Cardiac development is an intricate process involving the migration and assembly of multiple cell types from distinct lineages to form a mature heart. Cells of the proepicardium define one such lineage and have been receiving increasing attention given their role in the formation of the epicardium, and the epicardium’s central role as a signaling center during heart development and repair.1, 2 The proepicardium is a mesothelial structure in the wall of the pericardial cavity just dorsal to the developing heart tube that has a “cluster of grapes”-like appearance. It has been identified in human, mouse, chick and most recently in zebrafish embryos.3, 4 The transcription factors Tbx18, Wt-1, and Tcf21 (also known as Epicardin or Capsulin) are highly expressed in the proepicardium and serve as molecular markers of this structure, although none of these factors is expressed exclusively in the proepicardium. Cells of the proepicardium migrate onto the developing heart tube and completely cover it in an epithelial layer, the epicardium. Epicardial cells then undergo an epithelial-to-mesenchymal transition (EMT) and invade into the underlying myocardium to become fibroblasts and cells of the coronary vasculature. There are several examples of mutations in mice that disrupt epicardial development.1 The failure to form the epicardium leads to a reduction in cardiomyocyte proliferation, a disruption in cardiac morphogenesis, and embryonic demise due to heart failure.
Although the importance of the proepicardium for heart development is clear, we are just beginning to identify the signals that direct the formation of the proepicardium. The proepicardium is thought to arise from cells of the lateral plate mesoderm.3 Consistent with this notion, murine proepicardial cells have been shown to be derived from progenitors that express Nkx2.5 and Islet-1, both of which are highly expressed in the lateral plate mesoderm during early embyrogenesis.5 Further, mice deficient in Nkx2.5 do not develop a proepicardium.5 Genes involved in left-right asymmetry have also been shown to influence the development of the proepicardium.6 In chick, the proepicardium forms from bilaterally symmetric structures that develop asymmetrically, leading to a proepicardium just on the right side of the pericardial cavity. Disruption of right-sided gene expression determinants such as fgf8 or snai1 lead to the failure of proepicardial specification.6 In mice, the dependence of proepicardial specification on the left-right asymmetry machinery is unclear, as both right and left proepicardial precursors develop equally and fuse in the midline to generate the proepicardium.7
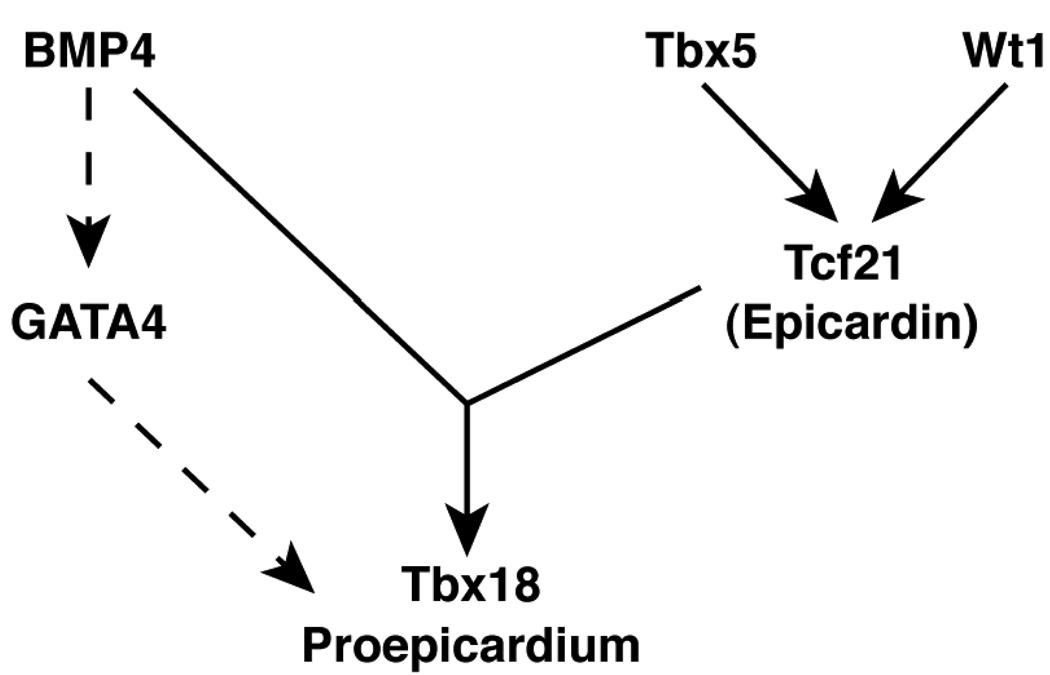
Previous work by Serluca had revealed the existence of the proepicardium in zebrafish and demonstrated that proepicardial tcf21 expression is dependent on wt1.3 In this issue of Circulation Research, Liu and Stainier provide further insights into the signals specifying the zebrafish proepicardium.8 Previous work in chick has suggested the importance of BMP signaling for proepicardial development, as implantation of beads coated with Noggin (a BMP antagonist) in chick led to a loss of proepicardial marker gene expression.9 Given these results, Liu and Stainier first focused on the BMP signaling pathway during zebrafish proepicardial development. They found that blocking BMP signaling using an inducible, dominate-negative BMP led to a significant reduction in tbx18 and tcf21 gene expression. Also, they found that proepicardial marker gene expression was completely blocked in zebrafish mutants lacking the type I BMP receptor acvr1l. Interestingly, gain of function studies using an inducible BMP2 expression system resulted in induction of ectopic tbx18 expression, but not tcf21, suggesting that BMP signaling alone was insufficient to completely commit cells to the proepicardial fate. Overexpression of Tcf21 could also induce tbx18, but this required acvr1l expression, consistent with a model in which two signals are required for proepicardial specification (see Figure 1). The role of BMP signaling in proepicardial specification appears distinct from a recently reported role of BMP signaling in directing the differentiation of proepicardial cells to the myocardial lineage10, suggesting that BMPs may have multiple roles during the specification and development of the proepicardium.
Figure 1. A Molecular Pathway directing Proepicardial Specification.
Shown above is schematic of the genes involved in proepicardial development. Dashed arrows indicate pathways not yet directly demonstrated in zebrafish.
As an approach toward identifying other factors involved in inducing the proepicardium, previous work from Ishii and colleagues using a tissue transplantation technique in chick embryos demonstrated the ability of the liver bud, but not the lung bud, to induce the proepicardial marker genes tbx18, wt1, and tcf21 in adjacent mesoderm.11 To test the importance of the liver in proepicardial induction in zebrafish, Liu and Stainier took advantage of several mutant zebrafish lines in which liver formation is severely disrupted or absent.8 In these mutants, the proepicardium still formed, demonstrating that liver-derived signals are not necessary for proepicardial specification in zebrafish. Instead, Liu and Stainier found that BMP4, produced by the overlying heart tube, was important for proepicardial specification, as mutant zebrafish lacking BMP4 had reduced levels of tbx18 and tcf21 gene expression. The reasons for the difference between the chick and fish results are unclear, but warrant further investigation.
As the transcription factor Tbx5 had been previously shown to play a role in the migration of proepicardial cells to the myocardium,12 Liu and Stainier next used the zebrafish system to determine if Tbx5 had an earlier role in proepicardial specification. There are two tbx5 genes in the zebrafish genome, with only tbx5a being expressed in the lateral plate mesoderm.13 Liu and Stainier demonstrated that in zebrafish tbx5a mutants, tbx18 and tcf21 expression was lost, but bmp4 expression was not altered. Further, overexpression of a dominant negative form of Tbx5a during the early somite stage of development significantly reduced tbx18 and tcf21 expression, suggesting the requirement of Tbx5a activity in cells of the lateral plate mesoderm. Thus, this work is consistent with a model in which proepicardial progenitors become competent to commit to the proepicardial lineage in the lateral plate mesoderm through the action of tbx5a. Then, upon exposure to BMP4 from the developing heart tube, become committed to the proepicardial fate.
Like most good science, this work raises interesting questions that warrant further exploration. One such question involves the role of GATA4 in proepicardial specification. GATA4 is highly expressed in the murine proepicardium and targeted disruption of GATA4 in mice results in a complete absence of the proepicardium, despite normal development of the early heart tube.14 Further, BMP4 has been shown to induce GATA4 expression during mouse embryogenesis.15 Taken together with the results of Liu and Stainier, these observations are consistent with a model in which BMP4 directs proepicardial commitment in a GATA4-dependent manner (see Figure 1). In zebrafish, the dependence of proepicardial development on gata4 has not been directly examined, but ‘knock-down’ of GATA4 in zebrafish embryos results in block in cardiac development after formation of the heart tube.16 It is conceivable that this phenotype may in part be due to the failure to develop the epicardium and proepicardium in these morphants. Further work is needed to more precisely define GATA4’s role in proepicardial development, and the zebrafish model system seems ideally suited for it.
Acknowledgments
Sources of Funding
E.C.S. is supported by NIH grant # HL71063.
Non-standard Abbreviations and Acronyms
- BMP
Bone Morphogenetic Protein
- Tbx
T-Box Factor
- Nkx2.5
NK2 Transcription Factor Related 5
- GATA4
GATA Binding Protein 4
- Wt-1
Wilms Tumor 1
- Tcf21
Transcription Factor 21
- Acvr1l
Activin A receptor, Type 1 Like
- Snai1
Snail Homolog 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None.
References
- 1.Olivey HE, Svensson EC. Epicardial-myocardial signaling directing coronary vasculogenesis. Circ Res. 2010;106:818–832. doi: 10.1161/CIRCRESAHA.109.209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 3.Serluca FC. Development of the proepicardial organ in the zebrafish. Dev Biol. 2008;315:18–27. doi: 10.1016/j.ydbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Manner J, Perez-Pomares JM, Macias D, Munoz-Chapuli R. The origin, formation and developmental significance of the epicardium: a review. Cells Tissues Organs. 2001;169:89–103. doi: 10.1159/000047867. [DOI] [PubMed] [Google Scholar]
- 5.Zhou B, von Gise A, Ma Q, Rivera-Feliciano J, Pu WT. Nkx2-5-and Isl1-expressing cardiac progenitors contribute to proepicardium. Biochemical and Biophysical Research Communications. 2008;375:450–453. doi: 10.1016/j.bbrc.2008.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlueter J, Brand T. A right-sided pathway involving FGF8/Snai1 controls asymmetric development of the proepicardium in the chick embryo. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0811944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulte I, Schlueter J, Abu-Issa R, Brand T, Manner J. Morphological and molecular left-right asymmetries in the development of the proepicardium: a comparative analysis on mouse and chick embryos. Dev Dyn. 2007;236:684–695. doi: 10.1002/dvdy.21065. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Stainier DY. Tbx5 and Bmp Signaling Are Essential for Proepicardium Specification in Zebrafish. Circ Res. 2010;106 doi: 10.1161/CIRCRESAHA.110.217950. XXX - XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlueter J, Männer J, Brand T. BMP is an important regulator of proepicardial identity in the chick embryo. Developmental Biology. 2006;295:546–558. doi: 10.1016/j.ydbio.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 10.van Wijk B, van den Berg G, Abu-Issa R, Barnett P, van der Velden S, Schmidt M, Ruijter JM, Kirby ML, Moorman AFM, van den Hoff MJB. Epicardium and myocardium separate from a common precursor pool by crosstalk between bone morphogenetic protein- and fibroblast growth factor-signaling pathways. Circulation Research. 2009;105:431–441. doi: 10.1161/CIRCRESAHA.109.203083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishii Y, Langberg JD, Hurtado R, Lee S, Mikawa T. Induction of proepicardial marker gene expression by the liver bud. Development. 2007;134:3627–3637. doi: 10.1242/dev.005280. [DOI] [PubMed] [Google Scholar]
- 12.Hatcher CJ, Diman NYS-G, Kim M-S, Pennisi D, Song Y, Goldstein MM, Mikawa T, Basson CT. A role for Tbx5 in proepicardial cell migration during cardiogenesis. Physiol Genomics. 2004;18:129–140. doi: 10.1152/physiolgenomics.00060.2004. [DOI] [PubMed] [Google Scholar]
- 13.Albalat R, Baquero M, Minguillon C. Identification and characterisation of the developmental expression pattern of tbx5b, a novel tbx5 gene in zebrafish. Gene Expr Patterns. 2009;10:24–30. doi: 10.1016/j.gep.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci USA. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, Black BL. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development. 2005;132:3405–3417. doi: 10.1242/dev.01913. [DOI] [PubMed] [Google Scholar]
- 16.Holtzinger A, Evans T. Gata4 regulates the formation of multiple organs. Development. 2005;132:4005–4014. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]