Summary
The human gut microbiota is a metabolic organ whose cellular composition is determined by a dynamic process of selection and competition. To identify microbial genes required for establishment of human symbionts in the gut, we developed an approach (insertion-sequencing, or INSeq) based on a mutagenic transposon that allows capture of adjacent chromosomal DNA to define its genomic location. We used massively parallel sequencing to monitor the relative abundance of tens of thousands of transposon mutants of a saccharolytic human gut bacterium, Bacteroides thetaiotaomicron, as they established themselves in wild-type and immunodeficient gnotobiotic mice, in the presence or absence of other human gut commensals. In vivo selection transforms this population, revealing functions necessary for survival in the gut: we show how this selection is influenced by community composition and competition for nutrients (vitamin B12). INSeq provides a broadly applicable platform to explore microbial adaptation to the gut and other ecosystems.
Introduction
Our indigenous microbial communities play critical roles in shaping myriad features of our biology. The distal gut hosts the majority of our microbes; these include representatives of all three domains of life, plus their viruses. The density of organisms occupying this habitat is astonishing, exceeding 1012 cells/mL. Most phylogenetic types (phylotypes) observed in the guts of humans and other mammals belong to just two bacterial divisions (phyla) - the Firmicutes and the Bacteroidetes (Ley et al., 2008). Microbial community (microbiota) exchange experiments indicate that gut community members are dynamically selected: for example, transplantation of a Proteobacteria-dominated zebrafish gut microbiota into germ-free mice transforms this community so that it comes to resemble a mouse gut microbiota, while transplantation of a mouse microbiota into germ-free zebrafish has the opposite effect, yielding a community that has the phylum-level characteristics of the native zebrafish microbiota (Rawls et al., 2006).
Within the Firmicutes and Bacteroidetes, hundreds to thousands of phylotypes partition available niches (professions) to create a community able to maintain itself in this continuously perfused ecosystem despite shifts in host diet, regular ingestion of foreign bacteria, intense resource competition, high bacteriophage levels, and immune surveillance. Comparisons of the sequenced genomes of cultured representatives of major gut phylogenetic lineages provide a means for identifying genomic features potentially important for colonization and competition in the gut. However, the recent surge in microbial genome sequencing projects has far outpaced development of broadly applicable tools for directly testing the role of genes in determining fitness in this habitat. The paucity of tools is unfortunate, as fundamental questions connecting genome content to function remain unexplored. For example, how are the determinants of fitness related to nutrient availability and how closely do the genes required for fitness in vivo mirror those required for maximizing growth rate in vitro? Does community structure influence this map of genetic requirements, or is competition largely ‘within species’? Do the major recognized components of the host immune system play a dominant role in determining which genes are critical for symbiont fitness in vivo? To address these questions, we integrated a simple and broadly applicable genetic tool with second-generation DNA sequencers and gnotobiotic mouse models to identify fitness determinants in the genome of a human gut mutualist.
Mariner transposon mutagenesis is an attractive forward genetic strategy for connecting phenotype to gene because stable random insertions can be generated in a recipient genome without specific host factors: the ability of these transposons to serve as mutagenic agents is well established in members of all three domains of life (Lampe et al., 1996; Mazurkiewicz et al., 2006). After alignment of the inverted repeat (IR) sequences that delimit mariner family transposons, we noted that a single G–T transversion at a nonconserved position would create a recognition sequence for the Type IIs restriction enzyme, MmeI. When directed to this location, MmeI would cleave 16 bp outside of the transposon, capturing a genomic fragment that identifies the insertion site. Moreover, if the genome sequence of the recipient organism were known, the short genomic DNA sequences captured by this MmeI digestion would be sufficient to uniquely map transposon location. We reasoned that in a mixed population of transposon mutants produced in a given recipient bacterial species, the relative abundance of each MmeI-liberated genomic fragment, identified after limited PCR amplification and massively parallel sequencing, would in principle mirror the abundance of each mutant in the population. Putting this population under potentially selective conditions (e.g. colonization of the intestines of gnotobiotic mice) could then be used to highlight mutants that change in relative abundance, and thereby identify genes and pathways critical for fitness under these conditions (Figure 1A).
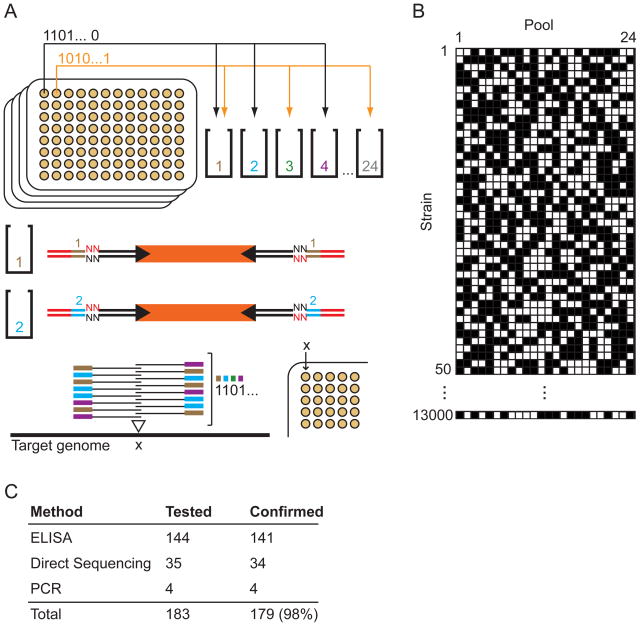
Figure 1. Mapping and quantifying tens of thousands of transposon insertion strains by high-throughput insertion sequencing (INSeq).
(A) A negative selection scheme for identification of genes required for colonization in vivo. Mutants in genes important for competitive growth (red) are expected to decrease in relative abundance in the output population. (B) Preparation of an INSeq library. Genomic DNA is extracted from the mutagenized bacterial population, digested with MmeI, and separated by polyacrylamide gel electrophoresis (PAGE). Transposon-sized fragments are appended with double-stranded oligonucleotide adapters by ligation. Limited cycles of PCR create the final library molecules for sequencing. (C) Map of insertion sites in the B. thetaiotaomicron genome. An arrow marks the origin of replication. (D) Reproducibility of library preparation and sequencing protocols. Technical replicates were prepared and sequenced from a single transposon mutant population. Each point represents the abundance of insertions in a single gene; the coefficient of determination, R2, on log-transformed abundance values is 0.92.
Using this insertion-sequencing (INSeq) approach, we subjected a mutagenized population of the prominent human gut symbiont, Bacteroides thetaiotaomicron, to varying selective pressures: in vitro growth in continuous flow chemostats, monoassociation of wild-type and knockout germ-free mice lacking major branches of their innate or acquired immune systems, and as one component of several defined communities of human gut-derived microbes. The results provide evidence that human gut symbionts, like their pathogenic counterparts, possess dedicated mechanisms critical for interaction with their host and each other. The relative importance of these mechanisms is not static, but instead shaped by other members of the microbiota. As a strategy for functional characterization of newly sequenced genomes in general, and of the human gut microbiome in particular, INSeq extends existing techniques in several important aspects.
Results
We constructed pSAM, a sequencing-adapted mariner transposon delivery vector with three major features: an antibiotic resistance cassette flanked by MmeI-modified mariner inverted repeats, a multiple cloning site immediately upstream of the himar1C9 mariner transposase (Lampe et al., 1999), and machinery for replication in the donor strain and transfer by conjugation (Figure S1A–C). B. thetaiotaomicron was chosen as the recipient species to test this approach for several reasons (Figure S2A). First, it is highly adapted to life in the distal human gut (Zocco et al., 2007). A prominent member of the gut microbiota, this mutualist is richly endowed with a broad arsenal of genes encoding glycoside hydrolases and polysaccharide lyases not represented in the human genome (Xu et al., 2003). These genes are incorporated together with genes encoding nutrient sensors and carbohydrate transporters into 88 polysaccharide utilization loci (PULs) representing 18% of organism’s genome (Martens et al., 2008). Thus equipped, B. thetaiotaomicron functions as a flexible forager of otherwise indigestible dietary glycans, as well as host glycans when dietary polysaccharides are not available (Martens et al., 2008). Second, >200 GeneChip datasets of B. thetaiotaomicron’s transcriptome have been collected during growth in vitro under a variety of conditions, after mono-colonization of germ-free mice fed different diets, as well as after co-colonization with another human gut bacterial or archaeal species (NCBI GEO archive). Third, functional genomic studies conducted in gnotobiotic mice have shown that monoassociation with B. thetaiotaomicron can recapitulate a number of host responses evoked by a complete mouse gut microbiota. Fourth, a limited number of in vivo competition experiments conducted in gnotobiotic mice colonized with isogenic wild-type and mutant B. thetaiotaomicron have identified a few fitness determinants that could serve as reference controls for the present study (Peterson et al., 2007).
Construction and characterization of a transposon mutant population by INSeq
We found that transfer of the MmeI-modified mariner transposon into the genome of B. thetaiotaomicron occurs with high efficiency (Figure S2). To identify the site of transposon insertion and the relative abundance of each mutant in an otherwise isogenic population, we developed a straightforward procedure to extract the two 16-bp genomic sequences adjacent to each transposon, append sequencing adapters to these fragments, and separate the desired molecules from genomic background (Figure 1B and Supplemental Experimental Procedures). Sequencing these tags using an Illumina Genome Analyzer II produced ~8 million raw reads from a single flowcell lane: ~90% of these reads contained the transposon (Figure S2D). We designed a software package, MapSAM, to filter out low-quality sequences, quantify and pair reads generated from either side of an insertion, and assign these paired reads to a specific location in the target genome (Figure 1C). Examination of technical replicates indicated that the library preparation, sequencing, and mapping strategies were highly reproducible (Figures 1D and S3). The proportion of reads that were unambiguously mapped (98%) matched predictions from an in silico model of random transposon insertion (Figure S4A); no insertion sequence bias beyond the expected ‘TA’ dinucleotide requirement (Bryan et al., 1990) was apparent (Figure S4B, C).
We first characterized a mutant population containing ~35,000 B. thetaiotaomicron transposon insertion strains. Insertions were well distributed across the genome at an average density of 5.5 insertions/kb (Figure 1C). After filtering out insertions in the distal (3′) 10% of any coding region (because such insertions could possibly still permit gene function) we found that 3435 of the 4779 predicted open reading frames in the genome (72%) had been directly disrupted in the mutant population. Inclusion of genes disrupted by upstream (polar) mutations in a predicted operon increased this number to 78%; rarefaction analysis suggested this population is approaching saturation (Figure S4D).
To identify B. thetaiotaomicron genes unable to tolerate transposon insertion, we generated and mapped a second, independent mutant population. We combined both data sets and applied a Bayesian model to account for the number of informative insertion sites in each gene (Lamichhane et al., 2003). The results yielded a conservative list of 325 candidate essential genes for growth under anaerobic conditions when plated onto rich (tryptone-yeast extract-glucose; TYG) medium (Table S1). These genes were significantly enriched (Benjamini-Hochberg corrected p<0.05) for Clusters of Orthologous Groups (COG) categories representing cell division (category D), lipid transport/metabolism (I), translation/ribosomal structure/biogenesis (J), and cell wall/membrane biogenesis (M). This is consistent with genome-wide mutagenesis studies of Escherichia coli and Pseudomonas aeruginosa (Baba et al., 2006; Jacobs et al., 2003). For non-essential genes, insertion frequency showed some correlation (R2 of log-transformed values = 0.33) with GeneChip-defined expression levels during mid-log phase growth of the parental wild-type strain in batch fermentors containing rich TYG medium (Figure S4E). The reason for this relationship is not known, although studies of mariner transposition in vitro suggest that the enzyme has a preference for bent or bendable DNA (Lampe et al., 1998).
Combinatorial mapping of individual insertion strains from an archived mutant collection
A mutant population of this complexity contains insertions in most of the coding potential of the genome and can facilitate forward genetic approaches for identifying genotypes connected with a phenotype of interest. These mixed populations, however, are less amenable to reverse genetics: specific genotypes are not individually retrievable. Arrayed transposon mutant collections, in which strains of known genotype are archived individually, provide an important avenue for retrieval and further study of specific strains of interest. To date, such collections have been created using a strain-by-strain procedure that typically consists of cell lysis, removal of cellular debris, multiple rounds of semi-random or single-primer PCR, DNA cleanup, and individual Sanger sequencing of each amplicon. As an alternative, we developed a combinatorial technique for simultaneously mapping thousands of individually archived transposon mutant strains in parallel (Figures 2A, S5, and Supplemental Experimental Procedures).
Figure 2. Mapping an archived strain collection by combinatorial pooling and high-throughput sequencing.
(A) Individual strains are archived in a 96-well format and then placed into a subset of 24 pools according to a unique 24-bit binary string assigned to each strain. Sub-libraries are prepared from each of these pools using one of 24 pool-specific barcoded dsDNA adapters in the ligation step. Sub-libraries are then combined and sequenced in a single run. Reads mapping to a specific insertion location are compiled and the associated pool-specific barcodes are identified to recreate the 24-bit string (and with it, the physical location of the corresponding strain in the archived collection). (B) Sample pool distribution patterns for archived strains. Strains are placed in pools designated with white boxes and omitted from pools marked with black boxes. The 13,000 patterns generated are each distinct in at least six positions and do not overlap to produce another pattern in the set. (C) Confirmation of strain-insertion assignments.
This approach consists of three basic steps: (1) culturing and archived storage of randomly picked mutant colonies in individual wells of 96-well (or higher density) plates; (2) placement of each of these strains into pools in unique patterns; and (3) sequencing of these pools by INSeq in order to associate each transposon insertion location with a strain in the original arrayed multi-well plates. Because n pools can contain 2n unique presence/absence patterns, a small number of pools can uniquely identify a large number of strains. To this end, a benchtop liquid handling robot was used to distribute archived transposon mutant strains across a subset of pools in a pattern selected to minimize the likelihood of mistaking one strain for another or incorrectly mapping clonal strains (Figure 2B and Supplemental Experimental Procedures). Sub-libraries were then prepared from each pool using the same method described in Figure 1B, except that a pool-specific, barcoded adapter (Table S2) was used in the ligation step. These sub-libraries were combined into a single sample that can be sequenced with an Illumina Genome Analyzer II using just one lane of the instrument’s 8-lane flow cell. Reads were first mapped to the reference B. thetaiotaomicron genome to determine insertion sites; to assign an insertion site to a specific archived strain, the pool-specific barcodes associated with a given insertion location on the chromosome were then matched with the patterns assigned to the strains in the original archived set of plates.
Using this strategy, we were able to identify the insertion coordinates for over 7000 individually archived B. thetaiotaomicron transposon mutant strains in parallel (Table S3). To verify the accuracy of these assignments, we first used ELISA to test strains predicted to have lost reactivity to two monoclonal antibodies of known specificity (Peterson et al., 2007). We also amplified transposon-genome junctions of test strains by semi-random PCR and sequenced the amplicons. In total, 179 of 183 strains tested (98%) produced the anticipated results (Figure 2C), confirming that combinatorial barcoding and INSeq can efficiently and economically generate archived, sequence-defined, mutant collections.
Identification of genes required for fitness in vitro
To identify genes that contribute to exponential growth in nutrient-rich conditions in vitro, we maintained a 35,000-strain mutant population in this growth phase (OD600 0.1–0.4), under anaerobic conditions, in chemostats that were continuously supplied with fresh TYG medium. Output populations were sampled from four independent chemostats; two after ~15h of continuous exponential growth and two after ~45h of growth. DNA was prepared from each of these populations and transposon-adjacent genomic fragments were identified by INSeq. After removing insertions in the 3′ 10% of each gene, a z-test was applied to identify genes that show a significantly altered representation from the overall distribution of output:input abundance ratios after q-value correction for multiple hypothesis testing (q<0.05; see Supplemental Experimental Procedures). To test this approach for identifying fitness determinants, we also examined insertions in intergenic ‘neutral loci’ (Figure S6A). None of these 80 control regions passed the statistical cutoff for underrepresentation (3 increased in abundance). In contrast, 477 genes (~14% of the genes represented in the input population) showed a statistically significant change in abundance after in vitro selection (265 underrepresented/212 overrepresented; Figure S6B and Table S4). Consistent with selection for maximal growth rate in rich medium, the list of factors important for fitness under these conditions was significantly enriched in genes annotated as being in COG category C (energy production and conversion).
As a proof-of-principle, we asked whether an observed enrichment in a broad functional group (COG category), represented among genes required for fitness, could be altered by manipulating environmental conditions. To do so, we harvested exponentially growing cells from mutant populations grown in minimal defined medium in the presence or absence of exogenous amino acids and quantified the abundance of transposon mutants by INSeq (n=4 replicate populations, each assayed independently). Gratifyingly, the set of genes required for fitness specifically in the amino acid-deplete condition was most highly enriched (p<0.0005) in COG category E (amino acid transport/metabolism) (Figure S7 and Table S5).
Genes required for establishment of B. thetaiotaomicron within the distal gut (cecum) of mono-associated gnotobiotic mice
To survey the B. thetaiotaomicron genome for genes critical for fitness in a mammalian gut ecosystem, we colonized germ-free mice with a single gavage of approximately 108 colony-forming units (CFU) of the 35,000-strain mutant population (n = 15 animals representing 3 independent experiments, each involving a cohort of five 8–12 week old C57BL/6J males; experiments were performed ~3 months apart). The five animals in each cohort were caged individually, in a shared gnotobiotic isolator, and fed a standard, autoclaved, polysaccharide-rich, low fat chow diet ad libitum. The relative abundance of each mutant strain in the cecal bacterial population was defined at the time of sacrifice 14d after gavage; this interval between gavage and sacrifice encompassed several cycles of turnover of the mucus layer and the underlying gut epithelium, and is sufficient to allow mobilization of innate and adaptive immune responses (Peterson et al., 2007).
All recipients of the gavage harbored equivalent levels of B. thetaiotaomicron at the time of sacrifice (~1011–1012 CFU/mL cecal contents as quantified by plating and by qPCR). Moreover, the relative representation of mutants was consistent between the ceca of individual mice (Tables S9–S11), and for most genes reflected their abundance in the input population (Figure 3A). However, compared to the input population, mutants in 370 genes showed significantly (q<0.05) altered representation (90 overrepresented/280 underrepresented) in all three cohorts of mice (Table S6; note that the largest category of genes identified in this screen encode hypothetical or conserved hypothetical proteins). Only one of the 80 ‘neutral intergenic’ controls described above was significantly underrepresented in these populations (two were overrepresented).
Figure 3. Identification of genetic determinants of fitness in vivo.
(A) The transposon mutant population is largely stable in vivo. The relative abundance of mutations in each gene (points) was compared between input and output (median from wild-type monoassociated mice, n = 15) populations. Genes that show a statistically significant change (q<0.05) in representation in all three cohorts of mice are shown in red, others in gray. The relative abundances of 80 gene-sized ‘neutral loci’ are shown in black (no significant change) and green (three ‘neutral loci’ that pass the significance criteria). (B) Individual strains retrieved from the archived collection demonstrate that genes critical for fitness in vivo are dispensable in vitro. Each strain was cultured individually in TYG medium and doubling time was calculated from OD600 measurements. Mutants predicted by INSeq to have in vitro growth defects are marked with arrows. N/D, no growth detected. rnf, Na+-transporting NADH:ubiquinone oxidoreductase (BT0616-22); S-layer, putative S-layer locus (BT1953-7); nqr, Na+-translocating NADH-quinone reductase (BT1155-60); hly, hemolysin A (BT3459). (C) Insertions in the CPS4 locus (BT1338-58) show a consistent in vivo fitness defect. Individual insertion locations (open arrowheads) in a representative gene (BT1346; green arrow) are shown at top. Read counts for input (black) and output (orange) samples at each insertion location are indicated (output counts represent the median from the ceca of wild-type monoassociated mice, n = 15). Median output:input ratios for each gene (black/green arrows) across the CPS4 locus are shown below. Asterisks indicate the average FDR-corrected p-value (q) across 3 experimental cohorts (n = 5 mice/cohort): *q< 0.05; **q< 0.01.
While the smaller group of 90 genes that produce a competitive advantage in the cecum when mutated (highlighted in Table S6 in blue and green) are not significantly enriched in any broad predicted functional (COG) categories, only half can be explained by corresponding behavior in chemostats containing rich medium. The underrepresented genes show a similar trend: half (146) of the 280 genes critical for in vivo fitness can be predicted from growth defects in rich medium; the remainder (134/280), which are highlighted in Table S6 in yellow, do not show a defect after prolonged exponential growth in vitro. These include loci with diverse predicted functions, including assembly of polysaccharide- and protein- based surface structures (BT1339-55, BT1953-7), synthesis and utilization of vitamin B12-dependent cofactors (BT2090-1, BT2760), and an rnf-like oxidoreductase complex (BT0616-22) (e.g. Figure S8).
To confirm that the requirement for these genes in vivo could not be explained by general growth defects, we analyzed individual mutant strains retrieved from the archived strain collection. This collection contained sequence-defined, single-insertion transposon mutants in ~70% of the genes designated as critical for survival in the distal gut in vivo (~80% if predicted polar effects are included) (Table S3). After validating the site of selected transposon insertions by semi-random PCR and Sanger sequencing, we determined the exponential doubling time of representative strains individually (Figure 3B). Strains carrying transposon insertions in genes uniquely required in vivo had an in vitro growth rate similar to wild-type B. thetaiotaomicron, further suggesting that the critical function of these genes in vivo cannot be simply explained by a necessity for sustaining exponential growth in rich medium. In contrast, mutants that exhibited a competitive defect in the 35,000-strain population both in vitro and in vivo had a slower doubling time when cultured individually, suggesting that these genes play a basic role in bacterial cell physiology.
An earlier report from our group used a targeted mutagenesis strategy to disable expression of genes encoding capsular polysaccharide (CPS) 4 in this organism; this strain was rapidly displaced by wild-type B. thetaiotaomicron after initial inoculation as a 1:1 mixture into germ-free mice (Peterson et al., 2007). The 35,000-strain transposon mutant population recapitulated this observation (Figures 3C and S9). The transposon mutant population included over 1100 independent insertions across 143 genes that span all eight CPS loci encoded in the B. thetaiotaomicron genome. None of the other CPS loci were required for fitness in vivo, indicating that CPS4 plays a unique role for B. thetaiotaomicron in the gut environment of mono-associated gnotobiotic mice fed a standard plant polysaccharide-rich chow diet. Moreover, our observation that transposon inactivation of genes in any single PUL did not confer a competitive disadvantage in vivo is consistent with B. thetaiotaomicron’s capacity for adaptive foraging of a broad range of glycans present in this diet (a total of 5137 distinct transposon mutants, involving 810 genes in all 88 polysaccharide utilization loci).
The impact of host genotype and community structure on selection in vivo
We next asked whether a broad view of the mutant population as a whole could help address some basic questions in mammalian gut microbial ecology described in the Introduction: are the genetic determinants of fitness influenced by the bacterial community (microbiota) context; do inter- and intra-specific competition play distinct roles in shaping the selective pressures on a genome; is this selection primarily maintained by elements of the host immune system?
To explore these questions, we manipulated two features of the host habitat: (i) the immune system, by introducing the B. thetaiotaomicron mutant population into germ-free mice with genetically engineered defects in innate or adaptive immunity (Myd88−/− and Rag1−/− respectively) or (ii) microbial composition, by including this mutant population as one component of three different types of defined communities: one consisting of 6 other sequenced human gut-associated Bacteroidetes, another composed of 8 sequenced human gut-associated Firmicutes and Actinobacteria, and a third consisting of all 14 of these representatives of the human distal gut microbiota (n = 4–5 animals per treatment group; Figure 4A and Table S7). Animals were sacrificed 14d after gavage and their cecal contents harvested. qPCR assays of cecal DNA, using species-specific primers, revealed that gavage with a given multi-species input community yielded consistent cecal ‘output’ community compositional profiles (Figure 4B and Table S8).
Figure 4. Identification of environmental and microbial factors that determine the fitness landscape for B. thetaiotaomicron.
(A) Shifts in the population structure of the ~35,000-strain B. thetaiotaomicron mutant population reflect alterations in the selective pressures on its genome. The relative abundance of each mutant in the B. thetaiotaomicron population was evaluated in multiple host genotypes (wild-type C57Bl/6; Rag1−/−; Myd88−/−) and microbial contexts. (B) qPCR assays of cecal microbial community composition in gnotobiotic mice at the time of sacrifice. The ~35,000-strain B. thetaiotamicron population is indicated with arrows.
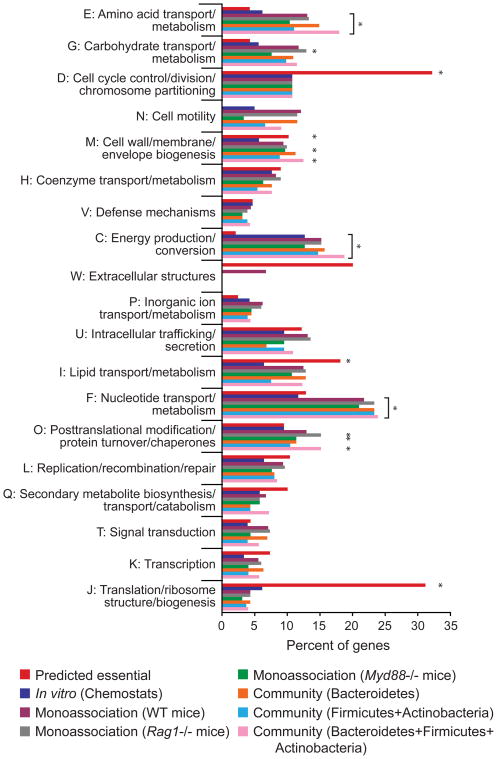
Each of these treatments shifted the B. thetaiotaomicron population from its input distribution (Tables S9–S12). To search for functional trends in these shifts, we assigned the genes underrepresented in output populations to functional (COG) categories. The in vivo fitness determinants were significantly enriched in different predicted functions compared to essential genes, or to the genes required for maximal exponential growth in rich medium in vitro (Figure 5). For example, the predicted essential genes are most prominently enriched in COG categories J (translation, ribosome structure/biogenesis) and D (cell cycle control and cell division), neither of which is enriched among the in vivo fitness determinants. Instead, the genes required in vivo are biased towards energy production/conversion (category C), and amino acid and nucleotide transport/metabolism (COG categories E and F, respectively). This represents an expansion beyond the single category (C) enriched after selection for maximal growth rate in vitro and is consistent across all in vivo treatment groups.
Figure 5. COG category-based classification of genes critical for fitness in vitro and in vivo.
Percent representation was calculated as (number of genes in a COG category/number of genes underrepresented in output population). Significant enrichments in specific COG categories, assessed by comparing these percentages to a null expectation based on the size of the gene list and the representation of a given category in the genome, are marked with asterisks (Benjamini-Hochberg corrected p<0.05).
Closer examination of enriched COGs and carbohydrate-active enzyme (CAZy) families highlight the role of polysaccharide synthesis for competitive fitness in this environment; overrepresented functions include UDP-glucose-4-epimerases and glycosyltransferases [specifically, GT2 and GT4 families (Cantarel et al., 2009)] (Table S13). Together, these observations suggest that at the level of statistical enrichment of broad functional groups, the in vivo fitness requirements were distinct from those derived in vitro, but that these enrichments were consistent across in vivo treatment conditions. Many of the annotated fitness determinants (e.g. CPS4 and the rnf-like oxidoreductase) followed this pattern: dispensable in vitro but critical across all in vivo conditions tested.
This functional category enrichment analysis depends critically on genome annotation (~50% of genes were not assignable to COG categories), while not accounting for differences in selection magnitude between treatments. To circumvent these limitations, we used unsupervised hierarchical clustering to evaluate the relationships between all in vitro and in vivo output populations (48 samples in total). In this analysis, samples are clustered based on the relative abundance of mutants in each gene independent of annotation. A dendrogram of the clustering results separates the in vitro populations from those shaped by in vivo selection (Figure 6A). The populations recovered from monoassociated wild-type, Rag1−/− and Myd88−/− mice formed shared branches on this tree of mutant population structures, suggesting that B. thetaiotaomicron may have multiple, redundant pathways for countering immune pressures, at least under the conditions of these experiments. In contrast, the transposon mutant populations recovered from mice that also harbored other Bacteroidetes species were fully separable from the population structures selected in B. thetaiotaomicron monoassociations or those resulting from the presence of a Firmicutes+Actinobacteria consortium. These patterns were recovered in >99% of bootstrapped dendrograms, and also by unsupervised hierarchical clustering of samples based only on the genes that showed a significantly altered output:input ratio by z-test (data not shown).
Figure 6. Clustering of B. thetaiotaomicron mutant population structures after manipulation of host environment or microbial context.
(A) Unsupervised hierarchical clustering of B. thetaiotaomicron mutant population structures in vitro and in vivo. Branches are colored by treatment. Pie charts indicate the microbial context of the mutant population in samples from different branches of the tree. Bootstrap support is indicated by a square at each node: >50% (white), >90% (yellow), >99% (black/unmarked). (B) Principal coordinates analysis based on the representation of transposon-disrupted genes.
To further evaluate the robustness of the clustering algorithm, we conducted a principal coordinates analysis on these 48 transposon mutant populations (Figures 6B and S10). Similar to the hierarchical clustering dendrograms, the first principal coordinate separates in vitro from in vivo mutant populations. The second coordinate separates these populations by microbial context: Bacteroidetes-containing communities shape a B. thetaiotaomicron mutant population that is distinct from that produced in monoassociations, or in the Firmicutes+Actinobacteria consortia, further indicating that although the same broad functional categories are enriched among B. thetaiotaomicron fitness determinants under a range of in vivo conditions, these mutant populations are additionally shaped by changes in microbial community composition.
We applied a random forest classifier (Breiman, 2001) to identify genes that were responsible for the observed separation of mutant populations in monoassociated mice from those mutant populations present in Bacteroidetes co-colonized mice. This machine-learning algorithm serves to estimate the importance of predictor variables (i.e. genes) for differentiating between classes (i.e. the monoassociation versus Bacteroidetes co-colonized groups, which were distinguished by unsupervised clustering and principal coordinates analysis). This approach identified a total of 220 genes as important for differentiating these groups (Table S14). Mutants in 144 of these predictor genes had lower output:input ratios in the monoassociations: in other words, these genes were more important when other Bacteroidetes were not present. Mutants in 76 genes had lower output:input ratios in mice co-colonized with other Bacteroidetes. These 76 genes, which provide a signature of functions under increased selection for B. thetaiotaomicron in the presence of other Bacteroidetes, are enriched for components of amino acid biosynthetic pathways, suggesting that although these functions are required in all in vivo conditions, other Bacteroidetes may outcompete B. thetaiotaomicron for exogenous amino acids.
The functional requirement for a vitamin B12-regulated locus is modulated by community composition
165 independent transposon insertions, mapping to five adjacent genes (BT1957-53), conferred a drastic fitness disadvantage during monoassociation of germ-free mice, yet had no impact on exponential growth in vitro. Moreover, their effect on fitness was influenced by community context: the Bacteroidetes-only community exacerbated the competitive defect while the Firmicutes+Actinobacteria consortia fully nullified the requirement for these genes. Introducing all 14 of these species resulted in an intermediate phenotype (Figure 7A, B).
Figure 7. Community context modulates the fitness requirement for a vitamin B12-regulated locus in B. thetaiotaomicron.
(A) Genetic organization and predicted annotation of the BT1957-49 locus. (B) The relative abundance of transposon insertion mutants in input and output mutant populations is dependent on community context. Asterisks indicate FDR-corrected p-value (q) for each cohort: *q< 0.05; **q< 0.01. Few insertions/reads were identified for BT1952-49; fitness defects were similar in wildtype, Rag1−/−, and Myd88−/− monoassociated animals (data not shown). (C) BT1956 gene expression is regulated by vitamin B12. Error bars represent one standard deviation based on triplicate qRT-PCR experiments. Similar results were observed for BT1954 (data not shown). Interestingly, BT1956 insertion mutants do not exhibit growth defects in medium containing either 37 nM or 0.37 nM B12, suggesting that multiple loci are involved in acquisition of this vitamin in vitro (data not shown). Consistent with this observation, other loci annotated as being potentially involved in B12 uptake were coordinately upregulated with BT1957-49 in the genome-wide transcriptional profiling experiments discussed above. (D) Pie charts showing that the defined microbial communities characterized in this study vary in their capacity for vitamin B12 biosynthesis. Color codes: dark blue, species with a predicted complete biosynthetic pathway (see Tables S16 and S17 for annotations) that are able to grow in defined medium lacking B12 (‘demonstrated prototrophs’); light blue, organisms with a predicted complete biosynthetic pathway able to grow on rich medium but not on defined medium with or without B12 (‘predicted prototrophs’); dark green, species without a complete pathway whose growth in defined medium requires B12 (‘demonstrated auxotrophs’); light green, species without a complete biosynthetic pathway unable to grow on defined medium with or without B12 (‘predicted auxotrophs’); black, species that do not grow on defined medium with or without B12 but that possess a B12-independent methionine synthase (the presence of such an enzyme implies the absence of a B12 requirement). The relative proportions of prototrophs and auxotrophs shown represent the average in each community in vivo as determined by species-specific qPCR of cecal contents. (E) The B. thetaiotaomicron fitness requirement for BT1957-3 correlates with levels of the B12-prototrophic species Ruminococcus obeum in the microbial community. Each point represents an individual mouse containing a defined multi-species microbiota in addition to the B. thetaiotaomicron transposon mutant population. Color code: blue, Firmicutes+Actinobacteria community; pink, Bacteroidetes+Firmicutes+Actinobacteria. The relative abundance of R. obeum (determined by qPCR analysis of cecal contents at the time of sacrifice) is plotted against the average output:input ratio of B. thetaiotaomicron transposon mutants in genes BT1957-3 in each individual.
Examination of B. thetaiotaomicron transcriptional profiles (NCBI GEO archive) disclosed that expression of genes in this locus (spanning BT1957-BT1949) is strongly upregulated in vivo compared to growth in vitro under a variety of conditions. Moreover, in two closely matched experiments conducted in defined minimal medium that differed in five components, expression was modulated >10-fold (Table S15). This observation was validated by qRT-PCR assays of BT1954 and BT1956 (data not shown). Systematic addition of each variable component revealed that locus transcription is induced in response to reduced levels of vitamin B12 (Figure 7C). Moreover, the ABC transporter encoded by BT1952-50 shares homology with the BtuFCD B12 acquisition system of S. typhimurium LT2 (23%, 35%, and 25% identity, respectively).
Vitamin B12 is critically involved in normal mammalian physiology, yet is synthesized exclusively by microbes (Krautler, 2005). Because the complete genome sequence of each member of the defined microbial communities used in our experiments was known, we were able to use BLAST to identify homologs to known B12 synthesis, transport, or utilization genes in the synthetic human gut microbiomes (Tables S16 and S17). As described for the related species Porphorymonas gingivalis (Roper et al., 2000), members of the Bacteroidetes community were missing some or all of the genes necessary for synthesis of B12 or its direct precursors, but encoded predicted transporters and likely have an obligate B12 requirement for growth. In contrast, the Firmicute/Actinobacteria group contained several members that harbored complete B12 biosynthetic pathways. To test these predictions, we attempted to culture each species on defined medium in the presence and absence of vitamin B12. R. obeum, a Firmicute predicted to require this vitamin for methionine synthesis, grew robustly without B12 in the medium, while the Bacteroidetes were auxotrophic (Table S16). These observations suggest that the capacity for vitamin B12 biosynthesis is determined by community structure (Figure 7D) and that B. thetaiotaomicron responds to changes in community membership via the gene products encoded by BT1957-49. Consistent with this observation, the fitness defect of BT1957-3 mutants correlated well with variation in R. obeum levels between mice in vivo (R2 of log-transformed values = 0.77; Figure 7E).
Discussion
By simultaneously profiling the relative abundance of tens of thousands of B. thetaiotaomicron mutants across multiple conditions, we identified hundreds of genes that are critical for the fitness of this prominent human gut symbiont in vivo. A large number of these genes are not determinants of exponential growth in nutrient-replete medium in vitro.
This approach (insertion sequencing, or INSeq) for functional genome-wide analysis of organisms for which a genome sequence (and possibly little else) is known, is generally applicable and extends existing techniques in several important ways. First, a single transposon replaces the sets of individually barcoded variants needed for signature-tagged mutagenesis (Hensel et al., 1995). Second, high-throughput sequencing provides a general alternative to the species-specific DNA microarrays required for hybridization-based mutant profiling (Mazurkiewicz et al., 2006). Third, this sequencing-based strategy identifies the precise genomic location and provides a ‘digital’ count-based abundance readout of individual insertions in both coding and noncoding regions. In this way, independent insertions with shared behavior serve to validate gene-level fitness effects. Finally, because mariner family transposon activity has been demonstrated in Bacteria, Archaea, and Eukarya, this method is generalizable. Further, the barcoded pooling strategy used to create a sequence-defined archived strain collection allows for retrieval of individual strains of interest for follow-up studies of the impact of individual gene disruptions on various microbial functions and adaptation. In this way, a forward genetic tool (a mutagenized cell population that can be screened for phenotypes en masse) can also serve as a platform for reverse genetics (a collection of isogenic, sequence-defined mutations in most of the coding potential of the target genome).
Surprisingly, mice lacking major branches of the immune system did not exhibit noticeably restructured B. thetaiotaomicron mutant populations compared to wildtype animals. It is possible that examination of microbial populations in closer contact with the host mucosa, or populations from host animals exposed to intentional immune stimulation, would aid the identification of genes differentially required for fitness in response to host immune surveillance. INSeq already can be applied to relatively small amounts of starting material: further decreases should help address the largely unexplored question of the relationship between the activities of the innate and/or adaptive immune systems and the biogeography of the microbiota.
Our study underscores how selection is shaped by microbial context. Because the INSeq strategy specifically targets transposon-adjacent chromosomal fragments, it is possible to monitor changes in the structure of a mutagenized population (in either wild-type or genetically manipulated gnotobiotic mice) even if this population constitutes a small fraction of a larger microbial community. For example, using unsupervised hierarchical clustering and principal coordinates analysis to evaluate the relationships between B. thetaiotaomicron mutant population structures in defined microbial communities in vivo, we observed that the presence of other Bacteroides was an important determinant of the selective pressures acting on the B. thetaiotaomicron genome. Additionally, we identified a mechanism by which B. thetaiotaomicron senses and responds to changes in gut microbial community structure: this species employs the products of BT1957-49 in response to variations in vitamin B12 levels that result from changes in community composition. These genes are dispensable in rich medium in vitro and become increasingly critical for fitness in vivo as the levels of B12 prototrophic species (such as R. obeum) are reduced. The mechanism by which R. obeum relieves this selective pressure, the role of the surface (S-layer) proteins encoded by this locus, and the consequences of microbial B12 competition on host physiology await further study.
In principle, INSeq can be extended to an analysis of communities of defined species composition introduced into gnotobiotic mice together with, before, or after, introduction of one or more transposon-mutagenized species. Subsequent INSeq-based time series studies of these deliberately constructed microbial populations offer the opportunity to address a wide range of unanswered questions about properties of our gut microbiota, ranging from the characteristics, determinants, and ecologic principles underlying its initial assembly, to the genomic correlates of niche partitioning among its members, to the genetic and metabolic factors that determine the persistence and impact of various probiotic and enteropathogen species (and degree to which the functions required for persistence of gut mutualists overlap with those of pathogens (Hendrixson and DiRita, 2004; Lalioui et al., 2005; Liu et al., 2008; Shea et al., 2000)].
In summary, INSeq can be applied to a variety of phylotypes to identify factors that shape their adaptations to myriad environments, and further, to readily retrieve mutants in the genes encoding such factors. As such INSeq, and the rapidly evolving capacity for massively parallel DNA sequencing that supports its application, should be a useful platform for microbial genetics, genomics and ecology.
Experimental Procedures
Bacterial culture conditions
Escherichia coli S-17 λ pir strains (Cowles et al., 2000) were grown at 37°C in LB medium supplemented with carbenicillin 50 μg mL−1 where indicated. B. thetaiotaomicron VPI-5482 (ATCC 29148) was grown anaerobically at 37°C in liquid TYG medium (Holdeman, 1977), or on brain-heart-infusion (BHI; Beckton Dickinson) agar supplemented with 10% horse blood (Colorado Serum Co.). Antibiotics (gentamicin 200 μg mL−1 and/or erythromycin 25 μg mL−1) were added as indicated. Other human gut-derived species were cultured in supplemented TYG(TYGs: see Supplemental Experimental Procedures).
Genetic techniques
DNA purification, PCR, and restriction cloning were performed using standard methods. Primer sequences are provided in Table S19. pSAM construction and mutagenesis protocols are described in Supplemental Experimental Procedures.
Preparation and sequencing of transposon population libraries
A detailed protocol is provided in Supplemental Experimental Procedures. Genomic DNA was purified, digested with MmeI, and separated by PAGE. Transposon-sized fragments were extracted from the gel and ligated to a double-stranded DNA adapter bearing a 3′-NN overhang. PAGE-purified adapter-ligated library molecules were PCR amplified for 18 cycles using a transposon-specific and an adapter-specific primer. The 125bp product was purified by PAGE and sequenced using an Illumina Genome Analyzer as described in the user’s manual. Sequence images were converted into raw reads using Illumina software with default settings. Filtered, normalized, and mapped sequencing results from all samples are provided in Tables S9–S12 and S18.
Gnotobiotic husbandry
All experiments using mice were performed using protocols approved by the animal studies committee of Washington University. Germ-free mice were maintained in gnotobiotic isolators and fed a standard autoclaved chow diet (B&K Universal, East Yorkshire, UK) ad libitum. Animals were sacrificed 14d after gavage and cecal contents frozen immediately at −80°C.
Supplementary Material
Acknowledgments
We thank David O’Donnell, Maria Karlsson, Sabrina Wagoner, Nicole Koropatkin, Daniel Peterson, Pankaj Pal, Laura Langton, Jessica Hoisington-Lopez, Xuhua Chen, Laura Kyro, and James Dover for assistance, plus Gary Stormo, Jay Shendure, Ryan Kennedy, and the Gordon laboratory for helpful suggestions. This work was supported by NIH grants DK30292 and 1F32AI078628-01 (to A.G.).
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–2008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- Bryan G, Garza D, Hartl D. Insertion and excision of the transposable element mariner in Drosophila. Genetics. 1990;125:103–114. doi: 10.1093/genetics/125.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CE, Nichols NN, Harwood CS. BenR, a XylS homologue, regulates three different pathways of aromatic acid degradation in Pseudomonas putida. J Bacteriol. 2000;182:6339–6346. doi: 10.1128/jb.182.22.6339-6346.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson DR, DiRita VJ. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol. 2004;52:471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- Holdeman LV, Cato ED, Moore WEC. Anaerobe Laboratory Manual. Blacksburg, VA: Virginia Polytechnic Institute and State University Anaerobe Laboratory; 1977. [Google Scholar]
- Jacobs MA, Alwood A, Thaipisuttikul I, Spencer D, Haugen E, Ernst S, Will O, Kaul R, Raymond C, Levy R, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautler B. Vitamin B12: chemistry and biochemistry. Biochem Soc Trans. 2005;33:806–810. doi: 10.1042/BST0330806. [DOI] [PubMed] [Google Scholar]
- Lalioui L, Pellegrini E, Dramsi S, Baptista M, Bourgeois N, Doucet-Populaire F, Rusniok C, Zouine M, Glaser P, Kunst F, et al. The SrtA Sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine. Infect Immun. 2005;73:3342–3350. doi: 10.1128/IAI.73.6.3342-3350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane G, Zignol M, Blades NJ, Geiman DE, Dougherty A, Grosset J, Broman KW, Bishai WR. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2003;100:7213–7218. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci U S A. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe DJ, Churchill ME, Robertson HM. A purified mariner transposase is sufficient to mediate transposition in vitro. Embo J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- Lampe DJ, Grant TE, Robertson HM. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, Lee SM, Vanlare JM, Kasper DL, Mazmanian SK. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci USA. 2008;105:3951–3956. doi: 10.1073/pnas.0709266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurkiewicz P, Tang CM, Boone C, Holden DW. Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nat Rev Genet. 2006;7:929–939. doi: 10.1038/nrg1984. [DOI] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper JM, Raux E, Brindley AA, Schubert HL, Gharbia SE, Shah HN, Warren MJ. The enigma of cobalamin (Vitamin B12) biosynthesis in Porphyromonas gingivalis. Identification and characterization of a functional corrin pathway. J Biol Chem. 2000;275:40316–40323. doi: 10.1074/jbc.M007146200. [DOI] [PubMed] [Google Scholar]
- Shea JE, Santangelo JD, Feldman RG. Signature-tagged mutagenesis in the identification of virulence genes in pathogens. Curr Opin Microbiol. 2000;3:451–458. doi: 10.1016/s1369-5274(00)00120-x. [DOI] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Zocco MA, Ainora ME, Gasbarrini G, Gasbarrini A. Bacteroides thetaiotaomicron in the gut: molecular aspects of their interaction. Dig Liver Dis. 2007;39:707–712. doi: 10.1016/j.dld.2007.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.