Abstract
It is widely accepted that the genome is regulated by histone modifications that induce epigenetic changes on the genome. However, it is still not understood how ubiquitously expressed chromatin modifying complexes are “guided” to specific genomic sites to induce intricate patterns of epigenetic modifications. Previously believed to represent “genome junk”, it is now becoming increasingly clear that large non-coding RNAs associate with chromatin modifying complexes. Here we explore an intriguing hypothesis that large non-coding RNA molecules might represent a molecular trafficking system that modulate chromatin modifying complexes to establish specific epigenetic landscapes.
Introduction
During development, the genome undergoes a complex choreography to establish distinctive gene expression patterns that define cellular identity. These changes are mediated through the presence of specific histone modifications and DNA methylation patterns, which are established by ubiquitously expressed chromatin modifying complexes with unknown specificity. However, what guides these complexes to distinct and specific sites under different cellular contexts is not understood. Almost 35 years ago a first clue came that perhaps RNA may play a role in this process based on the observation that chromatin structure was found to be associated with several unknown RNAs [1]. Two key studies further demonstrated that RNA was a critical component in the global localization of chromatin modifying complexes [2,3]. For example, depletion of single-stranded (ss)RNA, but not ssDNA was shown to be required for the localization of key histone modifications [2,3].
Indeed, several recent studies have begun to unravel the association of large non-coding RNAs with enzymatic complexes that establish these epigenetic landscapes [4, 5**,6**]. These studies suggest a potential role for large non-coding RNAs in regulating chromatin state. Specifically, large non-coding RNA molecules might be required for the specificity of chromatin formation across the genome [4, 5**,6**,7,8,9**]. Thus, expression patterns of non-coding RNAs may influence specific epigenetic states by interfacing with chromatin modifying complexes and thereby imparting specificity.
Although these examples suggest a key role for RNA in epigenetic regulation, it is not understood how RNA imparts specificity to otherwise ubiquitous chromatin modifying complexes. Although there is a wealth of information about small non-coding RNAs regulating chromatin [2,3,10–14], in this review we specifically focus on large non-coding RNAs in mammalian systems. Here, we discuss several recent studies that have gleaned insights into possible roles for large non-coding RNAs modulating the regulation of chromatin modifying complexes. By using examples of X inactivation, HOX gene regulation and imprinting, we propose putative models of how large non-coding RNAs could, in part, serve as a genetic trafficking system.
X Inactivation
X chromosome inactivation is a classic and dramatic example of RNA based establishment of epigenetic regulation. Briefly, X chromosome inactivation is a process in female mammalian cells in which one copy of the X chromosome is inactivated. This ensures that females produce the same dosage of X-linked genes as the male produces with only one X chromosome [15,16]. Remarkably, a multi-exonic, spliced, capped and poly-adenylated large non-coding RNA known as Xist (X inactive specific transcript), is expressed on only one female X chromosome and induces the entire chromosome to become transcriptionally inactive [17–23]. This RNA was the first example to illustrate the power of a large non-coding RNA to be required for epigenetic changes. The coating the future inactive X chromosome by Xist ultimately changes the epigenetic program of an entire chromosome [16,24–26].
Polycomb group complexes (PcG), such as PRC2 (comprised of the proteins EZH2, EED and SUZ12), are chromatin-modifying complex that are conserved from flies to mammals and are required for proper establishment of heterochromatic regions genome-wide [27]. As first demonstrated with the large non-coding RNA HOTAIR (see HOX section), PRC2 was also found to colocalize with Xist at the inactive X chromosome [4,5**,26,28]. This binding was shown to occur via a 1.6kb non-coding RNA encoded within Xist, named RepA [5**,29*]. Furthermore, a highly conserved double-stem loop structure in RepA was found to directly bind components of the PRC2 complex, most likely to Ezh2 [5**]. More recently, another study validated the binding between RepA and PRC2 and found that the full-length RepA (consisting of two double-stem loop structures) sequence binds to Suz12 via structural interactions between the stem-loop repeats and spacers that stabilize the binding interaction [29*]. Although it is too early to determine the exact interactions between RepA and PRC2, it is clear that PRC2 forms a ribonucleic-protein interaction with RepA that is critical for the deposition of histone 3 lysine 27 trimethylation (H3K27me3), leading to chromosome wide heterochromatin formation [5**].
A further layer of RNA dependent recruitment of PRC2 involves a large non-coding RNA transcribed antisense to Xist, namely Tsix [30]. In contrast to Xist, Tsix ceases to be expressed in the future inactive X chromosome and instead is increasingly expressed in the future active X chromosome [30]. Hence, it was also suggested that Tsix could compete for the binding of PRC2, and in this way might reduce PRC2 binding to RepA [5**]. One possibility is that Tsix prevents Xist function by reducing heterochromatin formation on the active X [5**]. All these findings demonstrate that physical association of large non-coding RNAs with RepA is required to bestow PRC2 localization on the future inactive X chromosome. Hence, X inactivation represents a powerful model of RNA dependent cis recruitment of chromatin modifying complexes.
HOX Gene Regulation
The Homeobox transcription factors (HOX genes) were famously discovered for their ability to transform the identities of body segments in fruit flies [31]. In mammals 39 HOX genes are encoded across four loci (HOX-A: HOX-D) on different chromosomes. The relative position of each HOX gene within a cluster is reflective of its spatial and temporal expression along the proximal-distal and anterior-posterior axes in developing embryos that define a unique positional cellular identity [32,33]. HOX gene expression is coordinately regulated by the dynamic interplay between PcG and Trithorax complexes that establish heritable collinear domains of heterochromatin and euchromatin respectively [34–36].
Interestingly, recent studies have revealed numerous large intergenic non-coding RNAs (lincRNAs) within the HOX clusters, which also exhibit similar spatial and temporal patterns of expression as the HOX genes [4,37,38]. It has been demonstrated that one of lincRNA encoded in the HOX-C cluster, termed HOTAIR, physically associates with PRC2. It is also required for the proper maintenance of a large domain of heterochromatin, but in a different cluster, namely HOX-D [4]. It is not know how HOTAIR interacts with HOX-D since it has very little sequence homology yet there are several possible models for how this regulation could occur. For example, HOTAIR could bind to PRC2 facilitating ribonucleic interactions with sequence specific adaptor proteins that recognize sequences in the HOX-D locus (Figure 2c). Consistent with this hypothesis HOTAIR was recently determined to interact with COREST (RCOR1) [7, M.C. Tsai and H.Y. Chang personal communication]; which serves a molecular beacon for silencing of neuronal specific genes several of which flank of HOX-D [39].
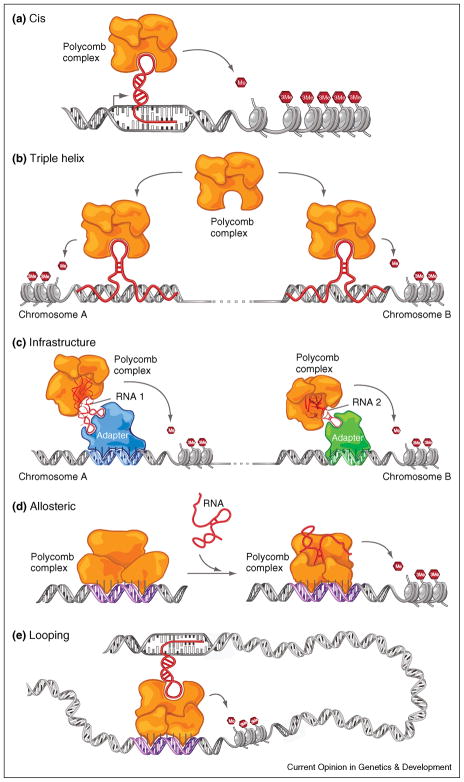
Figure 2.
Models how large non-coding RNAs could impart specificity to chromatin modifications (a) Cis model: at the site of RNA transcription, chromatin-modifying complexes could be recruited directly or indirectly and exert their function locally in cis. (b) Triple helix model: Formation of a RNA-DNA triplex could serve as a beacon to recruit directly or indirectly epigenetic modifying complexes to specific sites. (c) Infrastructure model: large non-coding RNAs could serve as an architectural scaffold that directly or indirectly links chromatin complexes to DNA binding proteins or transcription factors. (d) Allosteric model: RNA could bind directly or indirectly to chromatin remodeling proteins that change the shape of the protein or protein complex, activating/inactivating the complex and confer site specificity. (e) Looping model: RNA could serve as a scaffold to establish long range interactions via DNA looping and recruiting chromatin modifying complexes to distal sites of action in cis and or inter-chromosomal interactions (not shown).
Alternatively, HOTAIR could form triple helix interactions with the HOX-D cluster that are known to not follow the Watson-Crick base pairing [40] (Figure 2b). Regardless, loss of HOTAIR function results in decreased levels of H3K27me3 and PRC2 binding, leading to the activation of HOX-D genes in trans [4]. Hence, HOTAIR helps to maintain characteristic HOX gene expression pattern, and shows RNA based recruitment of PRC2 in trans, revealing a potentially new form of genomic crosstalk. It is likely that numerous other HOX large non-coding RNAs bind PRC2 and assist in the proper orchestration of HOX gene expression.
In addition to repressive polycomb protein regulation, HOX genes are also epigenetically regulated by trithorax proteins (TRX). TRX enzymatically modify histone H3 lysine 4 trimethylation (H3K4me3), thereby establishing the positional patterns of euchromatic HOX domains [41]. In one study it was demonstrated that a Drosophila large non-coding RNA Ultrabithorax (Ubx) binds to the methyl transferase trithorax (Ash1) and guides the complex to properly regulate the collinear expression of Ubx [42]. However, a contradicting observation was reported that this large non-coding RNA serves as a repressor for Ubx [43]. Nonetheless, both studies underscore the role of a lincRNA in establishment of epigenetic states. Another study found that the TRX protein Mll1 physically associates with two antisense HOX large non-coding RNAs Evx1as and HoxB5/6as during mammalian embryoid body (EB) differentiation [44]. Overall, these studies demonstrate that several large non-coding RNAs bind TRX group proteins in both flies and mammals.
Despite representing a fraction of the genome, HOX clusters encode a bounty of large non-coding RNAs. Many of these HOX large non-coding RNAs share a common role in interacting with chromatin modifying complexes such as PRC2 and TRX to coordinate a dynamic regulation of epigenetic states and establish proper domains of HOX gene expression.
Imprinting
In mammals, somatic cells possess two copies of a gene (alleles), one inherited from the mother and the other from the father. Most of the alleles are expressed simultaneously. However, a small fraction referred to as imprinted genes, are differentially expressed depending on whether the gene was maternally or paternally inherited [45,46]. Imprinted genes are regulated in cis by imprinting control regions (ICR), which can repress adjacent genes by utilizing large non-coding RNAs [47]. Despite regulating diverse genomic loci, they share an emerging mechanistic theme: antisense large non-coding RNAs that interact with chromatin modifying complexes result in silencing of maternal or paternal alleles. Recently, two large non-coding RNAs involved in imprinting were shown to bind chromatin modifying complexes and required for the proper localization of histone modifications [6**,8,9**].
For example, most of the genes near the Kcnq1 gene are expressed from the maternal chromosome, except the paternally expressed antisense non-coding RNA termed Kcnq1ot1 [48]. Recently, it was shown that Kcnq1ot1 represses the paternal allele of Kcnq1 by interacting with the histone methyltransferases G9a and PRC2 [6**,8,49]. A similar observation was reported with the large non-coding RNA Airn, which in the placenta imprints the Igf2R, Slc22a2 and Slc22a3 genes on the paternal allele [9**]. Interestingly, truncation of Airn results in the loss of G9a accumulation at the SLC22A3 promoter region, and in a decreased level of G9a recruitment and Slc22a3. This suggests that Airn helps to accumulate G9a at the promoter region to silence Slc22a3 [9**]. Although Airn and Kcnq1ot1 may or may not directly interact with the chromatin modifiers, it is clear that depletion of these large non-coding RNAs results in the loss of histone modifications at imprinted loci, presumably through displacement of key chromatin modifying complexes [4,6**,7].
Possible Mechanisms
Here we have surveyed several recent studies that demonstrate a common theme: large non-coding RNAs bind to chromatin modifying complexes such as PRC2, TRX and G9a and impart specific silencing of genomic loci both in cis and trans [4,8,9**,42,44]. Are these only distinct examples or can they be generalized to a common theme? A recent study demonstrated that numerous lincRNAs bind to PRC2 and multiple other chromatin modifiying complexes [7]. This suggests a more global role of lincRNAs, and we hypothesize that these might guide chromatin-modifying complexes to specific target sites. In support of this notion, the above study also demonstrated that lincRNAs are required for proper PRC2 mediated gene repression at gene loci that are normally repressed by PRC2 [7]. Collectively, these studies have revealed a global theme: large non-coding RNAs are required for the proper establishment of chromatin domains, possibly by steering epigenetic modifying complexes to their specific destinations. However, it is crucial to note that the models presented here do not exclude other proposed or plausible mechanisms. Here we only focus on a few possible RNA based mechanisms that may impart specificity to otherwise ubiquitous chromatin modifying complexes.
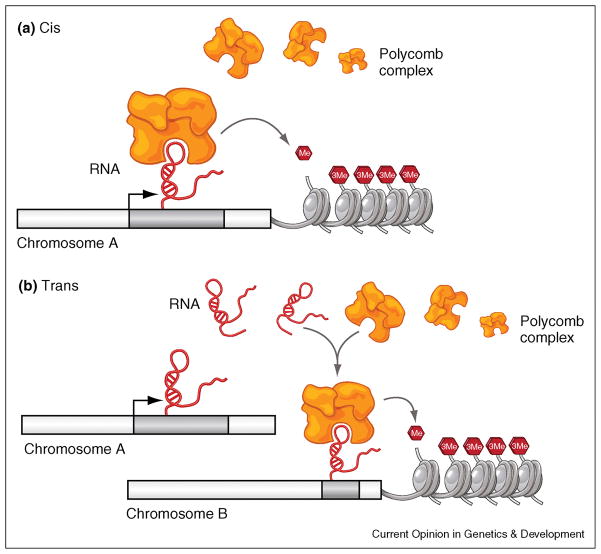
First, large non-coding RNAs could serve as RNA based tethers to recruit either directly or indirectly chromatin modifying complexes to the specific location of the RNA transcription [50]. Thus, a code of large non-coding RNAs could be expressed in a cell specific manner providing a map for proper epigenetic landscaping in cis (Figure 1, 2a). In contrast to the cis model, large non-coding RNAs could also act in trans, and could target multiple sites of action (Figure 1b).
Figure 1.
Model of large non-coding RNA function in cis versus trans (a) Cis model: large non-coding RNAs could serve as RNA based tethers to recruit either directly or indirectly epigenetic modifying complexes to the specific location of the RNA. (b) In trans, large non-coding RNAs could possibly target multiple sites of action.
There are several mechanistic models by which large non-coding RNAs could act in trans (Figure 2b–d). One possibility is the formation of a RNA-DNA triplex, which could serve as a beacon to recruit epigenetic modifying complexes either directly or indirectly (Figure 2b). This may be more likely than an alternative RNA-DNA duplex model: considering RNA sequence complementarity to DNA sequence has not yet been observed for trans acting large non-coding RNAs [7]. However, RNA-DNA triplexes do not rely on Watson and Crick base pairing [40] and may be alternative explanation of RNA-DNA interactions.
Another possibility of RNA based modulation of chromatin modifying complexes could occur by large non-coding RNAs serving as an architectural scaffold that directly or indirectly links chromatin complexes to DNA binding proteins or transcription factors (Figure 2c). Alternatively, RNA could bind directly or indirectly to chromatin remodeling proteins that change the shape of the protein or protein complex. This structural change might induce the complex in a particular way, confer site specificity or serve as an allosteric regulator (Figure 2d).
Finally, we propose that ribonucleic-protein interactions with chromatin modifying complexes such as PRC2 could mediate long-range interactions (both intra and inter-chromosomal) between DNA elements (Figure 2e). This raises an intriguing possibility that enhancer elements may contain large non-coding RNAs that mediate multi-lateral interactions with protein coding genes that result in “looping” of DNA to establish epigenetic control of neighboring genes. Consistent with this idea the interaction of Kcnq1ot1 with chromatin modifying complexes does not silence a continuous domain of neighboring genes. Since several genes are skipped perhaps Kcnq1ot1 mediates three-dimensional DNA-RNA-Protein interactions that precisely define regulated genes in the imprinted locus. It is important to note that if numerous other enhancer like control regions are transcribed, similar to Kcnq1ot1, Airn and Xist, all the mentioned studies demonstrate this is not due to passive transcription, rather the large non-coding RNA molecules themselves are required to facilitate the recruitment and silencing of these loci [5**,6**,9**].
Further investigation is required to resolve the mechanism by which large non-coding RNAs and protein coding genes such as chromatin-modifying complexes interact; and whether this interaction is important for the establishment of distinctive epigenetic states. However, it is clear that ncRNAs molecules play a key role in regulating the trafficking of epigenetic landscapes such as X inactivation, HOX gene regulation, imprinting and possibly numerous other biological processes.
Conclusion
In the 19th century, Lamarck’s idea how organisms inherit beneficial, environmentally acquired characteristics were diminished by Darwin’s theory of natural selection. However, the theory may still apply for non-coding RNA. Large non-coding RNAs might represent a way by which characteristics are propagated from mother to daughter cell and from generation to generation and perhaps in response to environmental cues. As we have summarized here, numerous large non-coding RNA molecules can attract chromatin-modifying complexes and might help establish heritable chromatin states. Thus, we envision the possibility that large non-coding RNA could be distributed between dividing cells and ensure epigenetic memory.
Consistent with this idea, it was recently demonstrated that the injection of a non-coding RNA (mir-124) into fertilized mouse eggs yielded a stable and heritable epigenetic change to the Sox9 locus that further lead to a transmissible growth phenotype across several generations of progeny [51]. Paramutation is another such example of RNA based genome molding, which describes the non-Mendelian interaction between two alleles of a single locus that results in heritable changes of one allele by the other allele [52–54]. Several studies have demonstrated that RNA is required to ensure that these changes can be inherited from one generation to the next [55,56]. Collectively these studies support our hypothesis of the importance large non-coding RNAs in genome plasticity and epigenetic memory.
By way of analogy, we propose an “RNA air traffic control” model to impart epigenetic inheritance. Daily, thousands of planes fly across the globe without inherent specificity, but are guided to their destination by air traffic control signals. Similarly, there are many distinct chromatin-modifying complexes and other proteins traveling around the nucleus, without inherent specificity. Perhaps large non-coding RNAs serve, at least in part, as an adaptable genetic air traffic control system to bring chromatin complexes to unique epigenetic destinations. If true, we could imagine reverse engineering the code of large non-coding RNAs to restore misregulated epigenetic states in disease models. Although, far from established, numerous recent studies are shedding new light on an emerging role of large non-coding RNAs in epigenetic regulation.
Acknowledgments
We would like to thank Sigrid Hart from the Broad Institute for the illustrations, M. Guttman, M. Cabili, M. Huarte, L. Goff, A.K. Khalil and J.S. Mattick for critical comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Paul J, Duerksen JD. Chromatin-associated RNA content of heterochromatin and euchromatin. Mol Cell Biochem. 1975;9:9–16. doi: 10.1007/BF01731728. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 4.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a shortrepeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. The study was the first to determine that RepA directly binds to the PRC2 chromatin modifying complex (likely through Ezh2) and is required for proper H3K27me3 localization on the inactive X chromosome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6**.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot11 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. The large non-coding RNA Kcnq1ot11 was shown to interact both with chromatin and the chromatin modifying complexes PRC2 and G9a. This study further demonstrates that loss of Kcnq1ot11 function prevents the accumulation of these complexes at the imprinted loci and biallelic expression of otherwise imprinted genes. [DOI] [PubMed] [Google Scholar]
- 7.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- 9**.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. This study showed that the large non-coding RNA Airn regulates imprinting of the Slc22a3 gene by recruiting the accumulation G9a, a H3K9 histone methyltransferase, to the promoter region of Slc22a3. These results suggest that Airn targets repressive histone modifying activities through molecular interaction with specific chromatin domains to epigenetically silence transcription. [DOI] [PubMed] [Google Scholar]
- 10.Gendrel AV, Colot V. Arabidopsis epigenetics: when RNA meets chromatin. Curr Opin Plant Biol. 2005;8:142–147. doi: 10.1016/j.pbi.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grewal SI, Rice JC. Regulation of heterochromatin by histone methylation and small RNAs. Curr Opin Cell Biol. 2004;16:230–238. doi: 10.1016/j.ceb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Djupedal I, Ekwall K. Epigenetics: heterochromatin meets RNAi. Cell Res. 2009;19:282–295. doi: 10.1038/cr.2009.13. [DOI] [PubMed] [Google Scholar]
- 14.Halic M, Moazed D. 22G-RNAs in transposon silencing and centromere function. Mol Cell. 2009;36:170–171. doi: 10.1016/j.molcel.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 16.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 17.Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 18.Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 19.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 20.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 21.Brown CJ, Lafreniere RG, Powers VE, Sebastio G, Ballabio A, Pettigrew AL, Ledbetter DH, Levy E, Craig IW, Willard HF. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991;349:82–84. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- 22.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 23.Herzing LB, Romer JT, Horn JM, Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 24.Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 25.Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 26.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 27.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 28.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 29*.Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 8:e1000276. doi: 10.1371/journal.pbio.1000276. This study used chemical and enzymatic probing to define the 2-dimensional structure of RepA. The authors conclude that RepA structure consists two long stem-loop structures each including four repeats. This structure is stabilized by intra-repeat interactions and contacts between spacers and the repeat structures. This study further employed Fluorescent Resonance Energy Transfer to demonstrate that RepA directly interacts with PRC2 via Suz12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 31.Scott MP. A rational nomenclature for vertebrate homeobox (HOX) genes. Nucleic Acids Res. 1993;21:1687–1688. doi: 10.1093/nar/21.8.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- 33.Duboule D, Tarchini B, Zakany J, Kmita M. Tinkering with constraints in the evolution of the vertebrate limb anterior-posterior polarity. Novartis Found Symp. 2007;284:130–137. doi: 10.1002/9780470319390.ch9. discussion 138–141, 158–163. [DOI] [PubMed] [Google Scholar]
- 34.Beuchle D, Struhl G, Muller J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development. 2001;128:993–1004. doi: 10.1242/dev.128.6.993. [DOI] [PubMed] [Google Scholar]
- 35.Busturia A, Morata G. Ectopic expression of homeotic genes caused by the elimination of the Polycomb gene in Drosophila imaginal epidermis. Development. 1988;104:713–720. doi: 10.1242/dev.104.4.713. [DOI] [PubMed] [Google Scholar]
- 36.Muller J, Bienz M. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 1991;10:3147–3155. doi: 10.1002/j.1460-2075.1991.tb04876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mainguy G, Koster J, Woltering J, Jansen H, Durston A. Extensive polycistronism and antisense transcription in the Mammalian Hox clusters. PLoS One. 2007;2:e356. doi: 10.1371/journal.pone.0000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sessa L, Breiling A, Lavorgna G, Silvestri L, Casari G, Orlando V. Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. RNA. 2007;13:223–239. doi: 10.1261/rna.266707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 40.Morgan AR, Wells RD. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J Mol Biol. 1968;37:63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- 43.Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surani MA, Barton SC. Development of gynogenetic eggs in the mouse: implications for parthenogenetic embryos. Science. 1983;222:1034–1036. doi: 10.1126/science.6648518. [DOI] [PubMed] [Google Scholar]
- 46.Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 47.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 48.Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, Yoshida MA, Niikawa N, Feinberg AP, Oshimura M. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet. 1999;8:1209–1217. doi: 10.1093/hmg/8.7.1209. [DOI] [PubMed] [Google Scholar]
- 49.Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, Nagano T, Cobb BS, Fraser P, Reik W. The long noncoding RNA Kcnq1ot11 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development. 2009;136:525–530. doi: 10.1242/dev.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grandjean V, Gounon P, Wagner N, Martin L, Wagner KD, Bernex F, Cuzin F, Rassoulzadegan M. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136:3647–3655. doi: 10.1242/dev.041061. [DOI] [PubMed] [Google Scholar]
- 52.Chandler VL. Paramutation: from maize to mice. Cell. 2007;128:641–645. doi: 10.1016/j.cell.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 53.Costa FF. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 54.Cuzin F, Grandjean V, Rassoulzadegan M. Inherited variation at the epigenetic level: paramutation from the plant to the mouse. Curr Opin Genet Dev. 2008;18:193–196. doi: 10.1016/j.gde.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 55.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 56.Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, Rassoulzadegan M. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]