Abstract
Peptide hormones and neuropeptides play important roles in endocrine and neural signaling, often using G protein-coupled receptor (GPCR)-mediated signaling pathways. However, the rate of novel peptide discovery has slowed dramatically in recent years. Genomic sequencing efforts have yielded a large number of cDNA sequences that potentially encode novel candidate peptide precursors, as well as hundreds of orphan GPCRs with no known cognate ligands. The complexity of peptide signaling is further highlighted by the requirement for specific posttranslational processing steps, and these must be accomplished in vitro prior to testing newly discovered peptide precursor candidates in receptor assays. In this review, we present historic as well as current approaches to peptide discovery and GPCR deorphanization. We conclude that parallel and combinatorial discovery methods are likely to represent the most fruitful avenues for both peptide discovery as well as for matching the remaining GPCRs with their peptide ligands.
KEY WORDS: deorphanization, GPCR, peptide hormones, proprotein convertases, screening
INTRODUCTION
Peptide hormones and neuropeptides play critical roles in diverse biological phenomena, often via G protein-coupled receptor (GPCR)-mediated signaling pathways. The successful sequencing of entire genomes of many different organisms has yielded a large number of cDNA sequences that potentially encode novel candidate peptide precursors, as well as hundreds of orphan GPCRs with no known cognate ligands. Because GPCRs already represent the largest class of target molecules for pharmaceutics, GPCR deorphanization of novel bioactive peptides has the potential to both open entirely new fields of research and to provide novel therapeutics. However, the genomic identification of novel bioactive peptides has been hampered by certain disadvantageous properties of peptides, such as their small size and the lack of definitive posttranslational processing information, particularly cleavage site usage.
In the first half of this review, we present a short discussion of the cellular mechanisms by which bioactive peptides are synthesized. We follow with a discussion of past methods of peptide discovery: classical and reverse-pharmacology methods; mass spectrometric-based methods; and bioinformatics. We then describe our recent approach to peptide discovery using prohormone convertases as screening agents. The second half of the review focuses on methods to de-orphanize GPCRs.
POSTTRANSLATIONAL MODIFICATIONS OF PEPTIDE HORMONES
Peptide hormones are initially synthesized as relatively large precursor proteins which gradually mature into specific bioactive forms via the sequential action of posttranslational processing enzymes. The proprotein convertases (PCs) are the initial processing enzymes responsible for precursor maturation; these enzymes cleave C-terminally to single or double basic sites (1). Furin and PACE4 are PCs which are distributed ubiquitously and contribute to cleavage of a wide variety of secreted proteins, for example growth factor precursors and blood proteins, within the constitutive secretory pathway (2,3). In contrast, the prohormone convertases PC1/3 and PC2 (4,5), expressed predominantly in neuroendocrine tissues, represent the processing enzymes required to produce neuropeptides and peptide hormones within the regulated secretory pathway. The products of convertase processing enzymes are then C-terminally trimmed, usually by carboxypeptidase E (6,7). Other precursor-specific posttranslational modifications such as C-terminal amidation, N- or O-acylation, tyrosine sulfation, N-glycosylation, phosphorylation, and even bromination can also take place; often, these modifications are required for bioactivity (reviewed in 8–14). Amidation is the most common modification (15,16); the other posttranslational modifications mentioned occur much less frequently on perhaps 10% of known neuropeptides and peptide hormones.
CLASSICAL AND REVERSE PHARMACOLOGICAL APPROACHES TO NOVEL PEPTIDE DISCOVERY
The discovery of enkephalins exemplifies the now-classical peptide discovery method, bioactivity assays coupled with peptide purification from specific tissues (17). A more recent approach, termed “reverse-pharmacology”, follows peptide activity against orphan GPCRs that are cloned into eukaryotic expression systems and coupled to various reporter assays (18–24). With this approach, several new peptide hormones involved in important physiological phenomena were discovered within the last decade; orexin/hypocretin, involved in sleep regulation and food intake; ghrelin, which mediates appetitive mechanisms; and kisspeptin, involved in reproduction (25–27). The major advantage of this method is the ability to couple peptide and receptor in an unbiased manner. However, it can be difficult to identify efficient reporter systems for an unknown receptor, since nothing is known a priori about second messenger coupling. There are certain methods that do not require such prior knowledge (see below), and these are gaining increasing use in deorphanization studies.
MASS SPECTROMETRY APPROACHES
Mass spectrometry (MS), a technique which can be both robust and rapid, has increasingly been used in recent years for peptide discovery. Fricker and colleagues reported the presence of a novel brain peptide precursor known as proSAAS (28) using mass spectroscopic analysis. This group has also published a comprehensive analysis of brain peptides in animals that do not express the precursor processing enzymes CPE and PC2 (29–35). Endocrine cell lines have also been used to identify novel peptides under specific stimulation conditions; for example, katacalcin, a novel peptide derived from the calcitonin precursor, has recently been identified by MS (36).
In a more functional approach, Hatcher and colleagues have used MS-based methods to determine the sequences of peptides involved in circadian rhythms (37). This study used MS to compare peptide products in the mouse brain under light and dark circadian phases. Several novel peptides were present (but not identifiable) as well as known small peptides derived from proSAAS (28). Brockmann et al. have reported an interesting MS methodology that connects the production of neuropeptides—both novel and unknown—with behavior in honeybees (38). These studies confirm that MS-based methods have the potential to reveal information about temporal and spatial expression of peptides, making this a powerful method for the study of peptide function. Limitations of the method include the need for high-end spectrometers and the need to handle enormous amount of information generated during MS runs, often requiring high capacity data handling facilities coupled with special informatics programs. It can also be extremely difficult to detect scarce peptides in the presence of large quantities of other proteins. Several different secretory granule proteomics projects have attempted to identify the major secreted species present within granules obtained from different tissues; however, no peptides derived from novel precursors have been identified to date, most likely due to these abundance issues as well as to their focus on proteins rather than peptides (39–41).
BIOINFORMATICS APPROACHES
With the successful revelation of the genomes of many species, bioinformatics has become an increasingly powerful tool to identify target peptide precursor molecules from enormous amounts of genomic information. Three different groups have used hidden Markov models to establish predictive algorithms for novel peptide precursors. Mirabeau and colleagues employed an algorithm to search for signal peptide-bearing proteins using evolutionary conservation of peptide sequences among species coupled with the presence of known sequences of prohormone convertase cleavage sites (42). This group, who concluded that there were likely to be few further undiscovered peptides, identified precursors to two novel peptides, termed “spexin”, expressed in esophagus and stomach, and “augurin”, expressed in endocrine tissues and choroid plexus (42; note that augurin had actually been previously discovered and given the name of “ecrg4”; 43). A second peptide discovery group used a somewhat similar bioinformatics analysis focused on peptide conservation across species to identify one of the same novel peptides (spexin; 44). Lastly, Gustincich analyzed the mouse transcriptome for nervous system-expressed proteins, searching for small open reading frames containing cleavage sites (45).
Unfortunately, it is clear that novel peptide precursor algorithms have not yet reached a satisfactory level of predictive ability. While Gustincich identified 90 hypothetical novel proteins, and Mirabeau over 200 potential precursor proteins, both lists contain many intron-containing proteins, cytokines, defensins, interleukins, and growth factors—all of which are unlikely to represent actual protein precursors processed within the secretory pathway. Indeed, from Mirabeau’s list of 300 genes of candidate peptide precursors, 200 genes are either cytokines or defensins, neither considered likely substrates for prohormone convertases. This low accuracy of prediction results in part from insufficiently detailed knowledge as to specific site requirements for PC cleavage (46). In this regard, it is useful to mention two online cleavage prediction programs. Southey et al. have developed the NeuroPred online program, available at the University of Illinois site (47). While we have found this to be a very useful program to predict prohormone cleavage sites, experimental data indicate that this prediction algorithm is insufficiently restrictive (48). By contrast, another online cleavage prediction program, ProP 3.0, tends to underestimate the predicted hormones (48,49) Some of this lack of predictive precision derives from the fact that these programs attempt to predict cleavages of both furin and PC cleavage sites, but due to the overlapping cleavage specificities of these enzymes, it is impossible solely from sequence data to establish which sequences are best cleaved by the constitutive processing enzyme furin, and which sequences are cleaved by the two prohormone convertases PC1/3 and PC2.
In summary, at the present time, it appears to be impossible to accurately predict all peptide hormones and neuropeptide precursors using only information derived from algorithms. However, the accuracy of prediction efforts in identifying PC-cleaved peptide hormones and neuropeptides may improve after algorithms are better trained using a sufficient number of experimentally-derived datasets.
COMBINATORIAL APPROACH: BIOINFORMATICS COUPLED WITH CONVERTASE SENSITIVITY
Together with coworkers at FivePrime Therapeutics, we have taken a combinatorial approach to the identification of novel peptide hormones. We begin with bioinformatics to identify novel peptide hormone candidates from the genome which possess certain sets of traditional peptide hormone properties. First, among these is the presence of pairs of basic residues conserved at least between rats and mice, but other criteria, such as typical exon number, overall size, and secondary structure characteristics surrounding cleavage sites are also used. We remove proteins from consideration that are ubiquitously expressed, since peptide hormones and neuropeptide precursors are known to exhibit restricted patterns of expression. We then perform prohormone convertase cleavage screening reactions on the remaining candidate peptide precursors, using our recombinant prohormone convertases (50,51), either in eukaryotic expression arrays or on bacterially-expressed proteins (unpublished results). Due to the highly restricted specificity of prohormone convertases, this step reduces the number of false-positive precursor proteins. As described above, prohormone convertases all cleave at motifs involving basic residues, mostly pairs of basic residues. Each convertase differs in its preferences regarding the residues surrounding these cleavage sites, though, as discussed below, some overlap between convertases exists (2,52).
Their exquisite specificity for only a subset of basic residue cleavage sites, which has led us to term the proprotein convertases “restriction proteases” (48), may be due to their recognition of primary and secondary structural elements that extend beyond the actual cleavage site (46,53–55). Using convertases to validate putative precursors in a secondary screen reduces the problem of overly generous bioinformatics predictions, in that proteins which contain double basic residues—even highly conserved double basics—but do not represent actual substrates are simply not recognized by these proprotein convertases. In support of this idea, we have observed many examples of proteins containing bioinformatically-predicted pairs of basics that do not undergo convertase cleavage in vitro. An additional benefit of this method is that we can use MALDI-TOF mass spectroscopy to obtain direct mass information on most of the actual peptide products of proprotein convertase cleavage (successful detection depends on chemical character and length). Disadvantages of this method arise from the overlapping specificities of the proprotein convertases mentioned above. For example, PC2 can cleave at certain furin sites in various precursors; however, if PC2 is not naturally co-expressed together with the candidate precursor, it will have no opportunity to perform this cleavage physiologically, even though it may do so efficiently in vitro. Therefore, it is possible to identify prohormone convertase cleavage sites that in fact may not occur naturally. A partial solution to this problem is to examine enzyme and precursor co-expression patterns for each precursor, since certain tissues (for example, liver) totally lack PC1 and PC2. However, actual coexistence of enzyme and substrate within the same cellular compartment must eventually be confirmed experimentally.
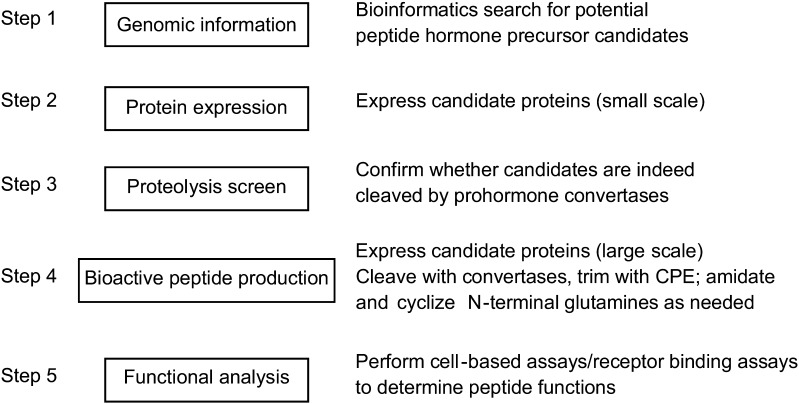
Once likely peptides are identified, function must be determined. We have used recombinant processing enzyme arrays (convertase [furin, PC1/3 or PC2], CPE, peptidyl amidating enzyme, and glutaminyl cyclase to generate 1-100 uM concentrations of natural peptide ligands from bacterially and mammalian-cell expressed precursors (48 and unpublished results; PAM and glutaminyl cyclase are added only when the primary sequence indicates that these modifications might take place). We have previously published on the efficacy of such an in vitro-generated POMC-derived peptide on receptor assays (48) and have now extended these results to proenkephalin and prodynorphin-derived peptides (unpublished results). Our workflow for generating known and novel peptide products is described in Fig. 1.
Fig. 1.
Workflow for generating known and novel peptides products
In vitro processed peptides can be studied using a range of bioassays, including testing with orphan GPCR assays as discussed later in this review. We expect that missing processing enzymes, such as sulfotransferases (required for CCK sulfation) and acylating enzymes (required for α-MSH and ghrelin synthesis), can be added in the future as they become available in recombinant form. While these modifications are rare, when present they appear to add greatly to bioactivity, and will be required to fully recapitulate natural peptide processing in vitro to produce functionally active ligands.
NOVEL PEPTIDE DISCOVERY: THE FUTURE
Historically-used peptide identification approaches have resulted in the discovery of a number of novel peptide precursors within the last two decades, namely the precursors to adrenomedullin, nociceptin/orphaninFQ, orexin/hypocretin, CART, AGRP, nesfatin, kisspeptin, pQRF/26RFamide, and neuropeptide S (18,20–22). However, the rate of identification of novel mammalian peptide precursors has slowed dramatically in recent years, with no new precursors identified in the last 2 years; this suggests that the approaches discussed above might have reached their intrinsic limits. For example, a low tissue abundance of certain peptides may preclude further reverse pharmacology approaches, and the low accuracy of prediction algorithms makes novel peptide identification solely by bioinformatics problematic. Combinatorial bioinformatics approaches that include definitive peptide properties—such as the ability to be made by convertases—and either functional and/or anatomical information are likely to represent the most fruitful approaches in the future. Mass spectroscopic identification can address certain issues of sensitivity, but still suffers from abundance issues when other proteins overwhelm scarce signals. Important novel peptides yet to be discovered include the many endogenous ligands of the orphan peptide GPCRs, as discussed below, as well as many other secreted signaling proteins likely to be involved in development, growth and control of metabolic function.
DEORPHANIZATION OF PEPTIDE GPCRs
The human genome contains genes for roughly 400 non-olfactory GPCRs (56,57); of these, approximately half are likely to be peptide GPCRs (we estimate 230). Of the GPCRs likely to be peptide receptors (based on homology with known peptide GPCRs), roughly half are orphans and our current estimate is 108. Thus, there remains a great deal to be accomplished in determining the ligands for these receptors as well as their potential role(s) in both normal physiology and disease. The peptide receptors, unlike some other subfamilies of GPCRs (e.g., the biogenic amine receptors), do not form a homogeneous group based on their sequences (see for example (58)), but are found among several major subgroups of GPCRs. The biology of these receptors is likely to be similarly diverse.
In the early history of cloning (typically by homology using degenerate oligonucleotides or low-stringency screening), almost every receptor discovered was an orphan receptor (e.g., G-21 which was subsequently demonstrated to be the 5-HT1A receptor (59)). The subsequent impact of the deorphanization of these receptors and their ligands is undeniable. While it could be argued that the law of diminishing returns may apply, it seems probable that continued deorphanization of GPCRs is likely to impact at least three fronts: (1) knowledge of the physiological role of these receptors; (2) discovery of their potential value as drug targets; and (3) elucidation of their potential to mediate off-target effects of both current and future therapeutics. Of these, the latter is likely to have the most immediate impact on human welfare and indeed new means to predict and test off-target effects of widely used GPCR ligands are already having considerable influence (60).
The primary challenge in deorphanization of GPCRs is, of course, that so little is known about them. Although their tissue distribution is well-described in various resources (e.g., (61), the Symatlas website [http://biogps.gnf.org/], the Allen Brain Atlas website [http://www.brain-map.org/], and others), and thus some preliminary inferences can be made on their possible biological roles, experimental design is complicated by lack of knowledge of their signaling pathways (including their G-protein partners), and the lack of positive controls for experimentation. However, the high degree of conservation of the sequences of these receptors among various species argues in favor of significant biological roles.
It is interesting that almost every new technology that is developed to measure GPCR activation and/or signaling has been suggested (almost immediately) to be suitable for deorphanization (see 62–67 for examples and 68–70) for review). However, despite the apparent suitability of many of these assays, relatively few studies have been published that use these assays in a systematic manner to actually deorphanize receptors. Examples of assays that have been used (with varying degrees of success), in addition to the now standard assays of radioligand binding, calcium mobilization, GTPγs binding, and modulation of cAMP levels in GPCR deorphanization are listed in Table I below. Some of these assays are made more practically useful by virtue of the fact that it is not necessary to know the “true” coupling partners of the receptors in order to carry out ligand discovery screens. It is our experience, for example, that the “best” Gα coupling partner of a GPCR expressed in yeast may very well not be the same as that seen in mammalian cells, or in native tissues, yet these GPCR-expressing yeast can be used to screen compound libraries for GPCR ligands (Kroeze & Roth, unpublished observations).
Table I.
Diversity of Methods and Tools Used to Study Orphan GPCRs
| Method | Reference |
|---|---|
| Arrestin translocation | (78) |
| Arachidonic acid release | (79) |
| Chimeric Gα proteins | (80) |
| Aequorin bioluminescence | (81) |
| Promiscuous Gα proteins | (82, 83) |
| Forskolin-stimulated luciferase | (82) |
| Receptor internalization | (84) |
| Gα13 and RhoA-dependent stress fiber formation | (85) |
| Stimulation of current in ion channel and GPCR co-transfected Xenopus oocytes | (86) |
| GPCR-Gα fusion proteins | (87) |
| Receptor chimeras | (88) |
| Phage display | (63) |
| Yeast fluorescent reporter assay | (64) |
| Yeast β-galactosidase assay | (66) |
| GPCR-promiscuous Gα fusion proteins | (89) |
| Scintillation proximity binding assay for GTPγs | (72) |
| β-lactamase reporter assay and inducible receptor expression | (74) |
| Pigment dispersion in Xenopus melanophores | (90) |
| Cytoskeletal rearrangement by surface plasmon resonance | (91) |
It can be predicted that additional newly developed assays, for example a high-throughput ERK phosphorylation assay (62) will also prove to be useful in GPCR deorphanization, but to our knowledge this assay has not yet been used for this purpose.
Many of the assays suggested for GPCR deorphanization require specialized (and expensive) instrumentation, and are thus unlikely to come into wide general use; however, some assays require little (if any) such instrumentation, for example, yeast growth assays, and yet have not been widely used for unknown reasons, despite having been established and validated more than a decade ago (65). Since the coupling specificity of these receptors is not known, it may be that more “universal” assays (e.g., 71) may prove to be valuable in deorphanization studies. Increasing miniaturization of assays (e.g., the 1,536-well format SPA assay of Johnson et al. (72)), or even the 3,456-well format β-lactamase reporter assay of Mishina et al., (73) will undoubtedly lead to “economies of scale” in large-scale deorphanization studies. The study of Bercher (74) is of particular interest; over-expression of the receptor (G2A, also known as GPR132), is toxic to cells, so an inducible system of receptor expression was devised that allowed for expansion of the cell line to be used in the absence of receptor, followed by induction of receptor expression the day before compound screening.
Although most of the assays listed in the table above are best suited for agonist discovery, constitutively active receptors, either naturally occurring or made so by mutagenesis, facilitate discovery of inverse agonists. In an interesting study, Xiao et al. (75) show that the constitutive activity of about 30 orphan GPCRs differs depending on the type of Gα protein with which they are co-expressed, and this information can easily be put to use in the development of assays for deorphanization; however, an additional 25 receptors showed no constitutive activity when co-expressed with 7 different Gα proteins, so this approach will not be useful for all receptors, or in all cell types. The arrestin recruitment assay work published by Yin et al. (76), although focused on orphan lipid receptors, shows the value of screening many orphan receptors in parallel, and it is to be hoped that similar approaches with peptide receptors in the future will be similarly fruitful. Given the increasing understanding of the importance of functional selectivity or biased agonism in receptor function (77), it seems clear that it may be useful to attempt deorphanization of GPCRs using a number of different functional screening methods in parallel. In some cases, where discovery of ligands for orphan receptors proves difficult, it may be of value to use knockout mice to establish biological functions for orphan receptors, and then to use these studies to further guide ligand discovery efforts. Of course, it is conceivable that orphan peptides such as those described here could have non-GPCR targets as well as the GPCRs, e.g., the peptide-gated ion channels and perhaps others yet unknown, and in such cases different approaches to discover their targets may need to be taken.
CONCLUSION
The combination of approaches described here for discovery and characterization of novel peptides, as well as for deorphanization of their receptors, holds much promise for meaningful investigation in the near future, and it is to be hoped that the results of studies such as these will accelerate the development of useful therapeutics in the longer term.
ACKNOWLEDGMENT
This work was supported by NIH grant 1R01DA027170-01 to IL and BR.
Contributor Information
Iris Lindberg, Phone: +1-410-7064778, Email: ilind001@umaryland.edu.
Bryan Roth, Phone: +1-919-9667535, Email: bryan_roth@med.unc.edu.
REFERENCES
- 1.Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2:31–9. doi: 10.1016/S1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 2.Nakayama K. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem J. 1997;327(Pt 3):625–35. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mains RE, Berard CA, Denault JB, Zhou A, Johnson RC, Leduc R. PACE4: a subtilisin-like endoprotease with unique properties. Biochem J. 1997;321(Pt 3):587–93. doi: 10.1042/bj3210587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smeekens ST, Steiner DF. Identification of a human insulinoma cDNA encoding a novel mammalian protein structurally related to the yeast dibasic processing protease Kex2. J Biol Chem. 1990;265:2997–3000. [PubMed] [Google Scholar]
- 5.Seidah NG, Gaspar L, Mion P, Marcinkiewicz M, Mbikay M, Chretien M. cDNA sequence of two distinct pituitary proteins homologous to Kex2 and furin gene products: tissue-specific mRNAs encoding candidates for pro-hormone processing proteinases. DNA Cell Biol. 1990;9:789. doi: 10.1089/dna.1990.9.789. [DOI] [PubMed] [Google Scholar]
- 6.Fricker LD. Methods for studying carboxypeptidase E. Peptidases and neuropeptide processing. Academic Press: San Diego; 1995. pp. 237–250. [Google Scholar]
- 7.Fricker L. Activation and membrane binding of carboxypeptidase E. J Cell Biochem. 1988;38(4):279–289. doi: 10.1002/jcb.240380407. [DOI] [PubMed] [Google Scholar]
- 8.Eipper B, Mains RE. Structure and biosynthesis of pro-adrenocorticotropin/endorphin and related peptides. Endocr Rev. 1980;1:1–27. doi: 10.1210/edrv-1-1-1. [DOI] [PubMed] [Google Scholar]
- 9.Murthy AS, Mains RE, Eipper BA. Purification and characterization of peptidylglycine alpha-amidating monooxygenase from bovine neurointermediate pituitary. J Biol Chem. 1986;261:1815–22. [PubMed] [Google Scholar]
- 10.Yang J, Brown MS, Liang G, Grishin NV, Goldstein JL. Identification of the acyltransferase that octanoylates ghrelin, an appetite stimulating peptide hormone. Cell. 2008;132:387–96. doi: 10.1016/j.cell.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Moore KL. The biology and enzymology of protein tyrosine O-sulfation. J Biol Chem. 2003;278:24243–6. doi: 10.1074/jbc.R300008200. [DOI] [PubMed] [Google Scholar]
- 12.Lee SN, Hwang JR, Lindberg I. Neuroendocrine protein 7B2 can be inactivated by phosphorylation within the secretory pathway. J Biol Chem. 2006;281:3312–20. doi: 10.1074/jbc.M506635200. [DOI] [PubMed] [Google Scholar]
- 13.Fujii R, Yoshida H, Fukusumi S, et al. Identification of a neuropeptide modified with bromine as an endogenous ligand for GPR7. J Biol Chem. 2002;277:34010–6. doi: 10.1074/jbc.M205883200. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka H, Yoshida T, Miyamoto N, et al. Characterization of a family of endogenous neuropeptide ligands for the G protein-coupled receptors GPR7 and GPR8. Proc Natl Acad Sci U S A. 2003;100:6251–6. doi: 10.1073/pnas.0837789100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukusumi S, Fujii R, Hinuma S. Recent advances in mammalian RFamide peptides: the discovery and functional analyses of PrRP, RFRPs and QRFP. Peptides. 2006;27:1073–86. doi: 10.1016/j.peptides.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 16.Southey BR, Rodriguez-Zas SL, Sweedler JV. Prediction of neuropeptide prohormone cleavages with application to RFamides. Peptides. 2006;27:1087–98. doi: 10.1016/j.peptides.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 17.Hughes J, Smith TW, Kosterlitz HW, Fothergill LA, Morgan BA, Morris HR. Identification of two related pentapeptides from the brain with potent opiate agonist activity. Nature. 1975;258:577–80. doi: 10.1038/258577a0. [DOI] [PubMed] [Google Scholar]
- 18.Civelli O. GPCR deorphanizations: the novel, the known and the unexpected transmitters. Trends Pharmacol Sci. 2005;26:15–9. doi: 10.1016/j.tips.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Nothacker HP, Wang Z, Bohn LM, Civelli O. Pharmacological characterization of a selective agonist for bombesin receptor subtype-3. Biochem Biophys Res Commun. 2009;387:283–8. doi: 10.1016/j.bbrc.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Civelli O, Zhou QY. Orphan G protein-coupled receptors and novel neuropeptides. Introduction. Results Probl Cell Differ. 2008;46:1–25. doi: 10.1007/400_2007_057. [DOI] [PubMed] [Google Scholar]
- 21.Chung S, Funakoshi T, Civelli O. Orphan GPCR research. Br J Pharmacol. 2008;153(Suppl 1):S339–46. doi: 10.1038/sj.bjp.0707606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Civelli O, Saito Y, Wang Z, Nothacker HP, Reinscheid RK. Orphan GPCRs and their ligands. Pharmacol Ther. 2006;110:525–32. doi: 10.1016/j.pharmthera.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Reinscheid RK, Xu YL, Okamura N, et al. Pharmacological characterization of human and murine neuropeptide s receptor variants. J Pharmacol Exp Ther. 2005;315:1338–45. doi: 10.1124/jpet.105.093427. [DOI] [PubMed] [Google Scholar]
- 24.Saito Y, Civelli O. G-protein-coupled receptor deorphanizations. Int Rev Neurobiol. 2005;65:179–209. doi: 10.1016/S0074-7742(04)65007-0. [DOI] [PubMed] [Google Scholar]
- 25.Sakurai T, Amemiya A, Ishii M, et al. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92:573–85. doi: 10.1016/S0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- 26.Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth hormone-releasing acylated peptide from stomach. Nature. 1999;402:656–60. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- 27.Roseweir AK, Millar RP. The role of kisspeptin in the control of gonadotrophin secretion. Hum Reprod Update. 2009;15:203–12. doi: 10.1093/humupd/dmn058. [DOI] [PubMed] [Google Scholar]
- 28.Fricker LD, McKinzie AA, Sun J, et al. Identification and characterization of proSAAS, a granin-like neuroendocrine peptide precursor that inhibits prohormone processing. J Neurosci. 2000;20:639–48. doi: 10.1523/JNEUROSCI.20-02-00639.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Che FY, Berezniuk I, Sonmez K, Toll L, Fricker LD. Peptidomics of Cpe(fat/fat) mouse brain regions: implications for neuropeptide processing. J Neurochem. 2008;107:1596–613. doi: 10.1111/j.1471-4159.2008.05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Decaillot FM, Che FY, Fricker LD, Devi LA. Peptidomics of Cpefat/fat mouse hypothalamus and striatum: effect of chronic morphine administration. J Mol Neurosci. 2006;28:277–84. doi: 10.1385/JMN:28:3:277. [DOI] [PubMed] [Google Scholar]
- 31.Lim J, Berezniuk I, Che FY, et al. Altered neuropeptide processing in prefrontal cortex of Cpe (fat/fat) mice: implications for neuropeptide discovery. J Neurochem. 2006;96:1169–81. doi: 10.1111/j.1471-4159.2005.03614.x. [DOI] [PubMed] [Google Scholar]
- 32.Che FY, Biswas R, Fricker LD. Relative quantitation of peptides in wild-type and Cpe(fat/fat) mouse pituitary using stable isotopic tags and mass spectrometry. J Mass Spectrom. 2005;40:227–37. doi: 10.1002/jms.742. [DOI] [PubMed] [Google Scholar]
- 33.Che FY, Fricker LD. Quantitative peptidomics of mouse pituitary: comparison of different stable isotopic tags. J Mass Spectrom. 2005;40:238–49. doi: 10.1002/jms.743. [DOI] [PubMed] [Google Scholar]
- 34.Che FY, Yuan Q, Kalinina E, Fricker LD. Peptidomics of Cpe fat/fat mouse hypothalamus: effect of food deprivation and exercise on peptide levels. J Biol Chem. 2005;280:4451–61. doi: 10.1074/jbc.M411178200. [DOI] [PubMed] [Google Scholar]
- 35.Che FY, Eipper BA, Mains RE, Fricker LD. Quantitative peptidomics of pituitary glands from mice deficient in copper transport. Cell Mol Biol (Noisy-le-grand) 2003;49:713–22. [PubMed] [Google Scholar]
- 36.Sasaki K, Satomi Y, Takao T, Minamino N. Snapshot peptidomics of the regulated secretory pathway. Mol Cell Proteomics. 2009;8:1638–47. doi: 10.1074/mcp.M900044-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatcher NG, Atkins N, Jr, Annangudi SP, et al. Mass spectrometry-based discovery of circadian peptides. Proc Natl Acad Sci U S A. 2008;105:12527–32. doi: 10.1073/pnas.0804340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brockmann A, Annangudi SP, Richmond TA, et al. Quantitative peptidomics reveal brain peptide signatures of behavior. Proc Natl Acad Sci U S A. 2009;106:2383–8. doi: 10.1073/pnas.0813021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wegrzyn J, Lee J, Neveu JM, Lane WS, Hook V. Proteomics of neuroendocrine secretory vesicles reveal distinct functional systems for biosynthesis and exocytosis of peptide hormones and neurotransmitters. J Proteome Res. 2007;6:1652–65. doi: 10.1021/pr060503p. [DOI] [PubMed] [Google Scholar]
- 40.Brunner Y, Coute Y, Iezzi M, et al. Proteomics analysis of insulin secretory granules. Mol Cell Proteomics. 2007;6:1007–17. doi: 10.1074/mcp.M600443-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Gauthier DJ, Lazure C. Complementary methods to assist subcellular fractionation in organellar proteomics. Expert Rev Proteomics. 2008;5:603–17. doi: 10.1586/14789450.5.4.603. [DOI] [PubMed] [Google Scholar]
- 42.Mirabeau O, Perlas E, Severini C, et al. Identification of novel peptide hormones in the human proteome by hidden Markov model screening. Genome Res. 2007;17:320–7. doi: 10.1101/gr.5755407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yue CM, Deng DJ, Bi MX, Guo LP, Lu SH. Expression of ECRG4, a novel esophageal cancer-related gene, downregulated by CpG island hypermethylation in human esophageal squamous cell carcinoma. World J Gastroenterol. 2003;9:1174–8. doi: 10.3748/wjg.v9.i6.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sonmez K, Zaveri NT, Kerman IA, et al. Evolutionary sequence modeling for discovery of peptide hormones. PLoS Comput Biol. 2009;5:e1000258. doi: 10.1371/journal.pcbi.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gustincich S, Batalov S, Beisel KW, et al. Analysis of the mouse transcriptome for genes involved in the function of the nervous system. Genome Res. 2003;13:1395–401. doi: 10.1101/gr.1135303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz TW. The processing of peptide precursors. ‘Proline-directed arginyl cleavage’ and other monobasic processing mechanisms. FEBS Lett. 1986;200:1–10. doi: 10.1016/0014-5793(86)80500-2. [DOI] [PubMed] [Google Scholar]
- 47.Southey BR, Amare A, Zimmerman TA, Rodriguez-Zas SL, Sweedler JV. NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 2006;34:W267–72. doi: 10.1093/nar/gkl161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ozawa A, Cai Y, Lindberg I. Production of bioactive peptides in an in vitro system. Anal Biochem. 2007;366:182–9. doi: 10.1016/j.ab.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duckert P, Brunak S, Blom N. Prediction of proprotein convertase cleavage sites. Protein Eng Des Sel. 2004;17:107–12. doi: 10.1093/protein/gzh013. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y, Lindberg I. Purification and characterization of the prohormone convertase PC1(PC3) J Biol Chem. 1993;268:5615–23. [PubMed] [Google Scholar]
- 51.Lamango NS, Zhu X, Lindberg I. Purification and enzymatic characterization of recombinant prohormone convertase 2: stabilization of activity by 21 kDa 7B2. Arch Biochem Biophys. 1996;330:238–50. doi: 10.1006/abbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- 52.Cameron A, Apletalina EV, Lindberg I. The enzymology of PC1 and PC2. The Enzymes. 2001;22:291–331. [Google Scholar]
- 53.Zhou A, Paquet L, Mains RE. Structural elements that direct specific processing of different mammalian subtilisin-like prohormone convertases. J Biol Chem. 1995;270:21509–16. doi: 10.1074/jbc.270.37.21509. [DOI] [PubMed] [Google Scholar]
- 54.Paolillo L, Simonetti M, Brakch N, et al. Evidence for the presence of a secondary structure at the dibasic processing site of prohormone: the pro-ocytocin model. EMBO J. 1992;11:2399–405. doi: 10.1002/j.1460-2075.1992.tb05304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brakch N, Rholam M, Simonetti M, Cohen P. Favourable side-chain orientation of cleavage site dibasic residues of prohormone in proteolytic processing by prohormone convertase 1/3. Eur J Biochem. 2000;267:1626–33. doi: 10.1046/j.1432-1327.2000.01154.x. [DOI] [PubMed] [Google Scholar]
- 56.Bjarnadottir TK, Gloriam DE, Hellstrand SH, Kristiansson H, Fredriksson R, Schioth HB. Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics. 2006;88:263–73. doi: 10.1016/j.ygeno.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 57.Vassilatis DK, Hohmann JG, Zeng H, et al. The G protein-coupled receptor repertoires of human and mouse. Proc Natl Acad Sci U S A. 2003;100:4903–8. doi: 10.1073/pnas.0230374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gloriam DE, Fredriksson R, Schioth HB. The G protein-coupled receptor subset of the rat genome. BMC Genomics. 2007;8:338. doi: 10.1186/1471-2164-8-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fargin A, Raymond JR, Lohse MJ, Kobilka BK, Caron MG, Lefkowitz RJ. The genomic clone G-21 which resembles a beta-adrenergic receptor sequence encodes the 5-HT1A receptor. Nature. 1988;335:358–60. doi: 10.1038/335358a0. [DOI] [PubMed] [Google Scholar]
- 60.Keiser MJ, Setola V, Irwin JJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–81. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Regard JB, Sato IT, Coughlin SR. Anatomical profiling of G protein-coupled receptor expression. Cell. 2008;135:561–71. doi: 10.1016/j.cell.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crouch MF, Osmond RI. New strategies in drug discovery for GPCRs: high throughput detection of cellular ERK phosphorylation. Comb Chem High Throughput Screen. 2008;11:344–56. doi: 10.2174/138620708784534806. [DOI] [PubMed] [Google Scholar]
- 63.Bikkavilli RK, Tsang SY, Tang WM, et al. Identification and characterization of surrogate peptide ligand for orphan G protein-coupled receptor mas using phage-displayed peptide library. Biochem Pharmacol. 2006;71:319–37. doi: 10.1016/j.bcp.2005.10.050. [DOI] [PubMed] [Google Scholar]
- 64.Overton HA, Babbs AJ, Doel SM, et al. Deorphanization of a G protein-coupled receptor for oleoylethanolamide and its use in the discovery of small-molecule hypophagic agents. Cell Metab. 2006;3:167–75. doi: 10.1016/j.cmet.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 65.Pausch MH. G protein-coupled receptors in Saccharomyces cerevisiae: high-throughput screening assays for drug discovery. Trends Biotechnol. 1997;15:487–94. doi: 10.1016/S0167-7799(97)01119-0. [DOI] [PubMed] [Google Scholar]
- 66.Bates B, Zhang L, Nawoschik S, et al. Characterization of Gpr101 expression and G protein coupling selectivity. Brain Res. 2006;1087:1–14. doi: 10.1016/j.brainres.2006.02.123. [DOI] [PubMed] [Google Scholar]
- 67.Suga H, Haga T. Ligand screening system using fusion proteins of G protein-coupled receptors with G protein alpha subunits. Neurochem Int. 2007;51:140–64. doi: 10.1016/j.neuint.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 68.Mertens I, Vandingenen A, Meeusen T, De Loof A, Schoofs L. Postgenomic characterization of G protein-coupled receptors. Pharmacogenomics. 2004;5:657–72. doi: 10.1517/14622416.5.6.657. [DOI] [PubMed] [Google Scholar]
- 69.Eglen RM, Bosse R, Reisine T. Emerging concepts of guanine nucleotide-binding protein-coupled receptor (GPCR) function and implications for high throughput screening. Assay Drug Dev Technol. 2007;5:425–51. doi: 10.1089/adt.2007.062. [DOI] [PubMed] [Google Scholar]
- 70.Wise A, Jupe SC, Rees S. The identification of ligands at orphan G protein coupled receptors. Annu Rev Pharmacol Toxicol. 2004;44:43–66. doi: 10.1146/annurev.pharmtox.44.101802.121419. [DOI] [PubMed] [Google Scholar]
- 71.Wang CJ, Hsu SH, Hung WT, Luo CW. Establishment of a chimeric reporting system for the universal detection and high-throughput screening of G protein-coupled receptors. Biosens Bioelectron. 2009;24:2298–304. doi: 10.1016/j.bios.2008.11.023. [DOI] [PubMed] [Google Scholar]
- 72.Johnson EN, Shi X, Cassaday J, Ferrer M, Strulovici B, Kunapuli P. A 1, 536-well. Assay Drug Dev Technol. 2008;6:327–37. doi: 10.1089/adt.2007.113. [DOI] [PubMed] [Google Scholar]
- 73.Mishina YM, Wilson CJ, Bruett L, et al. Multiplex GPCR assay in reverse transfection cell microarrays. J Biomol Screen. 2004;9:196–207. doi: 10.1177/1087057103261880. [DOI] [PubMed] [Google Scholar]
- 74.Bercher M, Hanson B, van Staden C, Wu K, Ng GY, Lee PH. Agonists of the orphan human G2A receptor identified from inducible G2A expression and beta-lactamase reporter screen. Assay Drug Dev Technol. 2009;7:133–42. doi: 10.1089/adt.2008.179. [DOI] [PubMed] [Google Scholar]
- 75.Xiao SH, Reagan JD, Lee PH, et al. High throughput screening for orphan and liganded GPCRs. Comb Chem High Throughput Screen. 2008;11:195–215. doi: 10.2174/138620708783877762. [DOI] [PubMed] [Google Scholar]
- 76.Yin H, Chu A, Li W, et al. Lipid G protein-coupled receptor ligand identification using beta-arrestin PathHunter assay. J Biol Chem. 2009;284:12328–38. doi: 10.1074/jbc.M806516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Urban JD, Clarke WP, von Zastrow M, et al. Functional selectivity and classical concepts of quantitative pharmacology. J Pharmacol Exp Ther. 2007;320:1–13. doi: 10.1124/jpet.106.104463. [DOI] [PubMed] [Google Scholar]
- 78.Barak LS, Ferguson SS, Zhang J, Caron MG. A beta-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. J Biol Chem. 1997;272:27497–500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- 79.Hinuma S, Habata Y, Fujii R, et al. A prolactin-releasing peptide in the brain. Nature. 1998;393:272–6. doi: 10.1038/30515. [DOI] [PubMed] [Google Scholar]
- 80.Saito Y, Nothacker HP, Wang Z, Lin SH, Leslie F, Civelli O. Molecular characterization of the melanin-concentrating-hormone receptor. Nature. 1999;400:265–9. doi: 10.1038/22321. [DOI] [PubMed] [Google Scholar]
- 81.Palyha OC, Feighner SD, Tan CP, et al. Ligand activation domain of human orphan growth hormone (GH) secretagogue receptor (GHS-R) conserved from Pufferfish to humans. Mol Endocrinol. 2000;14:160–9. doi: 10.1210/me.14.1.160. [DOI] [PubMed] [Google Scholar]
- 82.Elshourbagy NA, Ames RS, Fitzgerald LR, et al. Receptor for the pain modulatory neuropeptides FF and AF is an orphan G protein-coupled receptor. J Biol Chem. 2000;275:25965–71. doi: 10.1074/jbc.M004515200. [DOI] [PubMed] [Google Scholar]
- 83.Kotani M, Mollereau C, Detheux M, et al. Functional characterization of a human receptor for neuropeptide FF and related peptides. Br J Pharmacol. 2001;133:138–44. doi: 10.1038/sj.bjp.0704038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lenkei Z, Beaudet A, Chartrel N, et al. A highly sensitive quantitative cytosensor technique for the identification of receptor ligands in tissue extracts. J Histochem Cytochem. 2000;48:1553–64. doi: 10.1177/002215540004801112. [DOI] [PubMed] [Google Scholar]
- 85.Kabarowski JH, Feramisco JD, Le LQ, et al. Direct genetic demonstration of G alpha 13 coupling to the orphan G protein-coupled receptor G2A leading to RhoA-dependent actin rearrangement. Proc Natl Acad Sci U S A. 2000;97:12109–14. doi: 10.1073/pnas.97.22.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Borowsky B, Adham N, Jones KA, et al. Trace amines: identification of a family of mammalian G protein-coupled receptors. Proc Natl Acad Sci U S A. 2001;98:8966–71. doi: 10.1073/pnas.151105198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Takeda S, Yamamoto A, Okada T, et al. Identification of surrogate ligands for orphan G protein-coupled receptors. Life Sci. 2003;74:367–77. doi: 10.1016/j.lfs.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 88.Gupte J, Cutler G, Chen JL, Tian H. Elucidation of signaling properties of vasopressin receptor-related receptor 1 by using the chimeric receptor approach. Proc Natl Acad Sci U S A. 2004;101:1508–13. doi: 10.1073/pnas.0308250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tabata K, Baba K, Shiraishi A, Ito M, Fujita N. The orphan GPCR GPR87 was deorphanized and shown to be a lysophosphatidic acid receptor. Biochem Biophys Res Commun. 2007;363:861–6. doi: 10.1016/j.bbrc.2007.09.063. [DOI] [PubMed] [Google Scholar]
- 90.Suska A, Ibanez AB, Lundstrom I, Berghard A. G protein-coupled receptor mediated trimethylamine sensing. Biosens Bioelectron. 2009;25:715–20. doi: 10.1016/j.bios.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 91.Chen K, Obinata H, Izumi T. Detection of G protein-coupled receptor-mediated cellular response involved in cytoskeletal rearrangement using surface plasmon resonance. Biosens Bioelectron. 2009. [DOI] [PubMed]