds not only with the I-like domain (Fig. 2 A), but also with fibronectin (Fig. 2 B). A hydrogen bond was assigned when the distance of hydrogen and acceptor was <3.5 Å, and the angle of acceptor/donor/hydrogen was >150°. OH and NH groups were treated as donors, oxygen or nitrogen was defined as acceptor. With respect to the interactions between the glycans and the I-like domain, the mean and standard deviation of the number of hydrogen bonds obtained from the five independent pairs of MD simulations showed that desialylation of the glycans resulted in significantly decreased hydrogen bond formation between the glycans and the I-like domain (p < 0.05) (Fig. 2 A). With respect to the interactions between the glycans of the I-like domain and fibronectin, hydrogen bonds were formed only for the glycans attached to Asn-192 and Asn-343, which are located close to the binding region of the I-like domain with fibronectin (Fig. 1 C) (Fig. 2 B). The mean and standard deviation of the number of hydrogen bonds calculated from the trajectories of five sets of MD simulations showed that desialylation of the glycans resulted in significantly decreased hydrogen bond formation between the glycan at Asn-343 and fibronectin (p < 0.05) (Fig. 2 B). The change in interactions between the glycans and the I-like domain and fibronectin caused by altered sialylation could directly result in a change in allosteric regulation of the interaction between the I-like domain and fibronectin, which involves changes in the conformation of the proteins, changes in the degree of correlated motions between residues, and changes in residue motion within the residues of the proteins in the complex.
Figure 2.
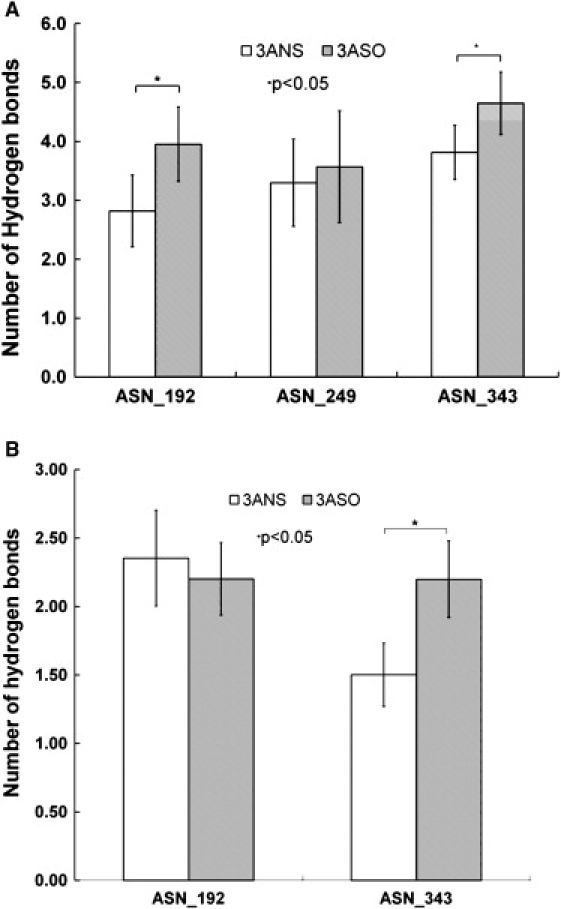
Hydrogen bonds formed between the glycans of the I-like domain with the (A) β1 I-like domain and (B) FN-III9-10. 3ANS, glycosylation without sialic acid; 3ASO, glycosylation with sialic acid.
Conformational changes in the β1 I-like domain
We calculated the mean and standard deviation for the root mean-square deviation (RMSD) of the critical functional sites of the β1 I-like domain, including the SDL, the metal ion-dependent adhesion site containing a DLSYS motif, and other critical residues (Asn-224, Asp-226, Glu-229, Asp-233, Asp-267, and Asp-295) (15–19) for the systems with and without sialic acid (Table S3). The reference structure for RMSD calculations was the initial structure of the I-like domain, and the average and standard deviations of the RMSD for the key functional sites were calculated using the trajectories of the five sets of independent MD simulations. Results showed that altered sialylation of the β1 I-like domain resulted in significant conformational changes for half of the key functional sites of the β1 I-like domain (Student's t-test, p < 0.05). Experimental studies have shown that movement of the α1 and α7 helices is directly involved in integrin activation (18,38,39). Analysis of the movement of the centers of mass of the α1 and α7 helices showed that altered sialylation of the β1 I-like domain caused significant movement of the α1 and α7 helices (p < 0.05) (Fig. 3). These conformational changes in the I-like domain might result from the changes in interactions between glycans and the I-like domain-fibronectin complex or changes in allosteric regulation caused by glycans with altered sialylation. The calculated results regarding the conformational changes in the β1 I-like domain caused by altered sialylation were consistent with the results from our previous study (20) and could contribute to the experimentally observed changes in binding between β1 integrin and fibronectin (11,13).
Figure 3.
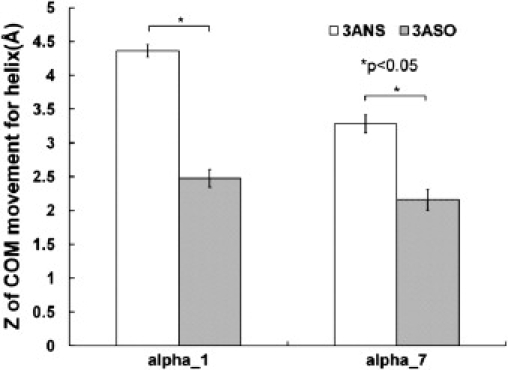
Z-coordinates of the center of mass for α1 and α7 helices.
Conformational changes in fibronectin
RGD conformational change
The RGD loop is the specific binding region of fibronectin for integrin (21). Experimental studies show that the conformation of RGD directly affects the binding specificity of RGD in cell adhesion (40–42). Furthermore, the distance from the RGD region to the FN-III10 core could affect the accessibility of RGD to integrin for integrin activation (43,44). The crystal structure of fibronectin shows that the RGD loop is located at the apex of a hairpin β-turn connecting strands F and G (28). We calculated the conformation of the RGD-containing loop for the glycosylated I-like domain-fibronectin complex systems with and without sialic acid using the five sets of independent MD simulations trajectories to determine the effect of altered sialylation on RGD conformational changes. Similar to previous studies quantifying the conformation of the RGD-containing loop under external force (44,45), we measured the distance from the RGD segment to the FN-III10 core by calculating the vertical distance from the Ca atom of Asp-1495 (at the apex of RGD loop) to the plane defined by the residues Val-1487 and Ala-1489 on the F strand and the residue Arg-1448 on the C strand (the C and F strands serve as the substrate surface of the RGD loop in fibronectin FN-III10) (Fig. 4, A and C). We also calculated the bend angle of the RGD region defined by the Ca atoms of residues Asp-1495 and Arg-1493 in RGD and residue Thr-1491 in the F strand (Fig. 4, B and D). The results showed that desialylation of the I-like domain in the glycosylated I-like domain-fibronectin complex caused an increase in distance between the RGD region and the FN-III10 core from 13.5 ± 1.8 Å to 16.1 ± 0.6 Å (Fig. 4 C). Experimental results have shown that distances ranging from 11 Å to 3.0 Å between RGD and the FN-III10 core are optimal for RGD binding (43). The calculated distances between RGD and the FN-III10 core for the systems with and without sialic acid were all in the allowable binding range suggested by experimental results (43). However, the increased distance between RGD and the FN-III10 core caused by desialylation of the I-like domain could increase the accessibility of RGD to membrane-bound integrin and enhance their interaction. The desialylation of the I-like domain also resulted in a changed bend angle of the RGD loop from 151 ± 2.4° to 146 ± 2.3° (Fig. 4 D). The conformational changes in RGD caused by altered sialylation of the I-like domain in the glycosylated I-like domain-fibronectin complex could contribute to differences in binding of fibronectin to integrin observed in experimental systems (11,13). The calculated conformational changes in the RGD loop caused by altered sialylation were also observed in the force-regulated conformational changes of RGD (44,45). These observations suggest that functional mediation of cell adhesion with external force might also be implemented through regulation of sialylation of the β1 I-like domain.
Figure 4.
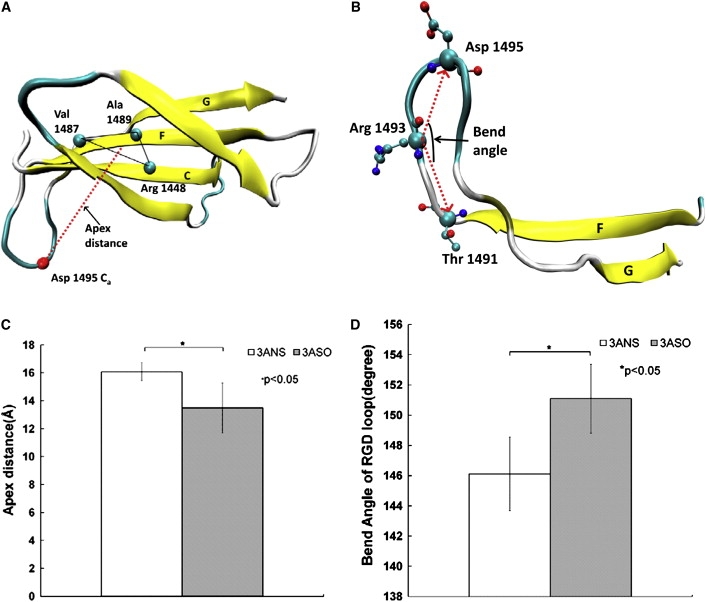
(A) Definition of the distance of the RGD segment to the FN-III10 core, measured from the Ca atom of Asp-1495 to the plane defined by the residues Val-1487 and Ala-1489 on the F strand and the residue Arg-1448 on the C strand. (B) Definition of the bend angle of RGD: angle formed by the Ca atoms of residues Asp-1495 and Arg-1493 in RGD and residue Thr-1491 in the F strand. (C) Distance of the RGD segment to the FN-III10 core. (D) Bend angle of RGD.
Interactive conformational changes in the RGD-synergy site
α5β1 integrin binds to both the RGD and the synergy sites of fibronectin, and the synergy site allows α5β1 integrin to bind to fibronectin preferentially over other RGD-containing matrix proteins (23–27). Experimental studies have suggested that the relative conformational changes between the RGD loop and synergy site play a critical role in the interaction between α5β1 integrin and fibronectin (46–48). We calculated the relative rotational orientation and the distance between the RGD loop and synergy site for the systems with and without sialic acid based on the five pairs of MD simulation trajectories. Similar to the definition of the relative rotational orientation and the distance between the RGD and synergy sites in a previous study (49), we defined the relative rotational orientation as a dihedral angle formed by the Ca atoms of residues Arg-1379 in FN-III9, Arg-1493 in FN-III10, and the centers of mass of FN-III9 and FN-III10. The distance between RGD and the synergy site was defined to be the distance between the Ca atoms of Arg-1379 on the synergy site and Arg-1493 on the RGD loop in fibronectin (Fig. 5 A). The calculated results showed that desialylation of the I-like domain in the glycosylated I-like domain-fibronectin complex caused a significant increase in the torsion angle from 25.9 ± 2.8° to 32.6 ± 3.1° (Fig. 5 B) (Student's t-test, p < 0.05), and resulted in a significant decrease in distance between RGD and the synergy site from 39.2 ± 2.2 Å to 34.6 ± 1.2 Å (Student's t-test, p < 0.05). The crystal structure of FN-III7-10 shows that Arg-1379 on the synergy site and Arg-1493 on the RGD loop are separated by ∼32 Å (28), which is optimal for fibronectin binding to α5β1 integrin (23,47). Furthermore, Arg-1379 in the synergy site is a key residue for binding to integrin (48). The calculated distance between the RGD loop and the synergy region for the system without sialic acid is closer than the system with sialic acid to the experimentally observed optimal distance for fibronectin binding to α5β1 integrin, consistent with experimental results regarding the effect of desialylation of β1 integrin on enhancement of β1 integrin binding to fibronectin (11,13). Experimental results also showed that the spatial positioning of the RGD loop and synergy site, including the distance between RGD and the synergy site and the twist and tilt angles between FIII9 and FIII10, is critical for fibronectin binding to integrin to signal cell adhesion (23). Altered sialylation of β1 integrin has been shown to affect β1 integrin binding to fibronectin and further influence cell adhesion (11,13). The results of the calculated changes in distance and orientation of the RGD loop relative to the synergy site caused by altered sialylation of the β1 I-like domain may contribute to the experimentally observed changes in fibronectin binding to β1 integrin caused by altered sialylation of β1 integrin (11,13). These results are consistent with experimental observations regarding the critical role of spatial positioning of the RGD loop and synergy site in cell adhesion (23). The calculated interactive conformational changes of the RGD and synergy site, which are important for β1 integrin binding to fibronectin, were also observed in the force regulation of integrin binding to the RGD and synergy site of fibronectin (49,50). This indicates that altered sialylation of the β1 I-like domain may play a similar role to external force in regulation of the β1 integrin/fibronectin interaction and mediation of cellular adhesion events.
Figure 5.
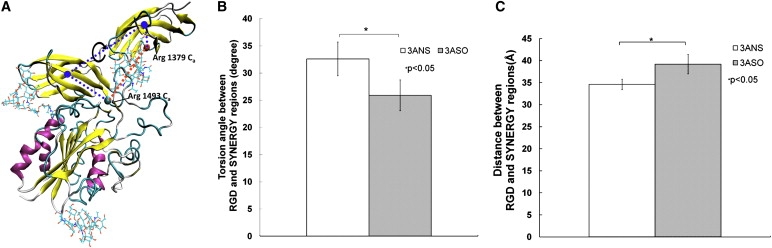
(A) Definition of the distance and torsion dihedral angle between the RGD loop and the synergy site. Distance was defined by the Ca atoms of Arg-1379 on the synergy site and Arg-1493 on the RGD loop. The torsion angle was defined by the Ca atoms of Arg-1379, Arg-1493, and the center of masses of FN-III9 and FN-III10. (B) Relative rotational orientation between the RGD loop and the synergy site. (C) Distance between the RGD loop and the synergy site.
Motion changes in the β1 I-like domain and fibronectin
Dynamical cross-correlation maps investigate the degree of correlated motions between the residues in the protein, which could provide further insight into the effects of the altered sialylation of the I-like domain on β1 integrin binding to fibronectin (Fig. 6). Fig. 6 A shows the cross-correlation maps for the I-like domain without sialic acid (top left half) and with sialic acid (bottom right half). The maps showed that desialylation of the I-like domain resulted in an overall decreased degree of correlated motion, and an increased degree of anticorrelated motion within the residues of the I-like domain. Specifically, desialylation of the I-like domain caused a decreased degree of correlated motion between residues 170–210 and residues 260–320, and between residues 220–260 and residues 260–320 (squared in Fig. 6 A). These regions included residues in key functional sites in the I-like domain, such as residues in the SDL region (residues 170–197) and α7 helix (residues 344–355), and binding-related critical residues N224, D226, E229, D233 (within residues 220–260) and D267, D295 (within residues 260–320). Desialylation of the I-like domain also resulted in an increased degree of anticorrelated motion between residues 170–230 and residues 320–360, and between residues 260–320 and residues 320–360 (circled in Fig. 6 A). These results indicate that desialylation of the I-like domain directly affects the degree of correlated motions between residues, including critical functional sites, which may contribute to changes in the binding of β1 integrin to fibronectin.
Figure 6.
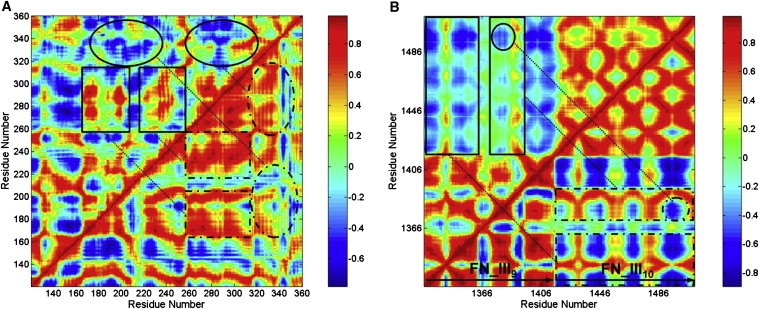
Dynamical cross-correlation maps for the degree of correlated motion of the residues in the glycosylated I-like domain-fibronectin complex. (A) I-like domain without sialic acid (top left) compared to the I-like domain with sialic acid (bottom right) (squared regions show the correlated motion between residues 170–210 and residues 260–320, and between residues 220–260 and residues 260–320; circled regions show the correlated motion between residues 170–230 and residues 320–360, and between residues 260–320 and residues 320–360). (B) Fibronectin for the complex system without sialic acid (top left) compared to the system with sialic acid (bottom right) (squared regions show the degree of correlated and anticorrelated motions between residues 1326–1360 and residues 1416–1509, between residues 1370–1400 and residues 1416–1509; circled regions show the degree of correlated motion between regions around the RGD loop (residues 1493–1495) in FN-III10 and the synergy site in FN-III9 (residues 1376–1380)). (For color map, red, correlation; blue, anticorrelation. For map in grayscale, white, correlation; black, anticorrelation.)
Dynamical cross-correlation maps for fibronectin within the glycosylated I-like domain-fibronectin complex without sialic acid (top left half) and with sialic acid (bottom right half) are shown in Fig. 6 B. In the complex lacking sialylation, the degree of correlated motion between residues within FN-III9 and within FN-III10 was not significantly changed, but the correlated and anticorrelated motion between residues in FN-III9 and residues in FN-III10 was significantly changed. Specifically, the degree of correlated and anticorrelated motions between residues 1326–1360 and residues 1416–1509, and also between residues 1370–1400 and residues 1416–1509 (squared) was significantly decreased, including the degree of correlated motion between regions surrounding the RGD-loop in FN-III10 (residues 1493–1495) and the synergy site in FN-III9 (residues 1376–1380) (circled). The RGD and synergy site cooperate with one another for recognition by α5β1 integrin (23–27). The change in degree of correlated motions between residues in FN-III9 and residues in FN-III10 caused by the altered sialylation of the I-like domain in the glycosylated I-like domain-fibronectin complex, particularly for the residues between the RGD-loop and synergy site, could directly contribute to changes in the binding between fibronectin and α5β1 integrin observed in previous experiments (11,13).
We also analyzed the dynamics of the system using principal component analysis (PCA), which uses a few nanoseconds of MD simulation trajectories to assess the primary motions of the system that occurred at a longer timescale (51). The primary motion of the system is usually represented by the relatively lower frequency modes obtained from PCA, and the first PCA mode is usually interpreted as the direction of the largest conformational fluctuation of the system during MD simulations (52,53). Each PCA mode is associated with an eigenvalue that corresponds to the amplitude of fluctuations along that mode, and each eigenvalue normalized by the sum of all eigenvalues can represent the relative contribution of a mode to the dynamical motion of the selected atoms observed in the MD simulation (54). Based on the five pairs of independent MD simulation trajectories, the distribution of the relative contribution of the first ten PCA modes of the I-like domain with and without sialic acid is shown in Fig. S6 A. The result indicated that the contribution of each PCA mode to the motion of the I-like domain was not significantly changed by desialylation. However, the PCA analysis for fibronectin showed that the desialylation of the I-like domain resulted in an increased contribution of the first mode to the dynamical motion of fibronectin from 57.8% (with sialic acid) to 67.1% (without sialic acid) (Fig. S6 B). The motion directions of the first PCA mode for the systems with and without sialic acid were shown on the atomic structure of the glycosylated I-like domain-Fn complexes (Fig. S7). The comparison of the direction and amplitude of the first PCA mode between the systems without sialic acid (Fig. S7 A) and with sialic acid (Fig. S7 B) showed that the direction and amplitude of the first PCA mode in I-like domain did not exhibit obvious difference. However, differences of the motion amplitude and direction of the first PCA mode in fibronectin were observed, particularly in the FN_III9 module, the first PCA mode in the system with sialic acid (3ASO) showed a larger motion amplitude and more dispersive motion direction compared to the system without sialic acid (3ANS). These motion changes are expected to be functionally relevant to the experimental observation that altered sialylation of β1 integrin affects the binding of fibronectin to β1 integrin (11,13).
Conclusions
In this study, we characterized the conformation of a relatively simple biantennary complex carbohydrate using the enhanced sampling technique, and constructed a validated model of the glycosylated β1 I-like domain- FN-III9-10 complex. We carried out five pairs of independent MD simulations for the glycosylated I-like domain-fibronectin complexes with and without sialic acid in the N-linked glycans of the I-like domain, with different initial orientations of the N-linked glycans relative to the β1 I-like domain. The calculated results about the positive effect of the desialylation of β1 I-like domain on the β1 integrin-fibronectin binding were qualitatively consistent with the experimental observation about the positive effect of hyposialylation of α5β1 integrins on its binding to fibronectin (11,13). Altered sialylation of the β1 I-like domain resulted in significant changes in interactions of N-glycans of the I-like domain with both the I-like domain and fibronectin. Altered sialylation of the β1 I-like domain caused significant changes in the conformations of key functional sites of the β1 I-like domain and the RGD loop in FN-III10, and also changes in the relative orientation between the RGD loop and the synergy site in FN-III9, which is critical for β1 integrin binding to fibronectin. A change in the degree of correlated motions between residues in the I-like domain and residues in fibronectin, and a change in the degree of motion of FN-III9-10 caused by altered sialylation of the I-like domain were also observed. These results provide important structural insights into the effect of altered sialylation of the β1 I-like domain on the binding of β integrin to fibronectin observed in experimental systems. Although the constructed model of a relatively simple biantennary complex carbohydrate was representative and limited by the lack of exact structural information on glycans, results from this study provide clear structural information regarding the effects of altered sialylation on the interaction between β1 integrin and fibronectin. These results also represent a potential avenue for the identification of novel strategies and drugs that perturb the integrin glycan profile, regulating β1 integrin binding to fibronectin, and potentially modulating cell adhesion in pathologic conditions such as cancer and cardiovascular diseases.
Supporting Material
Methods, three tables, and seven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00430-3.
Supporting Material
Acknowledgments
The authors thank S. L. Bellis for helpful discussion and the anonymous reviewers for their helpful remarks. D.P. acknowledges the Ireland Tuition Scholarship.
This work was supported by the National Science Foundation (grant MCB080078 to Y.H.S.).
References
- 1.Humphries M.J., Travis M.A., Mould A.P. Mechanisms of integration of cells and extracellular matrices by integrins. Biochem. Soc. Trans. 2004;32:822–825. doi: 10.1042/BST0320822. [DOI] [PubMed] [Google Scholar]
- 2.George E.L., Georges-Labouesse E.N., Hynes R.O. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- 3.Plopper G.E., Domanico S.Z., Quaranta V. Migration of breast epithelial cells on Laminin-5: differential role of integrins in normal and transformed cell types. Breast Cancer Res. Treat. 1998;51:57–69. doi: 10.1023/a:1006086218174. [DOI] [PubMed] [Google Scholar]
- 4.Melchiori A., Mortarini R., Albini A. The alpha 3 beta 1 integrin is involved in melanoma cell migration and invasion. Exp. Cell Res. 1995;219:233–242. doi: 10.1006/excr.1995.1223. [DOI] [PubMed] [Google Scholar]
- 5.Weber C. Novel mechanistic concepts for the control of leukocyte transmigration: specialization of integrins, chemokines, and junctional molecules. J. Mol. Med. 2003;81:4–19. doi: 10.1007/s00109-002-0391-x. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu Y., Rose D.M., Ginsberg M.H. Integrins in the immune system. Adv. Immunol. 1999;72:325–380. doi: 10.1016/s0065-2776(08)60024-3. [DOI] [PubMed] [Google Scholar]
- 7.Isaji T., Sato Y., Gu J. N-glycosylation of the I-like domain of beta1 integrin is essential for beta1 integrin expression and biological function: identification of the minimal N-glycosylation requirement for alpha5beta1. J. Biol. Chem. 2009;284:12207–12216. doi: 10.1074/jbc.M807920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isaji T., Sato Y., Gu J. N-glycosylation of the beta-propeller domain of the integrin alpha5 subunit is essential for alpha5beta1 heterodimerization, expression on the cell surface, and its biological function. J. Biol. Chem. 2006;281:33258–33267. doi: 10.1074/jbc.M607771200. [DOI] [PubMed] [Google Scholar]
- 9.Gu J., Taniguchi N. Regulation of integrin functions by N-glycans. Glycoconj. J. 2004;21:9–15. doi: 10.1023/B:GLYC.0000043741.47559.30. [DOI] [PubMed] [Google Scholar]
- 10.Bellis S.L. Variant glycosylation: an underappreciated regulatory mechanism for beta1 integrins. Biochim. Biophys. Acta. 2004;1663:52–60. doi: 10.1016/j.bbamem.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Semel A.C., Seales E.C., Bellis S.L. Hyposialylation of integrins stimulates the activity of myeloid fibronectin receptors. J. Biol. Chem. 2002;277:32830–32836. doi: 10.1074/jbc.M202493200. [DOI] [PubMed] [Google Scholar]
- 12.Sato Y., Isaji T., Gu J. An N-glycosylation site on the beta-propeller domain of the integrin alpha5 subunit plays key roles in both its function and site-specific modification by beta1,4-N-acetylglucosaminyltransferase III. J. Biol. Chem. 2009;284:11873–11881. doi: 10.1074/jbc.M807660200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seales E.C., Shaikh F.M., Bellis S.L. A protein kinase C/Ras/ERK signaling pathway activates myeloid fibronectin receptors by altering beta1 integrin sialylation. J. Biol. Chem. 2005;280:37610–37615. doi: 10.1074/jbc.M508476200. [DOI] [PubMed] [Google Scholar]
- 14.Lee J.O., Bankston L.A., Liddington R.C. Two conformations of the integrin A-domain (I-domain): a pathway for activation? Structure. 1995;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 15.Takagi J., DeBottis D.P., Springer T.A. The role of the specificity-determining loop of the integrin beta subunit I-like domain in autonomous expression, association with the alpha subunit, and ligand binding. Biochemistry. 2002;41:4339–4347. doi: 10.1021/bi016047u. [DOI] [PubMed] [Google Scholar]
- 16.Humphries M.J. Integrin structure. Biochem. Soc. Trans. 2000;28:311–339. [PubMed] [Google Scholar]
- 17.Xiong J.P., Stehle T., Arnaout M.A. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barton S.J., Travis M.A., Mould A.P. Novel activating and inactivating mutations in the integrin beta1 subunit A domain. Biochem. J. 2004;380:401–407. doi: 10.1042/BJ20031973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puzon-McLaughlin W., Takada Y. Critical residues for ligand binding in an I domain-like structure of the integrin beta1 subunit. J. Biol. Chem. 1996;271:20438–20443. doi: 10.1074/jbc.271.34.20438. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y., Pan D., Song Y. Effect of altered glycosylation on the structure of the I-like domain of beta1 integrin: a molecular dynamics study. Proteins. 2008;73:989–1000. doi: 10.1002/prot.22126. [DOI] [PubMed] [Google Scholar]
- 21.Pankov R., Yamada K.M. Fibronectin at a glance. J. Cell Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 22.Plow E.F., Haas T.A., Smith J.W. Ligand binding to integrins. J. Biol. Chem. 2000;275:21785–21788. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 23.Grant R.P., Spitzfaden C., Mardon H.J. Structural requirements for biological activity of the ninth and tenth FIII domains of human fibronectin. J. Biol. Chem. 1997;272:6159–6166. doi: 10.1074/jbc.272.10.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baron M., Main A.L., Campbell I.D. 1H NMR assignment and secondary structure of the cell adhesion type III module of fibronectin. Biochemistry. 1992;31:2068–2073. doi: 10.1021/bi00122a025. [DOI] [PubMed] [Google Scholar]
- 25.Corbett S.A., Schwarzbauer J.E. Requirements for alpha(5)beta(1) integrin-mediated retraction of fibronectin-fibrin matrices. J. Biol. Chem. 1999;274:20943–20948. doi: 10.1074/jbc.274.30.20943. [DOI] [PubMed] [Google Scholar]
- 26.Danen E.H., Aota S., van Muijen G.N. Requirement for the synergy site for cell adhesion to fibronectin depends on the activation state of integrin alpha 5 beta 1. J. Biol. Chem. 1995;270:21612–21618. doi: 10.1074/jbc.270.37.21612. [DOI] [PubMed] [Google Scholar]
- 27.Sechler J.L., Corbett S.A., Schwarzbauer J.E. Modulatory roles for integrin activation and the synergy site of fibronectin during matrix assembly. Mol. Biol. Cell. 1997;8:2563–2573. doi: 10.1091/mbc.8.12.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leahy D.J., Aukhil I., Erickson H.P. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- 29.Kirschner K.N., Yongye A.B., Woods R.J. GLYCAM06: a generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okur A., Wickstrom L., Simmerling C. Improved efficiency of replica exchange simulations through use of a hybrid explicit/implicit solvation model. J. Chem. Theory Comput. 2006;2:420–433. doi: 10.1021/ct050196z. [DOI] [PubMed] [Google Scholar]
- 31.Martí-Renom M.A., Stuart A.C., Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski R.A., MacArthur M.W., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 33.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hooft R.W., Vriend G., Abola E.E. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 35.Puklin-Faucher E., Gao M., Vogel V. How the headpiece hinge angle is opened: new insights into the dynamics of integrin activation. J. Cell Biol. 2006;175:349–360. doi: 10.1083/jcb.200602071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- 37.Imberty A., Pérez S. Structure, conformation, and dynamics of bioactive oligosaccharides: theoretical approaches and experimental validations. Chem. Rev. 2000;100:4567–4588. doi: 10.1021/cr990343j. [DOI] [PubMed] [Google Scholar]
- 38.Takagi J., Springer T.A. Integrin activation and structural rearrangement. Immunol. Rev. 2002;186:141–163. doi: 10.1034/j.1600-065x.2002.18613.x. [DOI] [PubMed] [Google Scholar]
- 39.Takagi J., Petre B.M., Springer T.A. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 40.Pierschbacher M.D., Ruoslahti E. Influence of stereochemistry of the sequence Arg-Gly-Asp-Xaa on binding specificity in cell adhesion. J. Biol. Chem. 1987;262:17294–17298. [PubMed] [Google Scholar]
- 41.Scarborough R.M., Naughton M.A., Charo I.F. Design of potent and specific integrin antagonists. Peptide antagonists with high specificity for glycoprotein IIb-IIIa. J. Biol. Chem. 1993;268:1066–1073. [PubMed] [Google Scholar]
- 42.Nowlin D.M., Gorcsan F., Cardarelli P.M. A novel cyclic pentapeptide inhibits alpha 4 beta 1 and alpha 5 beta 1 integrin-mediated cell adhesion. J. Biol. Chem. 1993;268:20352–20359. [PubMed] [Google Scholar]
- 43.Beer J.H., Springer K.T., Coller B.S. Immobilized Arg-Gly-Asp (RGD) peptides of varying lengths as structural probes of the platelet glycoprotein IIb/IIIa receptor. Blood. 1992;79:117–128. [PubMed] [Google Scholar]
- 44.Krammer A., Lu H., Vogel V. Forced unfolding of the fibronectin type III module reveals a tensile molecular recognition switch. Proc. Natl. Acad. Sci. USA. 1999;96:1351–1356. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao M., Craig D., Schulten K. Identifying unfolding intermediates of FN-III(10) by steered molecular dynamics. J. Mol. Biol. 2002;323:939–950. doi: 10.1016/s0022-2836(02)01001-x. [DOI] [PubMed] [Google Scholar]
- 46.García A.J., Vega M.D., Boettiger D. Modulation of cell proliferation and differentiation through substrate-dependent changes in fibronectin conformation. Mol. Biol. Cell. 1999;10:785–798. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamburger Z.A., Brown M.S., Bjorkman P.J. Crystal structure of invasin: a bacterial integrin-binding protein. Science. 1999;286:291–295. doi: 10.1126/science.286.5438.291. [DOI] [PubMed] [Google Scholar]
- 48.Redick S.D., Settles D.L., Erickson H.P. Defining fibronectin's cell adhesion synergy site by site-directed mutagenesis. J. Cell Biol. 2000;149:521–527. doi: 10.1083/jcb.149.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krammer A., Craig D., Vogel V. A structural model for force regulated integrin binding to fibronectin's RGD-synergy site. Matrix Biol. 2002;21:139–147. doi: 10.1016/s0945-053x(01)00197-4. [DOI] [PubMed] [Google Scholar]
- 50.Craig D., Krammer A., Vogel V. Comparison of the early stages of forced unfolding for fibronectin type III modules. Proc. Natl. Acad. Sci. USA. 2001;98:5590–5595. doi: 10.1073/pnas.101582198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lange O.F., Grubmuller H. Can principal components yield a dimension reduced description of protein dynamics on long time scales? J. Phys. Chem. 2006;110:22842–22852. doi: 10.1021/jp062548j. [DOI] [PubMed] [Google Scholar]
- 52.Hayward S., Kitao A., Go N. Effect of solvent on collective motions in globular protein. J. Mol. Biol. 1993;234:1207–1217. doi: 10.1006/jmbi.1993.1671. [DOI] [PubMed] [Google Scholar]
- 53.Berendsen H.J., Hayward S. Collective protein dynamics in relation to function. Curr. Opin. Struct. Biol. 2000;10:165–169. doi: 10.1016/s0959-440x(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 54.Bradley M.J., Chivers P.T., Baker N.A. Molecular dynamics simulation of the Escherichia coli NikR protein: equilibrium conformational fluctuations reveal interdomain allosteric communication pathways. J. Mol. Biol. 2008;378:1155–1173. doi: 10.1016/j.jmb.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


