Abstract
In this study, polyethylenimine (PEI) binding to DNA was examined by isothermal titration calorimetry. Two types of binding modes were found to describe the interactions between these polyelectrolytes in buffers and in water. One type of binding involves PEI binding to the DNA groove because the enthalpy change of this binding mode is positive, and PEI is deprotonated to bind to DNA. Another likely binding mode involves external binding of PEI to the DNA phosphate backbone, accompanied with DNA condensation. The enthalpy change is negative and PEI is protonated when it binds to DNA in this mode. The intrinsic enthalpy change of first binding mode is 1.1 kJ/mol and −0.88 kJ/mol for the second binding mode. This result implies that the PEI is rearranged from the groove to the phosphate backbone of DNA when DNA is condensed. The mechanism of DNA condensation caused by PEI is discussed in this study.
Introduction
Polyethylenimine (PEI) is one of the most effective polymers for nonviral gene delivery (1). The effective transfection ability of PEI is attributed to its capability to condense the string-like DNA molecule into nanoparticles and make the DNA suitable for cellular uptake (2). Several reports examined the details of PEI-DNA complexation; for example, Dunlap et al. have observed the DNA condensate with PEI by using atomic force microscopy (AFM) (3). Although useful for analyzing the final state of the particles, it is difficult to elucidate the DNA condensation mechanism in detail by AFM and similar imaging techniques. Zhou and Li (4) have investigated the PEI-DNA interactions by using ethidium bromide as a fluorescent probe. According to Wiethoff et al. (5), the ability to displace a reporter probe from a DNA will vary among the species of the probes, so that the information gained from such an analysis will depend on the choice of the fluorescent probe. The fluorescent probes also became inadequate for binding analysis because the intensity of the fluorescence changes with the extent of probe burial in complexes. In addition, PEI-DNA complex is known to aggregate (6), making spectroscopic analysis of the PEI-DNA interactions technically difficult and not reliable (7,8).
Isothermal titration calorimetry (ITC) is a powerful technique for analyzing interactions of biomolecules in solution because it does not require a reporter probe and it is not susceptible to solution turbidity. Despite its promise, only two studies investigated PEI-DNA interactions by using ITC. Choosakoonkriang et al. (9) have studied the effects of PEI binding to DNA in various buffer conditions, but they could not obtain the binding constant of this interaction because the complexation process was limited by aggregation of the complexes. The enthalpy change for PEI binding to DNA was calculated in an unusual way by using only part of the ITC data generated in that study. Ikonen et al. (10) were able to obtain a binding constant for PEI-DNA interaction from ITC; however, their ITC data were unsatisfactory on the following grounds; i), the dispersion of the data during the titration is too large to accurately determine the thermodynamic parameters, their data points were obtained from single run, and it would have been possible to fit a straight line instead of sigmoid fit in the heat integration data; ii), several peaks at the onset of titration were deleted without explanation (from the 39 titration peaks reported, only 32 were integrated), and the integration values of the peaks ignored would have been greater than two times larger than the other peaks, and; iii), no blank data were reported. The integration curve of PEI titrated into DNA was expected to be convex upward from their raw data. The blank data should be unusual because their integrated data subtracted blank data were sigmoidlike.
The ITC analysis has been used to probe DNA condensation mechanism as well (11,12). However, some studies ignored the DNA condensation for fitting ITC data have been reported (13–15). The reported fitting methods would have been inadequate because the regions at which the enthalpy change (ΔH) began to decrease and the heat change became constant are crucial to determine the thermodynamic parameters. To better elucidate the thermodynamics of ligand binding to DNA in the presence of DNA condensation, Kim et al. (16) have adeptly described a model, in which a ligand binding to DNA (i.e., first binding mode) was correlated to a second binding mode that included DNA condensation as well.
In this study, we obtained reliable thermodynamic parameters that describe the binding of PEI to DNA. The model proposed by Kim et al. (16) was used to describe the thermodynamics of the interaction, distinguishing clearly between the two different modes of interactions. We believe this is the first report to determine the thermodynamic parameters of different binding modes of PEI to DNA and to correlate the second binding mode with DNA condensation.
Materials and Methods
A 600 Da branched PEI (Polysciences, Worthington, PA) and salmon testes DNA (Sigma, St. Louis, MO) were dissolved in buffer (0.1 M HEPES or 0.1 M MES) or water (with or without 0.1 M NaCl). A phosphate buffer was also used to initially dissolve DNA, but the solution became cloudy when PEI was added to the buffer without DNA and, consequently, phosphate-containing buffers were not used in this study. For calorimetry experiments, 1.5 mM DNA solution (equivalent to phosphate concentration) was applied into the ITC cell (950 μL; Nano-ITC from TA instruments, New Castle, DE) and 20 mM PEI solution (equivalent to nitrogen concentration) in 250 μL syringe was injected into the DNA solution in 20 portions of 12.5 μL volume at 30-min intervals. All solutions were degassed before use. The ITC syringe was stirred at 250 rpm and the cell was equilibrated at 25°C before titrations. The 20 mM PEI solution was also injected into the buffers or water as a blank titration.
To describe binding, we used the model proposed by Kim et al. (16) because it is highly flexible for the two-variable binding constant system. Briefly, this model is based on the single set of identified site (SSIS) model (17). The equation of SSIS model is:
| (1) |
where Q is the total heat content of the solution contained in the sample cell, N is the stoichiometric number, Mt is the total concentration of macromolecule, Xt is the total concentration of ligand, ΔH is the molar heat of the ligand binding, V0 is the cell volume, and K is the binding constant. The heat released ΔQi from the ith injection is given by:
| (2) |
The normalized heat, NDH(i), is calculated by dividing the ΔQi with the number of moles in the ith injected volume. In the model proposed by Kim et al. (16) the fraction of bound ligands is described as the absolute value of NDH divided by ΔH. NDH1 was used for the first binding mode with SSIS model involving the parameters of N1, K1, and ΔH1. The sigmoidal NDH3 curve was used to describe the fraction of the occupied site on DNA during the second binding mode. The definition of parameters for NDH3 is basically the same as those for the NDH1, but only N3 is variable when considering the delay of the second binding mode. The NDH2 curve for the second binding mode was also generated based on N2, K2, and ΔH2. The second stage of the ITC curve was fitted by the hypothetical ITC curve NDH2′ and the increased sigmoidal curve ABS((ΔH1 − NDH3)/ΔH1). The NDH2′ that corresponds to a standard ITC curve without the initial binding mode is generated by the SSIS model, indicating that the NDH2′ curve is defined by N2′, K2′, and ΔH2′. The product was defined as NDH2 where N2′ − N3, K2′, and ΔH2′ are selected for N2, K2, and ΔH2, respectively. Finally, a complete ITC curve was obtained by summing NDH1 and NDH2.
It is known that the observed binding enthalpy (ΔHobs) is linearly dependent on the buffer ionization enthalpy (ΔHion) (9,18). The y-intercept of such a relationship corresponds to the buffer independent binding enthalpy (ΔH0) and the slope to the degree of protonation of the ligand (n), as defined by the following equation:
| (3) |
Positive n values indicate that PEI is protonated on binding to DNA. Baker and Murphy (18) described that the determination of n at a minimum of two experimental values is theoretically sufficient.
The pH measurements for the PEI titration of DNA was carried out 30 times of volume of ITC experiments, so that the pH electrode can be directly dipped into the solutions; i.e., 375 μL of 20 mM PEI solution was added into 28.5 mL of DNA solution and then 375 μL of aliquots were removed into a 1.5-mL tube. The concentrations of PEI and DNA corresponded to the concentrations used in the ITC experiment, but the temperature was slightly lower (room temperature, 23°C) during this experiment. The PEI injection and the extraction from the solution were repeated 20 times. The removed aliquots were centrifuged at 20, 000 × g for 1 min and the supernatant was assayed by measuring its absorbance at 260 nm using NanoVue (GE Healthcare, Morgan Boulevard, Quebec).
To determine the acid dissociation constant pKa of PEI under various conditions, a 15 μL of 0.1 M HCl was titrated into 28.5 mL of 1 mM PEI and the pH of the final solution was measured. The addition of HCl was repeated 25 times without removal of solution. The pH values were plotted against the volume of HCl added. The data were fitted using the model proposed by Suh et al. (19). They defined the deprotonation quotient P as the ratio of the fraction of unprotonated amino groups:
| (4) |
And logP is assumed to be linearly related to pH:
| (5) |
where α and β are constants.
The pKa can be represented as follows:
| (6) |
Results
Fig. 1 shows a typical raw data for titration of 20 mM PEI into 1.5 mM DNA in HEPES buffer (pH 7). After the first seven exothermic peaks, four endothermic peaks appeared (Fig. 1, blue line) in the case of PEI-DNA titration, whereas, all peaks were exothermic in the case of PEI addition to the buffer (i.e., blank titration, red line). The reproducibility among four experiments was very good (not shown). The raw data were integrated and the integrated data is shown in Fig. 2 a after subtracting the blank data. The thermodynamic parameters (Table 1) were obtained using the model proposed by Kim et al. (16). The parameters for the titration in MES buffer (pH 7) and in water (adjusted to pH 7 by 0.1 M NaOH) were obtained in a similar manner. The integrated titration data in MES buffer (Fig. 2 b) was similar to the data in HEPES buffer, whereas that of water was quite different. Based on the fitted parameters in Table 1, the enthalpies observed in buffers differed from the values obtained in water. We speculated that this was due to the protonation/deprotonation effect when the complex was formed. Fig. 3 is the observed enthalpy as a function of buffer ionization enthalpy. Based on the curve fit, 0.095 fraction of nitrogen of PEI is expected to be deprotonated when 1 fraction of nitrogen binds to DNA in the first binding mode (the slope of line in Fig. 3 a). On the other hand, 0.18 fraction of nitrogen of PEI is protonated when 1 fraction of nitrogen binds to DNA in the second binding mode (the slope of line in Fig. 3 b). The intrinsic enthalpy (the intersection of line on y axis in Fig. 3 a) at the first binding mode is 1.1 kJ/mol, and −0.88 kJ/mol at the second binding mode (the intersection of line on y axis in Fig. 3 b). The binding constant (K1) at first binding mode is 1.0 × 106 M−1 in either MES and HEPES buffer, but 1.0 × 105 M−1 in water. The stoichiometric number (N1) of first binding mode is 1.3 or 1.4 in the buffers, but 2.0 in water. The observed differences between the parameters obtained in the buffer and in water are likely due to pH change in the water, as discussed later. On the other hand, the stoichiometric number (N2) of the second binding mode was similar irrespective of the solvent. The binding constant (K2) at the second binding mode was decreased in MES buffer. Although this issue was not explored in detail, this may be due to increased Na+ concentration (i.e., the cation in NaOH used to adjust pH of solutions) present in this buffer.
Figure 1.
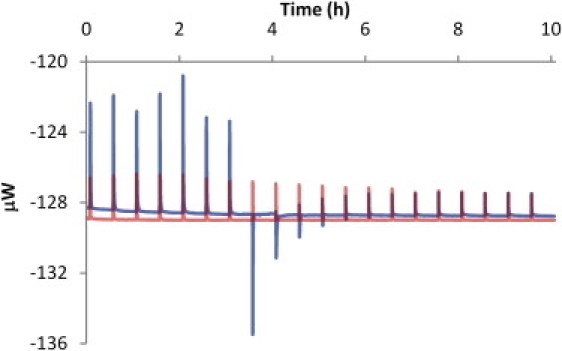
Calorimetric thermograms for titration of 20 mM PEI into 1.5 mM DNA in HEPES buffer (blue line) and into HEPES buffer alone without DNA (red line).
Figure 2.
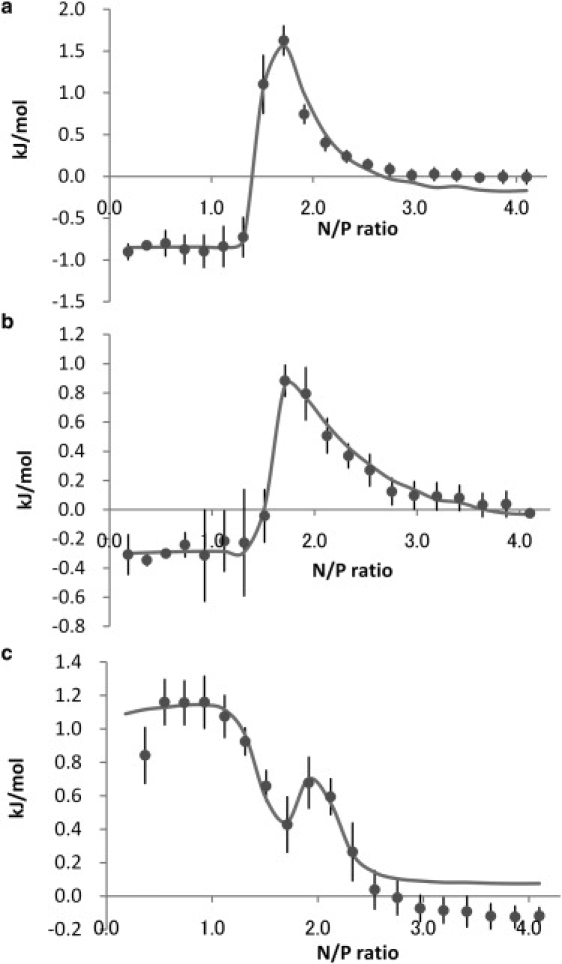
Integrated data obtained from titration of 20 mM PEI into 1.5 mM DNA (a) in 0.1 M HEPES buffer at pH 7, (b) in 0.1 M MES buffer at pH 7, and (c) in water adjusted to pH 7 by NaOH. The first point in c is out of figure (−1.4 ± 1.0 kJ/mol). The solid line represents the fitting data using the model proposed by Kim et al. (16). The data points are quadruplicate measurements.
Table 1.
Fitting parameters for the model proposed by Kim et al. (16)
| ΔH1 (kJ/mol) | K1 (M−1) | N1 | N3 | ΔH2 (kJ/mol) | K2 (M−1) | N2′ | N2 (N2′− N3) | |
|---|---|---|---|---|---|---|---|---|
| HEPES | −0.85 | 1,000,000 | 1.3 | 1.4 | 2.8 | 20,000 | 1.7 | 0.3 |
| MES | −0.30 | 1,000,000 | 1.4 | 1.5 | 1.8 | 5000 | 1.8 | 0.3 |
| H2O | 1.1 | 100,000 | 2.0 | 1.35 | −0.88 | 200,000 | 1.75 | 0.4 |
The errors are within 10%.
Figure 3.
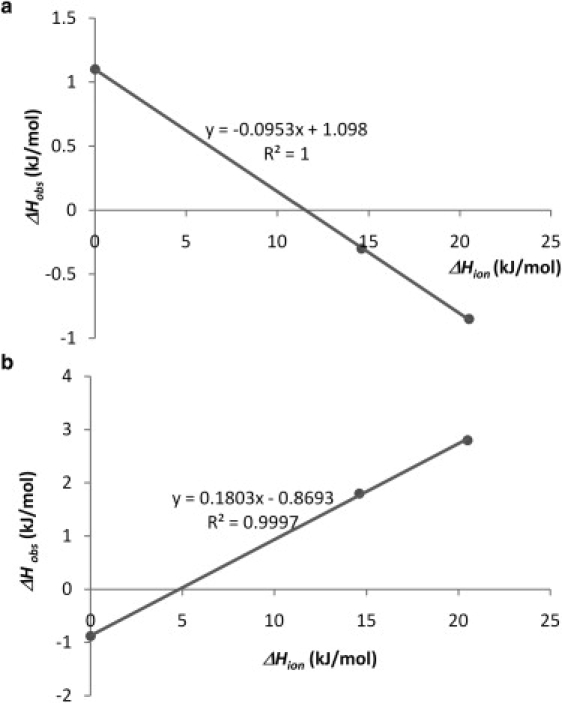
The observed enthalpy of (a) first and (b) second binding stage of PEI to DNA versus buffer ionization enthalpies that are 20.5 kJ/mol for HEPES buffer and 14.6 kJ/mol for MES buffer (33).
Fig. 4 shows the integrated data obtained from titrations of 20 mM PEI into 1.5 mM DNA in 0.1 M NaCl at different pH values. We used the SSIS model (17) for fitting these data because the heat of second binding site disappears in this condition. As shown in Table 2, the enthalpy change (ΔH1) seems to be pH independent. On the other hand, the binding constant (K1) and the stoichiometric number (N1) were dependent on the solution pH. K1 was decreased whereas N1 was increased as the pH was increased. Fig. 5 summarizes the pH change of the respective solutions during titration. The pH was increased initially, followed by a decrease. The pH change was relatively large when PEI was added to the DNA solution in water (i.e., without NaCl), whereas the change was less in the presence of NaCl. The point of pH decline seems to correlate with the N.
Figure 4.
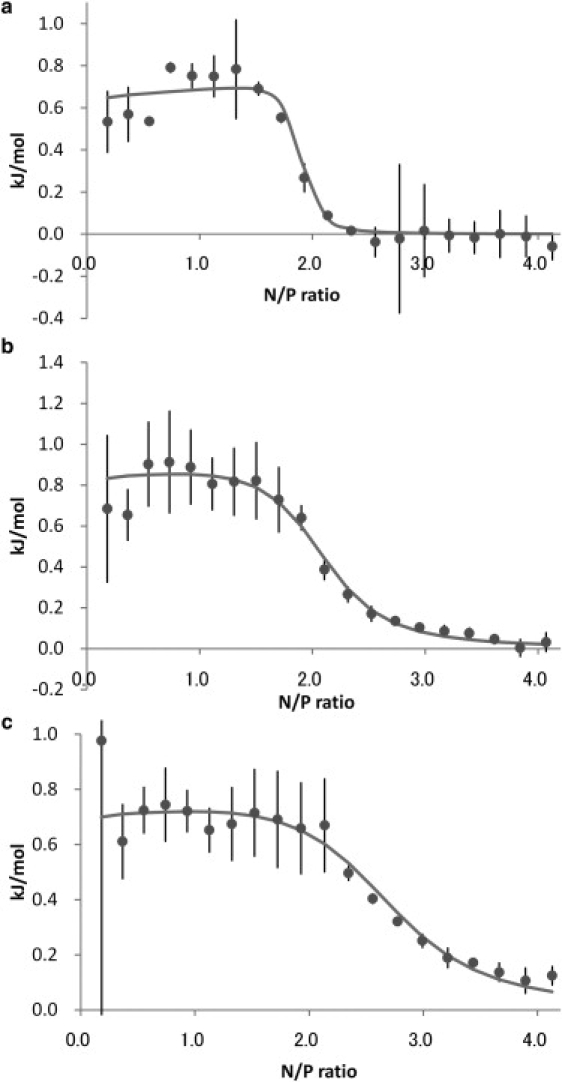
Integrated data obtained from titration of 20 mM PEI into 1.5 mM DNA in 0.1 M NaCl. The pH values of the PEI solutions are (a) 6, (b) 7, and (c) 8. The solid line represents the fitting data using the SSIS model. The data acquisition was repeated at least two times and the data are summarized as mean ± SD of at least duplicate measurements.
Table 2.
Fitting parameters for SSIS model
| ΔH1 (kJ/mol) | K1 (M−1) | N1 | |
|---|---|---|---|
| pH 6 | 0.65 | 200,000 | 1.8 |
| pH 7 | 0.85 | 18,000 | 2.0 |
| pH 8 | 0.72 | 10,000 | 2.6 |
The errors are within 10%.
Figure 5.
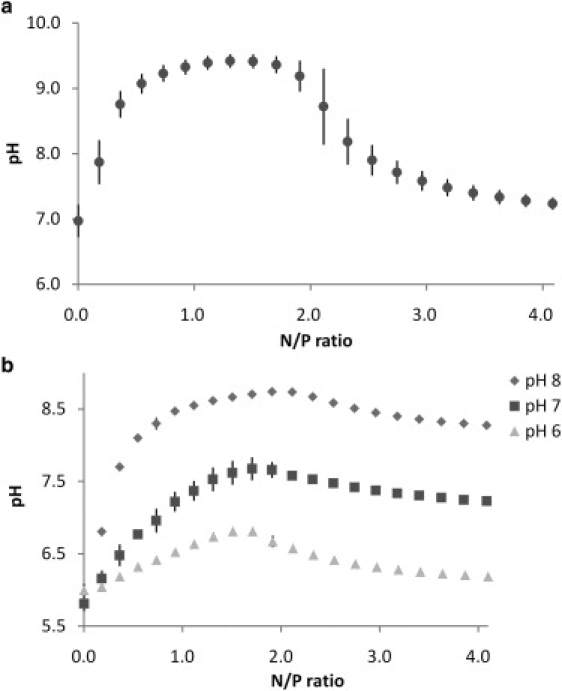
pH change obtained from titration of (a) 20 mM PEI (pH 7) into 1.5 mM DNA in water (pH7), and (b) 20 mM PEI (pH 6, 7, and 8) into 1.5 mM DNA in 0.1 M NaCl. The data points are mean ± SD of (a) four measurements and (b) duplicate measurements.
To independently verify changes in DNA solution during the titrations, a centrifugation assay was used to examine the amount of free DNA remaining in solution at different N/P ratio (Fig. 6). The data supported the correlation between the binding affinity and the pH; the PEI had a slightly greater tendency to form a complex with DNA at pH 6 as compared to the pH 7. On the other hand, more PEI molecules at pH 8 were needed to precipitate the DNA.
Figure 6.
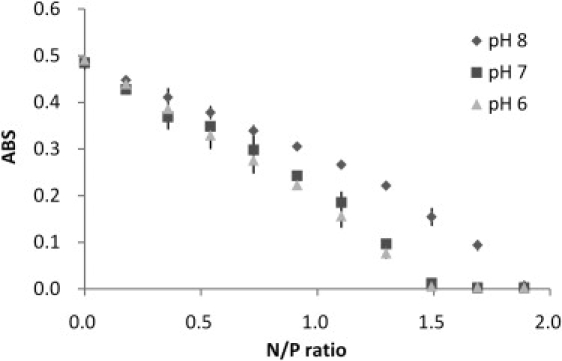
The free DNA concentration remaining in solution after PEI binding and centrifugation. The data points are duplicate measurements. The data points larger than N/P = 2 were not shown because their values were 0.
To elucidate the protonation ratio of PEI molecule at each pH, the titration of HCl into PEI solution was carried out (Fig. 7). The data were fitted with the model proposed by Suh et al. (19) using α = 0.260 and β = −1.51. The pKa and the fraction of protonated nitrogen of PEI molecule at pH 6, 7, and 8 were calculated using those parameters (Table 3). About 50% of nitrogens in PEI molecule were protonated at pH 6, whereas only ∼21% was protonated at pH 8.
Figure 7.
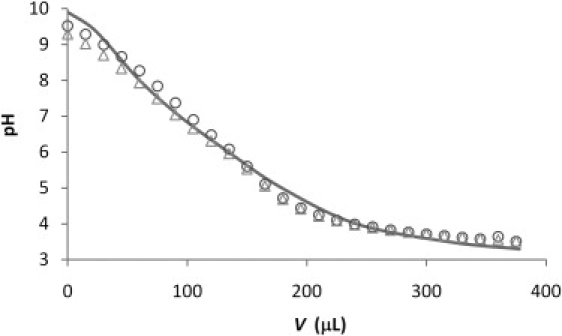
The pH value obtained as a function the volume (V) of 0.1 M HCl added to 1 mM PEI solution (28.5 mL). The solid line represents the fitting data using the model proposed by Suh et al. (19) for measurements carried out in duplicate.
Table 3.
pKa and the fraction of protonated nitrogen in PEI molecule calculated from the model proposed by Suh et al. (19)
| pH 6 | pH 7 | pH 8 | pH 9 | |
|---|---|---|---|---|
| pKa | 5.95 | 6.69 | 7.43 | 8.17 |
| Fraction of N+ (%) | 47 | 33 | 21 | 13 |
Discussion
It is known that the structural dispersity of commercial PEI in terms of in molecular weight and architecture is large. Previous studies using ITC to investigate PEI-DNA interactions used 2–750 kDa PEI without purification (9,10), so that the observed behavior is expected to be representative of a wide ensemble of molecular weights and architectures. To minimize the effect of heterogeneity, 600 Da PEI were used in this study, which is the smallest commercial product available. These results indicated that the PEI had at least two modes of binding to DNA. One mode was when the PEI is deprotonated on binding to DNA (the negative slope of line in Fig. 3 a). According to the result of FTIR spectra analysis by Zho and Li (4), PEI is capable of interacting with the DNA bases. Our study indicated that the intrinsic enthalpy change of this binding was positive (the intersection of line on y axis in Fig. 3 a), so that this interaction seemed to be entropy driven. This was likely to be attributed to the release of water molecules bound strongly to the groove of DNA (20). Such an interaction is analogous to the DNA binding of certain type of proteins, whose binding to the DNA groove was entropy driven (21). Considering all these factors, the first binding mode was likely to represent the groove binding by the PEI.
Another likely binding mode is the external binding of PEI to the phosphate backbone of DNA, through which DNA is condensed into a toroidal form. The intrinsic enthalpy change (the intersection of line on y axis in Fig. 3 b) for this interaction was found to be negative in this study, and PEI was protonated when PEI bound to DNA in this second mode (the positive slope of line in Fig. 3 b). The absolute value of the intrinsic enthalpy change of second binding mode (0.88 kJ/mol) was nearly equal to the value of first binding mode (1.1 kJ/mol). This may indicate that the interaction in second binding mode occurred in reverse of the first binding mode; i.e., the water molecule would move back into the groove of DNA. At this time, PEI would have moved from the groove to the phosphate backbone and became protonated. Rau and Parsegian (22) have proposed that the attractive hydration force was important to condense a DNA molecule. Although condensation occurs after the neutralization of DNA charges, the PEI binding to the phosphate backbone of DNA would be athermic because the enthalpy of electrostatic interaction is zero (23). It is known that the DNA condensation is caused when ∼89–90% of its charge is neutralized by counterions (24). Patel and Anchordoquy assume the datum at molar ratio 0.15 to represent DNA condensation caused by spermine (see Fig. 4 A in Patel and Anchordoquy (8)), but it would be due to the formation of basepairs of the region of single strand in supercoiled DNA (12,25). In fact, their electron microscopy results representing the DNA toroidal structure were not observed at this molar ratio (8).
Wiethoff et al. (5) have reported that bis-ethidium could rebind to PEI–DNA complex at N/P ratio >∼2 (see Fig. 3 in Wiethoff et al. (5)). This result supports our explanation. The bis-ethidium used in that study had an ethylenediamine linker between the chromophores (26–28). The linker region would be placed in the groove of DNA when the dye was bound to DNA. The dye was kicked out from the groove as the PEI was bound to the DNA initially. The dye returned to its original position when the PEI was rearranged accompanied with the condensation of DNA. The DSC data by Choosakoonkriang et al. (9) back up this explanation. The DNA had a melting peak between 95–110°C, where the peak disappeared at low N/P ratios and reappeared at high N/P ratios. It is known that the change of isobaric heat capacity that obtained from the DSC data is believed to arise purely from molecular solvation or desolvation associated with binding (8).
The lower K1 value obtained in water as compared to the K1 values in buffers should be attributed to more drastic change of pH in water. The pH was increased to >9 (Fig. 5 a), and the fraction of protonated nitrogen of PEI was <13% in water. Accordingly, a part of the PEI molecule would have bound electrostatically to the phosphates of DNA and the other part bound to the groove of DNA. Therefore, the decrease of positive charge of PEI (as a result of pH change) caused a reduction in the affinity of the polymer to the DNA.
The ΔH for the second binding mode (i.e., phosphate backbone binding accompanied with DNA condensation) almost disappeared under 0.1 M NaCl conditions. The possible reasons for this phenomenon are: 1), the screening of the negative charge of DNA phosphates by Na+ inhibits the rearrangement of PEI; 2), the placement of Na+ in the groove of DNA hinders the binding of PEI to the position; and/or 3), the number of water molecules entering the groove was reduced and the ions replaced the water at these sites. In any event, the reduced ΔH corresponded to smaller pH change under these conditions (Fig. 5). Although a slight heat change of this binding mode was observed, we could not obtain reliable thermodynamic parameters under the binding conditions because the experimental error was larger than the value itself (Fig. 4, b and c). It will be important to investigate the effect of NaCl on PEI-DNA interaction, but it was not possible to do this in this study due to unusual titration curves obtained when we attempted to use NaCl in a buffered environment (i.e., HEPES).
The different ΔHobs values obtained between in MES and HEPES buffers was due to the difference in buffer ionization enthalpy. Choosakoonkriang et al. (9) have proposed that the PEI should be protonated to bind to DNA. According to Fig. 5 B in Choosakoonkriang et al. (9), 10–20% of PEI nitrogens are protonated at pH 7.0. That seems to be corresponding to our second mode of binding where external PEI binding to the phosphate backbone was involved. However, we found that solutions become cloudy when PEI was added to phosphate buffers, which was used in that study. They might be indicative of weaker buffer concentration as compared to our conditions. In that case, the buffering effect of PEI may become larger and the values of ionic enthalpies of buffers are not available due to the effect of PEI buffering. Unlike the PEI, the DNA structure is not expected to be influenced by the pH change in the medium because the pKa of DNA phosphodiester bond is ∼3, and it will remain charged at all times under the experimental conditions.
We believe the binding features of PEI to DNA reported in this study will have direct implications in its transfection efficiency. As PEI-DNA complex is intemalized through endocytic pathway (1,2), the PEI will undergo a pH change from 7.2–7.4 on the cell surfaces to 5.5–6.0 in the endosomes, to ∼5.0 in acidic endosomes/lysosomes (29). The PEI binding to DNA is expected to be progressively tighter in this pathway, so that DNA could be protected from the nuclease attack in acidic endosomes. On endosomal release, most likely due to its proton-sponge effect (30), PEI might elevate the cytosolic pH by as much as 0.4 units (31) and manifest a lower affinity to its DNA cargo. The binding of PEI to DNA might be further weakened on nuclear uptake due to basic nuclear environment whose pH is 0.3–0.5 unit higher than the cytoplasm (32). Consequently, PEI dissociation from DNA and the readout of genetic information on exogenous DNA may become possible. The pH-dependent binding behavior of PEI to DNA on pH might be a key reason why PEI is one of the most effective transfection agents.
Conclusions
Based on analysis of isothermal titration calorimetry analysis, we showed that the PEI had two modes of binding to the DNA molecules. One of them was attributed to DNA groove binding, and the other to external phosphate backbone binding that also involved DNA condensation. The PEI was deprotonated in the first binding mode and protonated in the second binding mode. The absolute value of the intrinsic enthalpy change in the second binding mode was nearly equal to the value of first binding mode, however, whereas the first binding mode was endothermic, the second binding mode was exothermic. This heat may reflect the dissociation and association of water molecule to the groove of DNA. This result implies that the PEI may rearrange from the groove to the phosphate backbone of DNA when DNA is condensed.
Acknowledgments
The authors thank Ms. Vanessa Incani for assistance in these experiments. The ITC instrument used in this study was purchased through a grant from the Natural Sciences and Engineering Council of Canada.
This study was supported by the Institute of National College of Technology in Japan (K.U.), the Natural Sciences and Engineering Council of Canada, the Canadian Institutes of Health Research, and Alberta Advanced Education and Technology (to H.U.).
Contributor Information
Kuniharu Utsuno, Email: ukuniharu@yahoo.ca.
Hasan Uludag, Email: huludag@ualberta.ca.
References
- 1.Godbey W.T., Wu K.K., Mikos A.G. Poly(ethylenimine) and its role in gene delivery. J. Control. Release. 1999;60:149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 2.Godbey W.T., Wu K.K., Mikos A.G. Improved packing of poly(ethylenimine)/DNA complexes increases transfection efficiency. Gene Ther. 1999;6:1380–1388. doi: 10.1038/sj.gt.3300976. [DOI] [PubMed] [Google Scholar]
- 3.Dunlap D.D., Maggi A., Monaco L. Nanoscopic structure of DNA condensed for gene delivery. Nucleic Acids Res. 1997;25:3095–3101. doi: 10.1093/nar/25.15.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y.L., Li Y.Z. The interaction of poly(ethylenimine) with nucleic acids and its use in determination of nucleic acids based on light scattering. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2004;60:377–384. doi: 10.1016/s1386-1425(03)00243-9. [DOI] [PubMed] [Google Scholar]
- 5.Wiethoff C.M., Gill M.L., Middaugh C.R. A fluorescence study of the structure and accessibility of plasmid DNA condensed with cationic gene delivery vehicles. J. Pharm. Sci. 2003;92:1272–1285. doi: 10.1002/jps.10391. [DOI] [PubMed] [Google Scholar]
- 6.Sharma V.K., Thomas M., Klibanov A.M. Mechanistic studies on aggregation of polyethylenimine-DNA complexes and its prevention. Biotechnol. Bioeng. 2005;90:614–620. doi: 10.1002/bit.20444. [DOI] [PubMed] [Google Scholar]
- 7.Hellweg T., Henry-Toulme N., Roux D. Interaction of short DNA fragments with the cationic polyelectrolyte poly(ethylene imine): a dynamic light scattering study. Colloids Surf. A Physicochem. Eng. Asp. 2000;163:71–80. [Google Scholar]
- 8.Patel M.M., Anchordoquy T.J. Contribution of hydrophobicity to thermodynamics of ligand-DNA binding and DNA collapse. Biophys. J. 2005;88:2089–2103. doi: 10.1529/biophysj.104.052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choosakoonkriang S., Lobo B.A., Middaugh C.R. Biophysical characterization of PEI/DNA complexes. J. Pharm. Sci. 2003;92:1710–1722. doi: 10.1002/jps.10437. [DOI] [PubMed] [Google Scholar]
- 10.Ikonen M., Murtomäki L., Kontturi K. Controlled complexation of plasmid DNA with cationic polymers: effect of surfactant on the complexation and stability of the complexes. Colloids Surf. B Biointerfaces. 2008;66:77–83. doi: 10.1016/j.colsurfb.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Matulis D., Rouzina I., Bloomfield V.A. Thermodynamics of DNA binding and condensation: isothermal titration calorimetry and electrostatic mechanism. J. Mol. Biol. 2000;296:1053–1063. doi: 10.1006/jmbi.1999.3470. [DOI] [PubMed] [Google Scholar]
- 12.Utsuno K. Thermodynamics of DNA condensation caused by mn2+ binding. Chem. Pharm. Bull. (Tokyo) 2008;56:247–249. doi: 10.1248/cpb.56.247. [DOI] [PubMed] [Google Scholar]
- 13.Prevette L.E., Kodger T.E., Lynch M.L. Deciphering the role of hydrogen bonding in enhancing pDNA-polycation interactions. Langmuir. 2007;23:9773–9784. doi: 10.1021/la7009995. [DOI] [PubMed] [Google Scholar]
- 14.Prevette L.E., Lynch M.L., Reineke T.M. Correlation of amine number and pDNA binding mechanism for trehalose-based polycations. Langmuir. 2008;24:8090–8101. doi: 10.1021/la800120q. [DOI] [PubMed] [Google Scholar]
- 15.Prevette L.E., Lynch M.L., Reineke T.M. Amide spacing influences pDNA binding of poly(amidoamine)s. Biomacromolecules. 2010;11:326–332. doi: 10.1021/bm900824g. [DOI] [PubMed] [Google Scholar]
- 16.Kim W., Yamasaki Y., Kataoka K. Development of a fitting model suitable for the isothermal titration calorimetric curve of DNA with cationic ligands. J. Phys. Chem. B. 2006;110:10919–10925. doi: 10.1021/jp057554e. [DOI] [PubMed] [Google Scholar]
- 17.Frelre E., Mayorgal O.L., Straume M. Isothermal titration. Anal. Chem. 1990;62:950A. [Google Scholar]
- 18.Baker B.M., Murphy K.P. Evaluation of linked protonation effects in protein binding reactions using isothermal titration calorimetry. Biophys. J. 1996;71:2049–2055. doi: 10.1016/S0006-3495(96)79403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suh J., Paik H.-J., Hwang B.K. Ionization of poly(ethylenimine) and poly(allylamine) at various pH's. Bioorg. Chem. 1994;22:318–327. [Google Scholar]
- 20.Egli M., Tereshko V., Manoharan M. X-ray crystallographic analysis of the hydration of A- and B-form DNA at atomic resolution. Biopolymers. 1998;48:234–252. doi: 10.1002/(SICI)1097-0282(1998)48:4<234::AID-BIP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 21.Dragan A.I., Klass J., Privalov P.L. DNA binding of a non-sequence-specific HMG-D protein is entropy driven with a substantial non-electrostatic contribution. J. Mol. Biol. 2003;331:795–813. doi: 10.1016/s0022-2836(03)00785-x. [DOI] [PubMed] [Google Scholar]
- 22.Rau D.C., Parsegian V.A. Direct measurement of the intermolecular forces between counterion-condensed DNA double helices. Evidence for long range attractive hydration forces. Biophys. J. 1992;61:246–259. doi: 10.1016/S0006-3495(92)81831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson C.F., Record M.T. Salt-nucleic acid interactions. Annu. Rev. Phys. Chem. 1995;46:657–700. doi: 10.1146/annurev.pc.46.100195.003301. [DOI] [PubMed] [Google Scholar]
- 24.Wilson R.W., Bloomfield V.A. Counterion-induced condensation of deoxyribonucleic acid. a light-scattering study. Biochemistry. 1979;18:2192–2196. doi: 10.1021/bi00578a009. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka H., Mielke S.P., Kawai T. Visualization of the detailed structure of plasmid DNA. J. Phys. Chem. B. 2008;112:16788–16792. doi: 10.1021/jp804634s. [DOI] [PubMed] [Google Scholar]
- 26.Gaugain B., Barbet J., Le Pecq J.B. DNA bifunctional intercalators. I. Synthesis and conformational properties of an ethidium homodimer and of an acridine ethidium heterodimer. Biochemistry. 1978;17:5071–5078. doi: 10.1021/bi00617a001. [DOI] [PubMed] [Google Scholar]
- 27.Gaugain B., Barbet J., Le Pecq J.B. DNA bifunctional intercalators. 2. Fluorescence properties and DNA binding interaction of an ethidium homodimer and an acridine ethidium heterodimer. Biochemistry. 1978;17:5078–5088. doi: 10.1021/bi00617a002. [DOI] [PubMed] [Google Scholar]
- 28.Rentzeperis D., Medero M., Marky L.A. Thermodynamic investigation of the association of ethidium, propidium and bis-ethidium to DNA hairpins. Bioorg. Med. Chem. 1995;3:751–759. doi: 10.1016/0968-0896(95)00056-m. [DOI] [PubMed] [Google Scholar]
- 29.Asokan A., Cho M.J. Exploitation of intracellular pH gradients in the cellular delivery of macromolecules. J. Pharm. Sci. 2002;91:903–913. doi: 10.1002/jps.10095. [DOI] [PubMed] [Google Scholar]
- 30.Akinc A., Thomas M., Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 31.Ira M.Y., Krishnamoorthy G. DNA vector polyethylenimine affects cell pH and membrane potential: a time-resolved fluorescence microscopy study. J. Fluoresc. 2003;13:339–347. [Google Scholar]
- 32.Seksek O., Bolard J. Nuclear pH gradient in mammalian cells revealed by laser microspectrofluorimetry. J. Cell Sci. 1996;109:257–262. doi: 10.1242/jcs.109.1.257. [DOI] [PubMed] [Google Scholar]
- 33.Cooper A., Johnson C.M. Introduction to microcalorimetry and biomolecular energetic. In: Jones C., Mulloy B., Thomas A.H., editors. Microscopy, Optical Spectroscopy, and Macroscopic Techniques. Vol. 22. Humana Press; Totowa, NJ: 1994. pp. 109–124. [DOI] [PubMed] [Google Scholar]


