Abstract
The motor protein myosin II plays a crucial role in muscle contraction. The mechanical properties of its coiled-coil region, the myosin rod, are important for effective force transduction during muscle function. Previous studies have investigated the static elastic response of the myosin rod. However, analogous to the study of macroscopic complex fluids, how myosin will respond to physiological time-dependent loads can only be understood from its viscoelastic response. Here, we apply atomic force microscopy using a magnetically driven oscillating cantilever to measure the dissipative properties of single myosin rods that provide unique dynamical information about the coiled-coil structure as a function of force. We find that the friction constant of the single myosin rod has a highly nontrivial variation with force; in particular, the single-molecule friction constant is reduced dramatically and increases again as it passes through the coiled-uncoiled transition. This is a direct indication of a large free-energy barrier to uncoiling, which may be related to a fine-tuned dynamic mechanosignaling response to large and unexpected physiological loads. Further, from the critical force at which the minimum in friction occurs we determine the asymmetry of the bistable landscape that controls uncoiling of the coiled coil. This work highlights the sensitivity of the dissipative signal in force unfolding to dynamic molecular structure that is hidden to the elastic signal.
Introduction
Myosin plays a key role in muscle contraction. The heavy chain of skeletal myosin II consists of the motor domain (head), the myosin-light-chain-binding domain (neck), and the dimerization domain (tail). Two myosin heavy chains dimerize through the formation of a parallel two-stranded α-helical coiled-coil ∼150 nm long, known as the myosin rod. In myofibrils, the myosin dimers assemble into thick filaments and form the sarcomere together with actin (thin) filaments. These filaments are mutually connected by cross-bridges composed mainly of myosin head and neck domains and slide relative to each other during muscle contraction. The elasticity of the myosin cross-bridge has been thought to play an important role as an energy store to generate power without simultaneous displacement of all the myosin cross-bridges in the filaments (1). Not only the cross-bridge, but the actin and myosin filaments themselves, change their length during muscle contraction (2,3). The motion of the myosin rod region may be related to effective force generation and/or transduction within the myofibril.
Since physiological loads are time-dependent, the viscoelastic response should be characterized to clarify how the myosin rod responds to such loads. In previous studies, the viscoelasticity of muscle fibers (4–7) and myosin in bulk solution (8) have been investigated, but interpretation is difficult due to the complexity of the muscle assembly. To gain greater insight, it is necessary to investigate the viscoelasticity of individual components. To date, the elastic response has been characterized only in a static setting, using atomic force microscopy (AFM), and in that study, the force-extension profile of a single myosin rod was shown to exhibit a characteristic plateau, indicating force-induced unfolding of the coiled coil (9). Further studies have suggested that uncoiling of both the coiled-coil superhelix and the α-helices is involved in the unfolding transition (10).
Direct investigation of the viscoelasticity of single biomolecules is now possible with novel AFM-based techniques that use thermal fluctuations (11) or magnetically driven oscillation (12–15) of an AFM cantilever. The viscoelastic properties at each extension or force can be measured, although the frequency is limited to near the resonance frequency of the cantilever. Previous studies on single molecules of polysaccharides (11,13), poly-(ethylene glycol) (15) and unstructured, fully extended polypeptide chains (14,16) have revealed that 1), the viscosity of a single-molecule chain is dominated by the internal friction of the stretched molecule, and that the friction working between the surrounding solvent molecules and the stretched molecule is negligible; 2), the nontrivial behavior of the viscoelastic constants with force can be intimately linked to the energy landscape of the molecule. With dextran, for example, a minimum in the profile of internal chain friction versus applied force was shown to be a fingerprint of a large microscopic free-energy barrier (compared to kBT) for interconversion, such as in the chair-boat transition of the glucopyranose ring (17). Single-molecule viscoelasticity measurements of myosin molecules are thus expected to provide unique information about the dynamics of such processes, such as the uncoiling transition previously observed (9).
Materials and Methods
Sample preparation
Purified rabbit skeletal myosin II (9.4 mg/ml) was a generous gift from Prof. P. Knight (University of Leeds, Leeds, United Kingdom). (C47S C63S I27)5 is a concatenated pentameric variant of the I27 domain (or I91, using the alternative nomenclature) of the distal region of the I-band segment of human cardiac titin. (C47S C63S I27)5 was expressed in Escherichia coli and purified as described previously (18,19). Typically, 2 μl of myosin or I27 solution (1 or 0.05 mg/ml, respectively) (20) was dropped onto a freshly template-stripped gold surface. After incubation for 5 min, the gold surface was rinsed with the measurement buffer (600 mM sodium acetate, 600 mM KCl, 25 mM imidazole, 4 mM MgCl2, and 1 mM dithiothreitol, pH 7.4) to remove unbound proteins. All measurements were performed at room temperature (∼25°C).
Single-molecule viscoelasticity measurement
The apparatus and procedure for single-molecule viscoelasticity measurements using a magnetically driven oscillation of the AFM cantilever have been described previously (14). Fig. 1 shows a schematic diagram of the system. A magnetized AFM cantilever stretching a single myosin rod molecule is oscillated by an external AC magnetic field. The deflection, z-sensor signal, and outputs from a lock-in amplifier (7265, Signal Recovery, Oak Ridge, TN) were recorded by an auxiliary personal computer (PC) (Fig. 1, PC2) via a DAQ board (NI-6014, National Instruments, Austin, TX) with a sampling rate of 50 kHz and were time-averaged for 3 ms before analysis, as described below.
Figure 1.
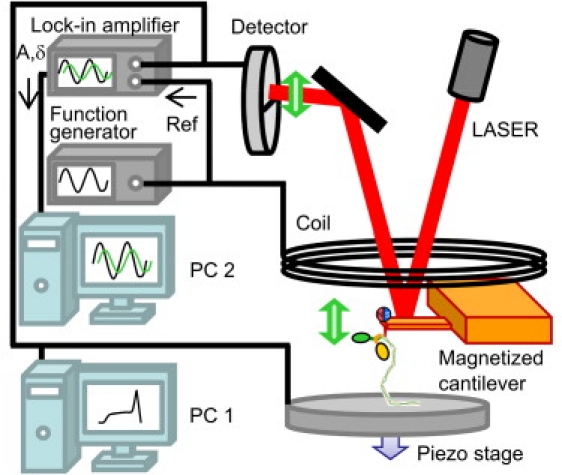
A scheme of the experimental setup for single-molecule viscoelastic measurement. An oscillatory force is applied to the cantilever through a magnetic bead attached at the tip of the cantilever. The AC signal (generated by a function generator) is analyzed by a lock-in amplifier and captured in PC2. PC1 controls the piezo stage to perform conventional single-molecule force spectroscopy.
We used a small, rectangular cantilever (BL-AC40TS-C2 Biolever-mini, Olympus, Tokyo, Japan) whose spring constant and resonance frequency are typically 0.1 N/m and 20 kHz (in fluid), respectively. An NdBFe particle was glued at the edge of the backside of the cantilever for strong magnetization. The use of a small cantilever facilitates measurement of the viscoelasticity of a single protein molecule with increased signal/noise ratio due to the resultant decrease in hydrodynamic drag of the cantilever. In the measurements, a part of myosin or (C47S C63S I27)5 was picked up by the cantilever through physisorption (nonspecific interaction). The attached molecule was extended at a constant speed of 100 nm/s. During the extension, an oscillatory force was applied constantly to the cantilever through an oscillating magnetic field.
The oscillatory motion of the free cantilever was analyzed by a force-damped simple harmonic oscillator model, described previously (14). The amplitude, A, and phase shift, δ, at driving frequency ν (in Hz) are written as
| (1) |
and
| (2) |
where F is the magnetic driving force; kc, mc, and ζc are the stiffness, mass, and friction coefficients of the free cantilever, respectively; and DC, DCδ, and a are instrumental parameters (the amplitude offset, an arbitrary constant phase shift, and a linear dependence of the phase on driving frequency, respectively) (14). We determined mc and ζc of the free cantilever, F, DC, DCδ, and a by fitting the amplitude and phase spectra of the free cantilever using Eqs. 1 and 2 with a fixed value of kc obtained previously from a thermal tune of the free cantilever (see Fig. S1 in the Supporting Material). With a molecule attached between the cantilever and the substrate, the stiffness and friction of the molecule-cantilever system change accordingly. With respect to motion of the cantilever tip, this results in a parallel mechanical arrangement (11) where the total frequency-dependent modulus is a sum of the moduli of the molecule and cantilever, respectively. We assume that the mass of the cantilever-molecule system is unchanged and equal to the free cantilever mass, mc, since the mass of a single molecule is negligible (11,13–15). If we assume that the dynamics of the myosin molecule are dominated by internal conformational rearrangements and thus have a single-mode response function of a spring and dashpot in parallel, Eqs. 1 and 2 become
| (3) |
and
| (4) |
respectively. Solving Eqs. 3 and 4 for a fixed oscillation frequency, ν, km and ζm are given by
| (5) |
and
| (6) |
We choose the frequency of the oscillation to be 22 kHz, which is close to the resonance frequency of the cantilever, and because at that frequency the phase shift is sensitive to the change of km or ζm. The advantage of measuring the response at a single frequency is that it significantly reduces the time needed to acquire the whole viscoelastic spectrum, which minimizes the effect of piezo drift. In addition, it is crucial here, since the lifetime of the bond between the AFM tip and the myosin is too short to allow acquisition of a full frequency spectrum for a single molecule (∼200 ms) (13). However, this means that when the molecular response is more complicated, the viscoelastic constants may be overestimated. In general, the response of a molecule is dominated by elastic processes at low frequency (A(ω) ∼ F/km) and frictional processes at high frequency (A(ω) ∼ F/ωζm), where 1/km = Σi1/ki and 1/ζm = Σi1/ζi, where ki and ζi are the elastic and friction constants of each mode. Thus, oscillation at a single frequency in each of those regimes will produce an accurate estimate of the elastic and friction constants.
The contour length, L, was determined by fitting force f versus extension x data using the wormlike chain (WLC) model, (f = (kBT/p)(1/{4(1 − x/L)2} − 1/4 + x/L)) with a fixed persistence length of p = 0.4 nm (16).
Results and Discussion
Viscoelasticity of single myosin rods at each extended state
Single myosin rods tethered between the substrate and cantilever were stretched by retracting the piezo stage at a speed of 100 nm/s with a periodic force applied simultaneously to the cantilever by a commercial magnetic driving system (see Fig. 1 for experimental setup and Fig. 2 a for a cartoon of the extension of a myosin rod). Fig. 2, b–d, shows typical profiles of the force, elastic constant, and friction constant, respectively, of a single myosin rod as a function of extension in the buffer condition previously shown to stabilize myosin II against thermal and mechanical unfolding (9). The force-extension profile seen in Fig. 2 b is similar to that reported previously for conventional AFM force-curve measurements (9,10) and comprises three phases. For the data shown, the myosin is extended to ∼175 nm before the protein-tip interaction is broken. As this interaction is nonspecific, it can, in principle, occur anywhere along the length of the rod (including the head region). However, similar experiments on headless myosin (by enzymatic removal using papain) gave identical results (data not shown), suggesting that tip attachment does not occur in the head region. Consistent with this, the total unfolded length of the coiled-coil domain is estimated to be 184 nm (∼540 amino acids), in accord with the contour lengths observed here. Schwaiger et al. (9) found that their data were modeled well by the stretching of a two-state semiflexible WLC. The three phases in Fig. 2 reflect the straightening of the coiled coil (I), (un)folding of the coiled coils (II), and extension of the denatured polypeptide chains (III). It should be noted that at intermediate extensions (phase II), the plateau at ∼50 pN, represents local and reversible hopping between coiled-coil and uncoiled states on the scale of a few nanometers.
Figure 2.
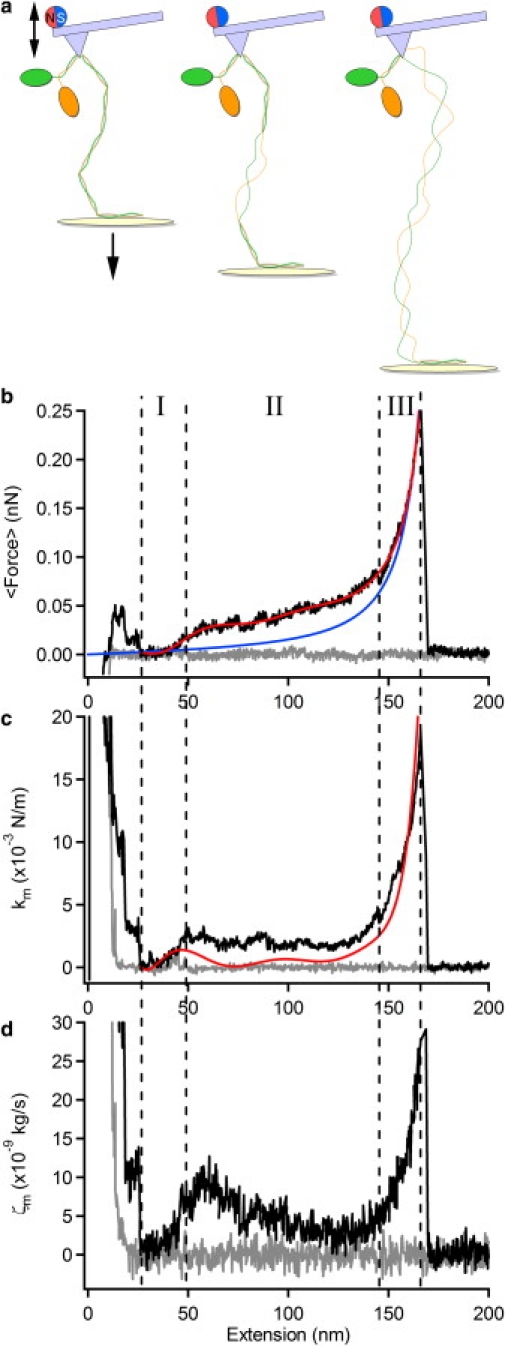
Viscoelasticity of a single myosin rod molecule. (a) A drawing representing each phase of extension of the myosin rod. Shown is the coiled-coil structure composed of a pair of α-helical strands in a superhelical arrangement. (b–d) Dependence of the force (b), elastic constant, km (c), and friction constant, ζm (d), on the extension of the rod. The approach and retraction phases are shown as gray and black lines, respectively. The elastic constant is also calculated from the derivative (dF/dx) of a polynomial fit of the force-extension profile (b, red line) and is overlaid in c as a red line. The blue line in b shows a fit to the WLC model. The cantilever was magnetically oscillated at 22 kHz and the stage was moved downward at a constant pulling speed of 100 nm/s.
The elastic and friction constants of the response of the myosin rod to an oscillatory force at a frequency of 22 kHz were calculated as described in Materials and Methods (Eqs. 5 and 6), and a typical single-molecule trace is shown as a function of extension in Fig. 2, c and d. The behavior of the elastic constant reflects the derivative of the force-extension curve, dF/dx (Fig. 2 b, red line), although, as we discuss below, there is a quantitative overestimation in the transition region, as shown by comparison to the derivative of the force-extension curve (Fig. 2 c, red line). However, the behavior of the friction constant is revealing: if solvent friction is the dominant mode of dissipation, then the measured friction should depend linearly on the extension (molecule length). This is clearly not the case, and we relate below the variation in friction with changes in dynamic flexibility of the molecule as it changes conformations under force.
As mentioned, the elastic constant is overestimated in the plateau region. As discussed in Materials and Methods, minimization of piezo drift and, in particular, the short lifetime of molecule-AFM tip bonds, necessitates measurement of the response at a single frequency. If the underlying response consists of more than a single mode (as is expected in the transition region, which consists of a series combination of coiled and uncoiled segments of the myosin rod plus a contribution from hopping between coiled and uncoiled states (17)), then it is simple to show that this will in general result in an overestimation of the total elastic and friction constants. As shown in Fig. S3, the value of the elastic constant for oscillation frequencies <5 kHz is found to decrease to a value close to that measured from the gradient of the force-extension curve; however, at 5 kHz the out-of-phase, and hence dissipative, signal would not be measurable. Meanwhile, the coincidence between the elastic constant obtained from the oscillatory measurement and that measured from the gradient of the force-extension curve, found in phase III of myosin (Fig. 2 c) and in unstructured polypeptide (see Fig. S4), indicates that in these regimes, the response is simpler and essentially single-mode at these frequencies. The elastic constant in the plateau region was mostly insensitive to changes in the amplitude of oscillation of the cantilever (2–8 nm (Fig. S3)), showing that the response measured is in the linear regime. However, we note from Fig. 2 that despite an overestimation of the elastic constant in the transition region, the extension and force at which the minimum occurs is robust.
Insights into the energy landscape of uncoiling
We performed the same experiment with a polyprotein ((C47S C63S I27)5). The force-extension curve of I27 (Fig. S4) shows a characteristic sawtooth pattern. Although the rising edges of the sawteeth at ∼60 and ∼90 nm represent the straightening of both folded and unfolded polypeptide chains, the last rising phase of the curve represents the force-extension profile of a fully unfolded polypeptide chain (Fig. S4), which fits well to a WLC model at almost all forces (Fig. S4 b). We thus use the final phase of the force-extension profile of (C47S C63S I27)5 as a model for an unstructured polypeptide, although at very low force, spontaneous refolding might somewhat affect the force-extension profile and the viscoelasticity. In Fig. 3, a and b, the elastic and friction constants, respectively, of five individual myosin rods (colored dots) and those of an unfolded polypeptide of I27 (C47S C63S I27)5 (crosses) are normalized by the contour length and plotted as a function of averaged applied force. The contour length was determined by a WLC fit of the fully unfolded region (Fig. 2 b, blue line). In addition, we note that there is good collapse of data for both myosin and I27, showing that solvent friction indeed is negligible (16,17). The red line in Fig. 3 a represents a fit of the elastic constant of the unfolded polypeptide to the WLC model (κwlc ∼ F3/2) and the red line in Fig. 3 b represents a fit of the friction constant to the frictional WLC model (16), which predicts that ζwlc ∼ F3/2. In the latter case, the monotonic increase in friction arises analogously to the elastic constant due to an increasing restriction of dynamical degrees of freedom of the chain; at the molecular scale, this friction arises from a local bending motion, including the rotation of dihedral angles of the unfolded polypeptide.
Figure 3.
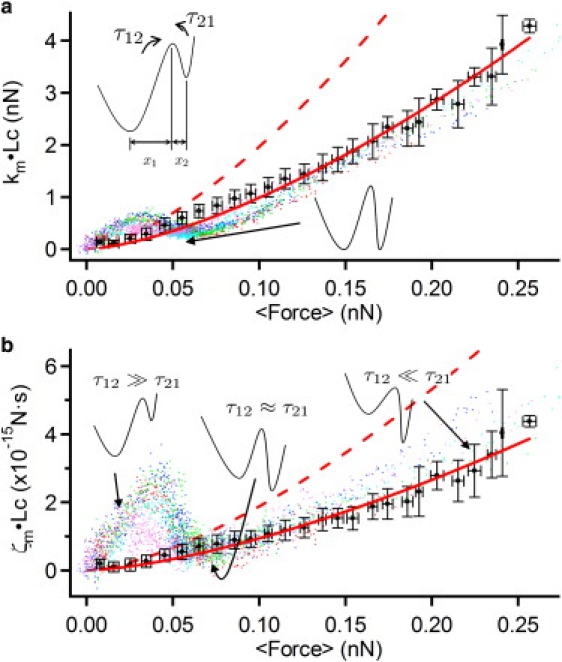
Force spectra of the elastic and friction constants of single myosin rods. The elastic (a) and friction (b) constants are normalized by the molecular length, i.e., km × Lc, where Lc is the contour length. The traces of five individual myosin rod molecules are overlaid as different-colored dots. Also shown are the elastic and friction constants of single unstructured polypeptides representing the mean ± SD of five individual molecules (solid black circles). The red dashed lines show the elastic (a) and friction (b) constants calculated for two independent unstructured polypeptide chains. The minimum in the elastic constant (a) arises when the free energies in the coiled and uncoiled states are equal, as shown schematically. At low (high) force, the friction (b) of the coiled (uncoiled) myosin rod dominates, as the large forward (backward) barrier to uncoiling, or large τ12 (τ21), freezes out dissipation due to hopping between coiled and uncoiled states. At intermediate forces, the barrier heights are reduced to the degree that hopping friction is smaller than the friction in either the coiled or uncoiled states of myosin. The force at which the minimum in friction occurs relative to the elastic constant determines the degree of asymmetry in the landscape: here, we see that the friction minimum is delayed relative to the elastic minimum which implies that the uncoiled state is closer to the transition state compared to the coiled state, as shown schematically by the bistable landscape with x1 > x2.
At low forces (0–30 pN), both the elastic and friction constants of the myosin rod increase rapidly as force increases and reaches values much higher than those for an unstructured polypeptide, indicating that the two-stranded coiled-coil structure is both statically and dynamically rigid; in other words, the large effective bending friction means that the structure responds slowly to applied forces, dissipating the energy of high-frequency motions.
As the force is further increased (from 30 pN), both parameters suddenly decrease for the myosin rod, showing minima at ∼60 pN for the elastic constant and ∼75 pN for the friction constant. The observation of a minimum in the friction constant, coincident with a plateau in the force-extension curve, is a signature of a force-induced bistable conformational transition, as has also been observed in dextran and poly-(ethylene glycol) (11,13,15). Friction on a bistable landscape with a large interconversion free-energy barrier is controlled by a characteristic time equal to the sum of the times to interconvert in the forward and backward directions (17). Force then changes the barrier heights and this modulates the friction constant, leading to a minimum at some critical force when the two interconversion times are approximately equal. The decrease in friction arises as friction due to hopping between states becomes smaller than that due to coiled-coil bending. If the landscape is asymmetric, the critical force at which the minimum in friction occurs depends on the degree of asymmetry of the bistable landscape; if x1 and x2 are the distances to the barrier from the two sides, then the excess force at which the minima occur in the friction constant compared to the elastic constant is ΔF = (kBT/Δx)ln(x1/x2), where Δx = x1 + x2 (17). Given that ΔF is positive, this determines that the distance from the coiled state to the transition state is larger than the corresponding distance from the uncoiled state. This is consistent with a picture in which the uncoiling of a number of helical turns is required before the onset of complete uncoiling at the transition state, which releases a relatively large amount of uncoiled peptide.
For F > 100 pN, although the uncoiled myosin rod consists of two polypeptide strands, the asymptotic viscoelastic properties tend toward that of a single unfolded polypeptide. This suggests that, although the coiled coil is denatured, the AFM tip is attached to only a single strand of the coiled coil. In addition, on closer inspection, there is a small increase in friction of the uncoiled myosin compared to a single unstructured polypeptide. We suggest that the origin of this increase may be due to the additional intermolecular friction between the two entangled strands in the myosin rod, still in superhelical topology (as shown schematically in Fig. 2 a). At the molecular scale, this friction would arise from local nonspecific association and dissociation between the two strands, as well as from specific interactions that stabilize the coiled coil in the native form. This demonstrates further that measuring the friction constant can be highly sensitive to extra dynamical structural detail at the molecular level, which is not available for the elastic constant.
Biological relevance and future directions
Does the myosin rod mechanically unfold in vivo? In the experimental conditions in this study (600 mM sodium acetate, pH 7.4), the plateau force of myosin rod unfolding was ∼50 pN, consistent with a previous measurement using conventional AFM in a protein unfolding study (9). In the absence of sodium acetate, unfolding forces decrease to 20–40 pN (9,10). Compared to the force required to induce unfolding, the force generated by a single power stroke of a myosin head is estimated to be 1–10 pN (21) and the unbinding force between a single myosin head and a single actin filament is 5–25 pN (22,23). Therefore, it is unlikely that unfolding of the myosin rod occurs during the normal muscle contraction cycle. In this case, the increased bending friction of the coiled coil could be useful in muscle function as a molecular shock absorber, whereby sudden loads are dissipated in the rod structure, preventing uncoiling of the coiled coil and thereby offering more robust muscle function. However, it is likely that myosin rods working in vivo may experience very high local and instantaneous tensile forces. For example, eccentric exercise, in which the sarcomere in the activated state is elongated by an opposing force greater than that generated by the muscle, damages sarcomere structure (24). With such extreme loads, local parts of the myosin rod, as well as of titin or other scaffold proteins, can partly unfold, increasing the length of the rod. After removal of the load, the rod refolds reversibly (9), i.e., the unfolding/refolding of the myosin rod might act as a secondary mechanism to maintain muscle function in the face of large unexpected loads, preventing irreversible fracture of key components. Here, the friction constant decreases with increasing tension, analogous to macroscopic shear-thinning of materials, for instance, tomato ketchup flowing when shaken or paint flowing when sheared with a brush. It is possible that this mechanism ensures that the coiled coil unfolds rapidly upon reaching a critical force, preventing local build-up of a detrimentally large stress. An alternative, or parallel, possibility is that decreased elastic and friction constants in the plateau may provide a signaling mechanism to induce cellular changes in the event of an applied mechanical stimulus. Indeed, titin kinase, at the sarcomeric M-band of myofibrils, has recently been shown to undergo mechanically modulated autoregulation (25). In contrast to the friction constant of the myosin coiled-coil rod, the free-energy barrier between coiled and uncoiled states may be used to fine-tune the dynamics and friction of the unfolding process. This suggests a target for point-mutation experiments to probe the dynamics of this transition and its molecular origin and thus further understand its possible role in muscle function.
These results hold promise that combining sensitive single-molecule measurement of dissipation with theory, computer simulation, and protein engineering experiments will offer novel insights into the role of molecular conformational change in mechanical processes in biology and shed light on important biological mechanisms such as muscle function and protein folding. More broadly, by clarifying the interplay between energy landscapes and their viscoelastic properties, such work can provide biological inspiration for the purposeful engineering design of molecular-scale devices for use in nanotechnology and nanomaterials.
Supporting Material
Four figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(10)00439-X.
Supporting Material
Acknowledgments
We thank Prof. P. Knight for his kind offer of the purified myosin sample.
This study was supported by the program Promotion of Environmental Improvement for Independence of Young Researchers under the Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and by the program Precursory Research for Embryonic Science and Technology (PRESTO) of the Japan Science and Technology Corporation (JST). Y.T. is a research fellow of the Japan Society for the Promotion of Science (JSPS).
References
- 1.Huxley A.F., Simmons R.M. Proposed mechanism of force generation in striated muscle. Nature. 1971;233:533–538. doi: 10.1038/233533a0. [DOI] [PubMed] [Google Scholar]
- 2.Wakabayashi K., Sugimoto Y., Amemiya Y. X-ray diffraction evidence for the extensibility of actin and myosin filaments during muscle contraction. Biophys. J. 1994;67:2422–2435. doi: 10.1016/S0006-3495(94)80729-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huxley H.E., Stewart A., Irving T. X-ray diffraction measurements of the extensibility of actin and myosin filaments in contracting muscle. Biophys. J. 1994;67:2411–2421. doi: 10.1016/S0006-3495(94)80728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang K., McCarter R., Ramirez-Mitchell R. Viscoelasticity of the sarcomere matrix of skeletal muscles. The titin-myosin composite filament is a dual-stage molecular spring. Biophys. J. 1993;64:1161–1177. doi: 10.1016/S0006-3495(93)81482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minajeva A., Kulke M., Linke W.A. Unfolding of titin domains explains the viscoelastic behavior of skeletal myofibrils. Biophys. J. 2001;80:1442–1451. doi: 10.1016/S0006-3495(01)76116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higuchi H. Viscoelasticity and function of connectin/titin filaments in skinned muscle fibers. Adv. Biophys. 1996;33:159–171. doi: 10.1016/s0065-227x(96)90031-3. [DOI] [PubMed] [Google Scholar]
- 7.Liu H., Miller M.S., Bernstein S.I. Paramyosin phosphorylation site disruption affects indirect flight muscle stiffness and power generation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2005;102:10522–10527. doi: 10.1073/pnas.0500945102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hvidt S., Nestler F.H., Ferry J.D. Flexibility of myosin rod determined from dilute solution viscoelastic measurements. Biochemistry. 1982;21:4064–4073. doi: 10.1021/bi00260a024. [DOI] [PubMed] [Google Scholar]
- 9.Schwaiger I., Sattler C., Rief M. The myosin coiled-coil is a truly elastic protein structure. Nat. Mater. 2002;1:232–235. doi: 10.1038/nmat776. [DOI] [PubMed] [Google Scholar]
- 10.Root D.D., Yadavalli V.K., Wang K. Coiled-coil nanomechanics and uncoiling and unfolding of the superhelix and α-helices of myosin. Biophys. J. 2006;90:2852–2866. doi: 10.1529/biophysj.105.071597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawakami M., Byrne K., Smith D.A. Viscoelastic properties of single polysaccharide molecules determined by analysis of thermally driven oscillations of an atomic force microscope cantilever. Langmuir. 2004;20:9299–9303. doi: 10.1021/la0486178. [DOI] [PubMed] [Google Scholar]
- 12.Humphris A.D.L., Tamayo J., Miles M.J. Active quality factor control in liquids for force spectroscopy. Langmuir. 2000;16:7891–7894. [Google Scholar]
- 13.Kawakami M., Byrne K., Smith D.A. Viscoelastic measurements of single molecules on a millisecond time scale by magnetically driven oscillation of an atomic force microscope cantilever. Langmuir. 2005;21:4765–4772. doi: 10.1021/la0469699. [DOI] [PubMed] [Google Scholar]
- 14.Kawakami M., Byrne K., Smith D.A. Viscoelastic study of the mechanical unfolding of a protein by AFM. Biophys. J. 2006;91:L16–L18. doi: 10.1529/biophysj.106.085019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawakami M., Byrne K., Smith D.A. Viscoelastic properties of single poly(ethylene glycol) molecules. ChemPhysChem. 2006;7:1710–1716. doi: 10.1002/cphc.200600116. [DOI] [PubMed] [Google Scholar]
- 16.Khatri B.S., Byrne K., McLeish T.C. Internal friction of single polypeptide chains at high stretch. Faraday Discuss. 2008;139:35–51. doi: 10.1039/b716418c. [DOI] [PubMed] [Google Scholar]
- 17.Khatri B.S., Kawakami M., McLeish T.C. Entropy and barrier-controlled fluctuations determine conformational viscoelasticity of single biomolecules. Biophys. J. 2007;92:1825–1835. doi: 10.1529/biophysj.106.097709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brockwell D.J., Beddard G.S., Radford S.E. The effect of core destabilization on the mechanical resistance of I27. Biophys. J. 2002;83:458–472. doi: 10.1016/S0006-3495(02)75182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zinober R.C., Brockwell D.J., Smith D.A. Mechanically unfolding proteins: the effect of unfolding history and the supramolecular scaffold. Protein Sci. 2002;11:2759–2765. doi: 10.1110/ps.0224602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi Y., Brockwell D.J., Kawakami M. The effect of temperature on mechanical resistance of the native and intermediate states of I27. Biophys. J. 2008;95:5296–5305. doi: 10.1529/biophysj.108.141275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tyska M.J., Warshaw D.M. The myosin power stroke. Cell Motil. Cytoskeleton. 2002;51:1–15. doi: 10.1002/cm.10014. [DOI] [PubMed] [Google Scholar]
- 22.Nishizaka T., Miyata H., Kinosita K. Unbinding force of a single motor molecule of muscle measured using optical tweezers. Nature. 1995;377:251–254. doi: 10.1038/377251a0. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima H., Kunioka Y., Ando T. Scanning force microscopy of the interaction events between a single molecule of heavy meromyosin and actin. Biochem. Biophys. Res. Commun. 1997;234:178–182. doi: 10.1006/bbrc.1997.6612. [DOI] [PubMed] [Google Scholar]
- 24.Agarkova I., Ehler E., Perriard J.C. M-band: a safeguard for sarcomere stability? J. Muscle Res. Cell Motil. 2003;24:191–203. doi: 10.1023/a:1026094924677. [DOI] [PubMed] [Google Scholar]
- 25.Puchner E.M., Alexandrovich A., Gautel M. Mechanoenzymatics of titin kinase. Proc. Natl. Acad. Sci. USA. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


