Abstract
The binding of RCC1 (Regulator of Chromosome Condensation) to chromatin is critical for cellular processes such as mitosis, nucleocytoplasmic transport and nuclear envelope formation because RCC1 recruits the small GTPase Ran to chromatin and sets up a Ran-GTP gradient around the chromosomes. However, the molecular mechanism by which RCC1 binds to nucleosomes, the repeating unit of chromatin, is not known. We have used biochemical approaches to test structural models for how the RCC1 β-propeller protein could bind to the nucleosome. In contrast to the prevailing model, RCC1 does not appear to use the β-propeller face opposite to its Ran-binding face to interact with nucleosomes. Instead, we find that RCC1 uses a conformationally flexible loop region we have termed the switchback loop in addition to its N-terminal tail to bind to the nucleosome. The juxtaposition of the RCC1 switchback loop to its Ran binding surface suggests a novel mechanism for how nucleosome-bound RCC1 recruits Ran to chromatin. Furthermore, this model accounts for previously unexplained observations for how Ran can interact with the nucleosome both dependent and independent of RCC1, and how binding of the nucleosome can enhance RCC1’s Ran nucleotide exchange activity.
Introduction
Mitosis, nucleocytoplasmic transport and nuclear envelope dynamics all rely on spatial coordination within the eukaryotic cell mediated by the small GTPase Ran (Ras-related nuclear) protein and its guanine-exchange factor, RCC1 (Regulator of Chromosome Condensation 1) 1; 2; 3; 4; 5; 6. RCC1 associates with chromatin and both recruits and activates Ran to create a concentration gradient of RanGTP (Ran in its GTP bound state) around the chromosomes. This RanGTP gradient is a key positioning signal within the cell for the aforementioned cellular processes and depends fundamentally on the binding of RCC1 to nucleosomes.
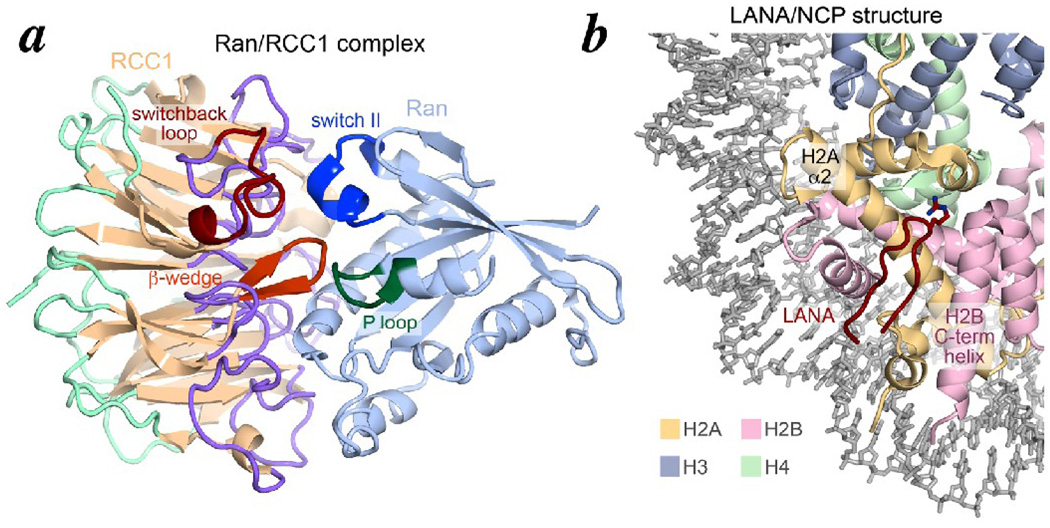
Previous biochemical and structural studies have determined the structures of RCC1 and Ran, and how they interact with each other. Human RCC1 is a 421 amino acid protein with an N-terminal tail of approximately 20 residues and a C-terminal seven-bladed β-propeller domain with strong structural similarity to WD40 repeat proteins such as the β-subunit of signal transduction G proteins (Fig. 1a) 7. Each blade in the β-propeller structure contains four antiparallel strands with loops between each strand. The equivalent loops which decorate the two faces of the β-propeller structure in the WD40 β-propeller cousin proteins frequently mediate protein-protein interactions with interacting proteins 8; 9; 10; 11; 12; 13; 14; 15; 16. In fact, the crystal structure of RCC1 bound to Ran shows that loops from all seven blades on one side of the RCC1 β–propeller interact with Ran in the complex (Fig. 1a) 17. A β-hairpin extension of RCC1 repeat 3, termed the β-wedge, mediates interactions with key regions of Ran such as the Ran nucleotide-binding P-loop and the GTPase switch II region 17. These interactions explain how RCC1 promotes the otherwise intrinsically slow exchange of guanine nucleotides in Ran, thus creating the RanGTP gradient around the chromosomes through the combination of this guanine exchange activity and RCC1’s association with the nucleosome.
Figure 1.
Ran/RCC1 and LANA/nucleosome core particle structures. (a) Ribbon representation of Ran/RCC1 crystal structure (PDB id 1I2M). The RCC1 β-strands are shown in beige, the loops on the Ran-interacting side in cornflower blue, the loops on the non Ran-interacting side in light green, the conformationally diverse switchback loop in red and the β-wedge in orange. The Ran protein is shown in light blue with its nucleotide interacting regions switch II in blue and the P-loop in green. (b) Ribbon and stick representation of the LANA/nucleosome core particle crystal structure (PDB id 1ZLA). The histone proteins H2A, H2B, H3 and H4 are shown in yellow, pink, blue and green respectively while the nucleosomal DNA is colored in grey. The backbone of the LANA 14 amino acids visible in the structure is shown in red, as is the Arg9 which makes key interactions with the histone dimer.
In contrast, we have relatively little structural information regarding how RCC1 binds to chromatin despite the importance of this interaction for the formation of the RanGTP gradient. Two different mechanisms have been proposed for how RCC1 binds to the nucleosome. Based on initial observations that RCC1 has DNA-binding activity as judged by binding to DNA-cellulose resin, RCC1 was proposed to bind to chromatin via contacts with DNA 18. Furthermore, an unusual post-translational modification of RCC1, N-terminal α-methylation, has been found to affect RCC1-chromatin association in vivo and to confer a two-fold increase in double-stranded DNA binding activity in vitro 19. And yet, another study has shown that RCC1’s N-terminal tail with its DNA-binding activity is not required for chromatin binding and may instead function as a nuclear translocation signal for RCC1 20. A second mechanism for RCC1-chromatin interactions emphasizes protein-protein interactions instead of protein-DNA interac tions. Glutathione S-transferase (GST) pulldown assays showed that RCC1 associates directly with nucleosomes via the histone H2A/H2B dimer, and that the RCC1 β-propeller domain (i.e. without the N-terminal tail) was sufficient for this interaction 21. Since removing the histone tails did not affect RCC1/nucleosome interactions, it appears that the surface exposed globular regions of the histone H2A/H2B mediate binding of RCC1 to the nucleosome. However, the molecular mechanism for this interaction is not known. Nor is it understood why binding of RCC1 to the nucleosome enhances RCC1’s guanine-exchange factor activity on Ran 21.
As the fundamental repeating unit of chromatin, the nucleosome has been the subject of intensive investigation in gene regulation and chromatin biology 22; 23. Although once considered to be a repressive structure that occludes nuclear factors from interacting with DNA in a eukaryotic cell, the nucleosome is now understood to be an active participant in gene regulatory processes and that a multitude of chromatin modification, remodeling and transcription factors function on a nucleosome template 24; 25; 26. However, we still lack a detailed understanding of how such chromatin factors and enzymes actually interact with their nucleosome substrate. The crystal structure of the 23 amino acid Kaposi’s sarcoma herpesvirius LANA (latency-associated nuclear antigen) peptide bound to the nucleosome core particle currently offers the only atomic view of a chromatin factor interacting with the nucleosome 27. The crystal structure (Fig. 1b) and corroborating biochemical data indicate that the LANA peptide interacts with a solvent accessible acidic patch created by the nucleosome H2A α-helix α2 and the H2B C-terminal helix.
In this study, we analyze how RCC1 binds to the nucleosome by employing biochemical methods to test structural models. We find that RCC1 targets a region of the nucleosomal H2A/H2B dimer recognized by the LANA peptide. Contrary to our expectations, the surface used by RCC1 to bind to the nucleosome does not appear to be the β-propeller face opposite to the Ran-binding face. Rather, we find that RCC1 uses a loop region adjacent to the Ran-interacting β-wedge to bind to the nucleosome. These considerations have allowed us to develop a model that not only explains how RCC1 binds to the nucleosome but also offers insight into how RCC1 might recruit Ran to the nucleosome and activate its guanine exchange activity.
Results
RCC1 binds the nucleosome in competition with the LANA peptide
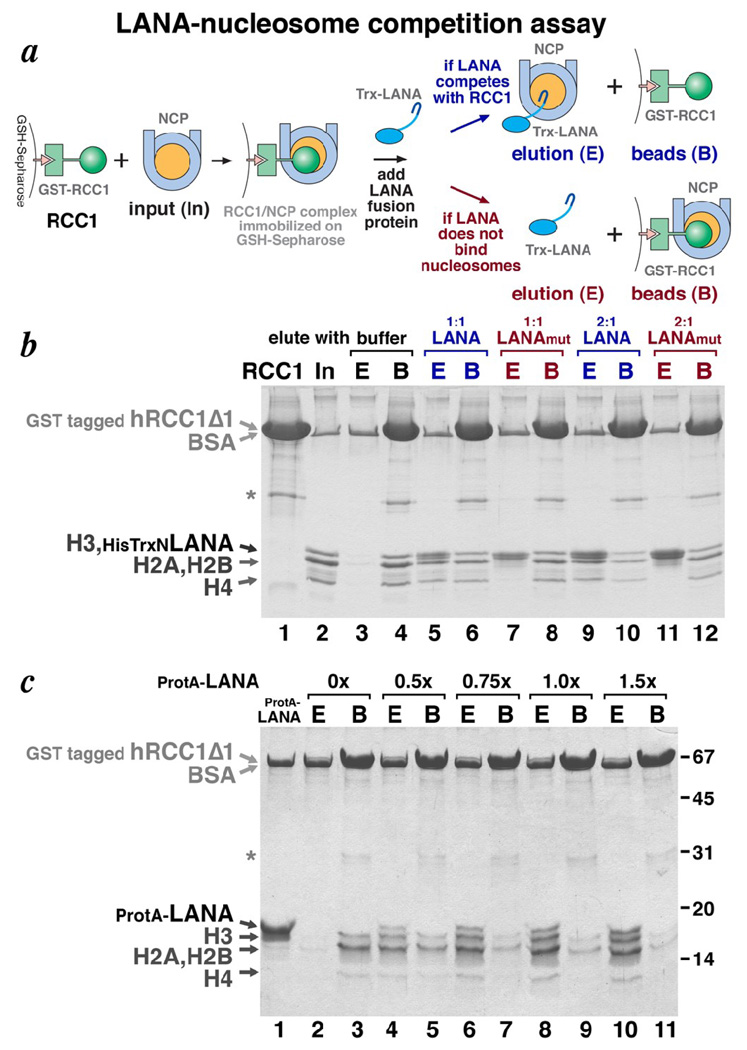
The 23 amino acid latency-associated nuclear antigen (LANA) peptide from Kaposi’s sarcoma herpesvirus binds to an acidic patch on the H2A/H2B histone dimer surface of the nucleosome (Fig. 1b) 27. Since RCC1 also has been shown to bind to the nucleosome via interactions with the H2A/H2B histone dimer 21, we asked if RCC1 is competitive or compatible with LANA binding to the nucleosome. We utilized a GST (glutathione S-transferase) competition pulldown assay to address this question (Fig. 2a). The β-propeller domain of human RCC1 (residues 21–421) was immobilized on glutathione-Sepharose via a GST fusion tag, and then allowed to bind recombinant nucleosome core particles (Fig. 2b, lanes 1 and 2). A LANA peptide-thioredoxin fusion protein was then added to the washed beads containing RCC1 and the bound nucleosome core particles. If LANA and RCC1 can simultaneously bind to nucleosomes, we would expect the beads to retain all three components. However, we find that adding increasing amounts of the LANA fusion protein causes the nucleosome core particles to elute from the RCC1 beads (Fig. 2b, lanes 5, 6, 9 and 10). To confirm that this release of nucleosomes is due to LANA displacing nucleosomes from RCC1, we repeated the experiment using a LANA mutant defective in binding the nucleosome 27. When the LANA LRS->AAA mutant-thioredoxin fusion protein (LANAmut) is used, the same additions of this mutant LANA-thioredoxin protein fail to dissociate nucleosome core particles from the RCC1 beads (Fig. 2b, lanes 7, 8, 11 and 12). These results indicate that RCC1 and LANA bind competitively to the nucleosome core particle, presumably due to overlapping binding sites on the histone dimer.
Figure 2.
LANA-nucleosome competition assay shows that RCC1 and LANA compete for binding to the nucleosome. (a) Schematic representation of the LANA-nucleosome competition assay. (b) Coomassie-stained SDS-PAGE gel for LANA-nucleosome competition assay using thioredoxin tagged LANA peptide. The recombinant GST tagged human RCC1Δ1 used is shown in lane 1. The asterisk marks the position of a contaminant. Lane 2 shows the input recombinant nucleosome core particles used in the experiment. The band that migrates at the same position as the tagged RCC1Δ1 corresponds to BSA present at 200 µg/ml in the assay. Samples eluted (E) by buffer or LANA peptide from nucleosome core particles bound to GST tagged RCC1Δ1 are shown in lanes 3, 5, 7, 9 and 11, while samples then remained bound before elution (B) from the GSH-Sepharose by glutathione are show in lanes 4, 6, 8, 10 and 12. Samples recovered with buffer are shown in lanes 3 and 4, with 1:1 molar ratio of LANA peptide in lanes 5 and 6, with 1:1 molar ratio of LANA LRS->AAA mutant peptide in lanes 7 and 8, with 2:1 molar ratio of LANA peptide in lanes 9 and 10, and with 2:1 molar ratio of LANA mutant peptide in lanes 11 and 12. (c) Coomassie stained SDS-PAGE gel for LANA-nucleosome competition assay using Protein A tagged LANA peptide. Samples eluted (E) by buffer (lane 2) or LANA peptide at 0.5, 0.75, 1.0 or 1.5 molar ratio of input nucleosome core particles (lanes 4, 6, 8 and 10) are shown, while the corresponding samples that remained bound before elution (B) from the GSH-Sepharose by glutathione are shown in lanes 3, 5, 7, 9 and 11.
This competitive binding of RCC1 and LANA to the nucleosome does not depend on the nature of the LANA fusion construct because a LANA fusion utilizing the IgG binding domains of Protein A instead of thioredoxin shows the same competitive binding, as demonstrated by the titration in Fig. 2c. Unlike the thioredoxin-LANA fusion, the Protein A-LANA fusion is well resolved from the histone H3 band, allowing us (a) to verify that the Protein A-LANA fusion does bind to the nucleosome core particle (data not shown), and (b) to determine that the Protein A-LANA elutes with nucleosome core particle released from immobilized RCC1 but does not bind to nucleosome core particles that remain bound to the immobilized RCC1 (Fig. 2c, lanes 2–11). The latter observation is somewhat unexpected because we anticipate two RCC1 binding sites per nucleosome core particle (one binding site for each histone dimer). If we assume that the immobilized RCC1 protein binds the nucleosome core particle through one of the two binding sites, the LANA peptide might be expected to bind the other site. A possible explanation for our finding that Protein A-LANA does not bind to nucleosome core particles bound by GST-RCC1 itself immobilized on GSH-Sepharose is that the second nucleosome binding site presented in this manner is not accessible to the Protein A-LANA fusion protein due to steric effects.
RCC1/NCP binding model 1: binding via a β-propeller face
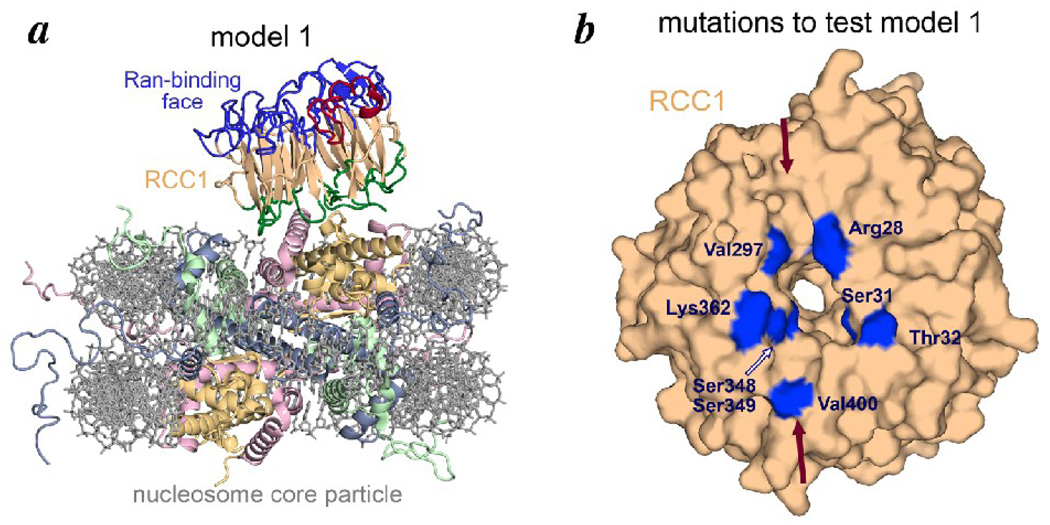
The β-propeller structure of RCC1 provides multiple loops which connect the seven β-sheets arranged like spokes in a wheel. Several studies have shown that the loops between β-strands that form the two opposite faces of β-propeller domains often mediate protein-protein interactions. RCC1 uses one of these faces to bind to its guanine exchange factor partner, Ran, (Fig. 1a) and the other face is expected to bind the nucleosome 19; 28. We observed that the non-Ran binding face of RCC1 contains a groove (highlighted by solid arrows in Fig. 3b) that could accommodate the H2B C-terminal helix exposed somewhat prominently on the nucleosome surface (Fig. 1b). These considerations led us to a model for how RCC1 might use its β-propeller face to interact with the nucleosome (Fig. 3a). The main feature of this model is that multiple loops of RCC1 interact with the exposed H2B C-terminal helix and neighboring H2A residues on the nucleosome surface.
Figure 3.
Model 1 for how RCC1 might bind the nucleosome. (a) Model 1 for RCC1-nucleosome complex in ribbon and stick representation. The RCC1 crystal structure (from PDB id 1I2M) was manually positioned on the nucleosome core particle structure (PDB id 1KX5) based on considerations described in the text. In this model, the loops on the non-Ran binding face of the RCC1 β-propeller interact with histone H2A/H2B surface residues. (b) Residues on the RCC1 non-Ran binding face mutated to test model 1 are highlighted in blue on a surface representation of the RCC1 molecule. The RCC1 groove conjectured to bind to the H2B C-terminal helix is indicated by the red arrows.
We tested this binding model by mutating RCC1 residues along the putative nucleosome binding surface, and assaying these point mutants by a nucleosome pulldown assay. In particular, recombinant nucleosome core particles hexahistidine tagged at the H2B N-termini were immobilized on Talon metal affinity resin and allowed to interact with target RCC1 molecules (the same β-propeller domain used in the previous LANA competition pulldown experiment). The RCC1 residues analyzed are shown in Fig. 3b as well as in Table 1. We find that none of the seven mutants we created changing a total of 8 residues affected binding of RCC1 to nucleosome core particles. We therefore conclude that RCC1 does not require these residues to bind to the nucleosome core particle, and that RCC1 does not appear to use its non-Ran binding face to interact with the nucleosome.
Table 1.
Summary of hRCC1Δ1 mutations and effect on nucleosome binding. + indicates the hRCC1Δ1 mutant bound to nucleosomes. − indicates that the hRCC1Δ1 mutant did not bind to nucleosomes.
| hRCC1Δ1 mutations | location of mutation | nucleosome pulldown results |
|---|---|---|
| R28A | non-Ran binding face | + |
| S31A,T32A | non-Ran binding face | + |
| V297Y | non-Ran binding face | + |
| S348A,S349A | non-Ran binding face | + |
| V400W | non-Ran binding face | + |
| T32Y | non-Ran binding face | + |
| K362E | non-Ran binding face | + |
| E209A,L210A,F211A | switchback loop | + |
| R217A,Q218A | switchback loop | − |
| R217E,Q218A | switchback loop | − |
| T277A,E278A | adjacent to switchback loop | + |
| R217A | switchback loop | − |
| Q218A | switchback loop | + |
| R214A | switchback loop | − |
RCC1/NCP binding model 2: binding via a β-propeller edge
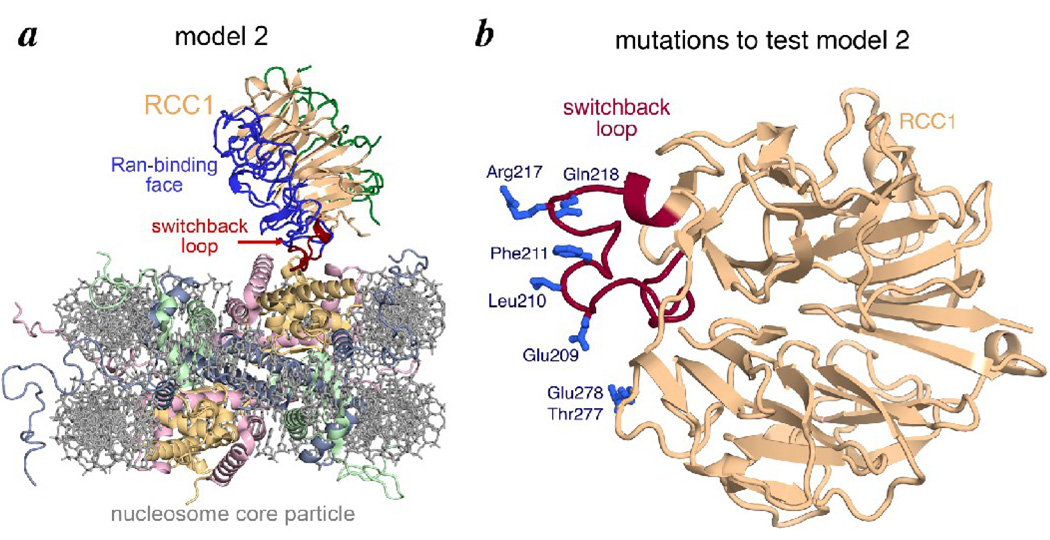
In our search for other ways by which RCC1 could bind to the nucleosome, we considered a particular RCC1 loop region found to be conformationally diverse in previous structural determinations (see below). This loop region, which joins RCC1 strands 4C and 4D and which juts out from the β-propeller domain, reverses direction several times like a switchback hiking trail. We therefore termed this region the switchback loop (Fig. 4b). The RCC1 switchback loop (residues 198–223) presents an arginine residue (Arg217) into solution which reminded us of a critical LANA peptide arginine residue which inserts itself into the histone H2A/H2B acidic patch 27 (this arginine residue 9 is the “R” in the LRS to AAA triple mutation defective in nucleosome binding and is depicted in Fig. 1b). We used model building to manually position the RCC1 molecule via its switchback loop and Arg217 on the nucleosome structure to mimic how the LANA peptide and its arginine residue bind the nucleosome. As one might expect, there are multiple ways to position RCC1 on the nucleosome with so few structural constraints. Nevertheless, our modeling exercise led us to binding model 2 where RCC1 binds to the nucleosome via its β-propeller edge (Fig 4a), in contrast to the face-binding model 1.
Figure 4.
Model 2 for how RCC1 might bind to the nucleosome. (a) Model 2 for RCC1-nucleosome complex in ribbon and stick representation. Arg217 within the RCC1 switchback loop was manually positioned on the nucleosome core particle structure to mimic LANA Arg9’s interaction with the nucleosome. (b) Residues on and around the RCC1 switchback loop (red) mutated to test model 2 are show in blue.
We used the same immobilized nucleosome pulldown assay and site-directed RCC1 mutants to test this edge-binding model (Fig. 5a). Mutations in Glu209, Leu210 and Phe211 within the switchback loop did not affect RCC1 binding the nucleosome, but both the R217A,Q218A and the R217E,Q218A double mutants did not bind nucleosomes (Fig. 5b, Table 1 and data not shown). We examined the roles of each of these residues by creating the individual R217A and Q218A RCC1 mutants. Analysis of these individual mutants indicate that Arg217 and not Gln218 is required for RCC1 to bind to nucleosomes since the Q218A but not the R217A mutant binds nucleosomes (Fig. 5b). The inability of the R217A mutant to bind to nucleosome is not due to aggregation because this mutant protein shares similar monodisperse characteristics to the wild type protein as determined by dynamic light scattering (polydispersity of 13.6% for wild type protein, 11.4% for R217A mutant protein). We also analyzed the Thr277 and Glu278 residues in an adjacent loop and found that mutations of both of these residues to alanine did not affect RCC1’s ability to bind nucleosomes (Table 1 and data not shown). These results identify the switchback loop and Arg217 in particular as critical for RCC1’s β-propeller domain to interact with nucleosomes.
Figure 5.
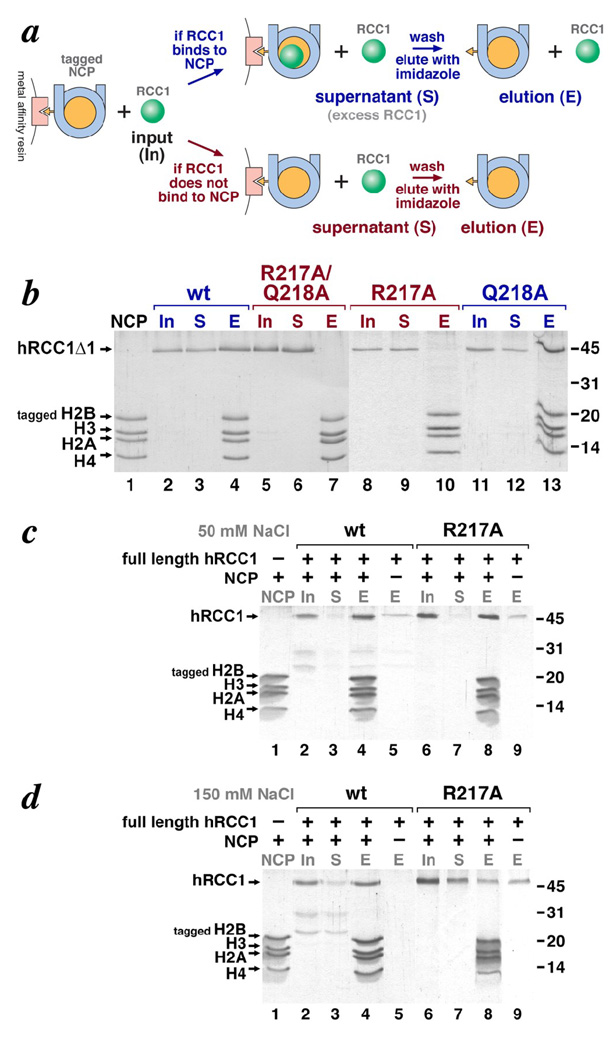
Immobilized nucleosome pulldown assays show that Arg217 is required for RCC1 β-propeller domain to bind to nucleosomes, and further that the RCC1 N-terminal tail is additionally involved in nucleosome binding. (a) Experimental scheme for the immobilized nucleosome pulldown assay. (b) Coomassie-stained SDS-PAGE gel for immobilized nucleosome pulldown assay using the RCC1 β-propeller domain . Input nucleosome core particles (tagged on histone H2B) are shown in lane 1. The input hRCC1Δ1 = hRCC1(21–421) proteins are shown in lanes 2, 5, 8 and 11. Proteins that did not bind to the immobilized nucleosomes and remained in the supernatant (S) are shown in lanes 3, 6, 9 and 12, while proteins that were eluted (E) from the beads are shown in lanes 4, 7, 10 and 13. The positions of molecular weight markers are shown on the right. The results for experiments using wild type human RCC1Δ1, the R217A/Q218A double mutant, the R217A mutant and the Q218A mutant are shown in lanes 2–4, 5–7, 8–10 and 11–13 respectively. (c) Coomassie-stained SDS-PAGE gel for immobilized nucleosome pulldown assay using full length RCC1 in 50 mM NaCl. Similar comments as for panel (b). Proteins that bound to the metal affinity resin in the absence of nucleosomes are shown in lanes 5 and 9 for wild type and R217A mutant respectively. (d) Coomassie-stained SDS-PAGE gel for immobilized nucleosome assay using full length RCC1 in 150 mM NaCl. Same comments are for panel (c).
Role of RCC1’s N-terminal tail in nucleosome binding
Since RCC1 contains an N-terminal tail implicated in DNA binding, we also investigated the binding of recombinant, full length human RCC1 (residues 2–421) to nucleosomes using the immobilized nucleosome pulldown assay. As expected, we find that full length RCC1 binds nucleosomes under the same 50 mM NaCl conditions used for our studies with the RCC1 β-propeller domain (residues 21–421, lacking the N-terminal tail) (Fig. 5c, lanes 2–5). However, we find that the R217A mutation that eliminates binding of RCC1(21–421) to nucleosomes does not affect binding of full length RCC1 to the same nucleosomes (Fig. 5c, lanes 6–9). Given the proposed role of RCC1’s N-terminal tail in DNA-binding and both the previous and our current findings that the RCC1 β-propeller domain binds to histone proteins, it seems likely that full length RCC1 associates with nucleosomes via both RCC1-DNA and RCC1-histone contacts. Thus mutations in only one interface, such as the R217A mutation in the RCC1-histone interface, might be masked by the additional interactions provided by a second interface.
To test this hypothesis, we repeated the pulldown binding assay under increasing NaCl concentrations to weaken charged interactions such as those expected to mediate protein-DNA interactions. Similar results are obtained using 100 mM as for 50 mM NaCl (data not shown), but we find that while full length RCC1 binds to nucleosomes at 150 mM NaCl (Fig. 5d, lanes 2–5), the same protein containing the R217A mutation shows significantly reduced nucleosome binding activity (Fig. 5d, lanes 6–9). Although some RCC1(R217A) protein remains bound to the immobilized nucleosome core particles after incubating and washing with 150 mM NaCl, a similar amount of protein binds the beads in the absence of nucleosomes presumably due to non-specific binding or aggregation on the beads (Fig. 5d, lanes 8 and 9). Such non-specific binding was reproducibly detected for full length RCC1(R217A) at 150 mM NaCl, but was either not detected or was present at much lower levels for the wild type, full length RCC1 at 50 or 150 mM NaCl (Fig. 5c, lanes 8 and 9, Fig. 5d, lanes 8 and 9) or for either wild type or mutated RCC1 β-propeller domain (data not shown). These results suggest that RCC1 uses both its N-terminal tail and β-propeller domain to bind to the nucleosome, and that a mutation critical for the β-propeller domain to interact with nucleosomes also affects binding of the full length protein when assayed under higher salt conditions that might weaken the tail-nucleosome interactions. Consistent with this interpretation, we find the wild type RCC1 β-propeller domain is unable to bind to nucleosomes at 200 mM NaCl presumably due to loss of the RCC1 N-terminal tail-DNA interactions (data not shown).
The RCC1 switchback loop is not sufficient for binding to nucleosomes
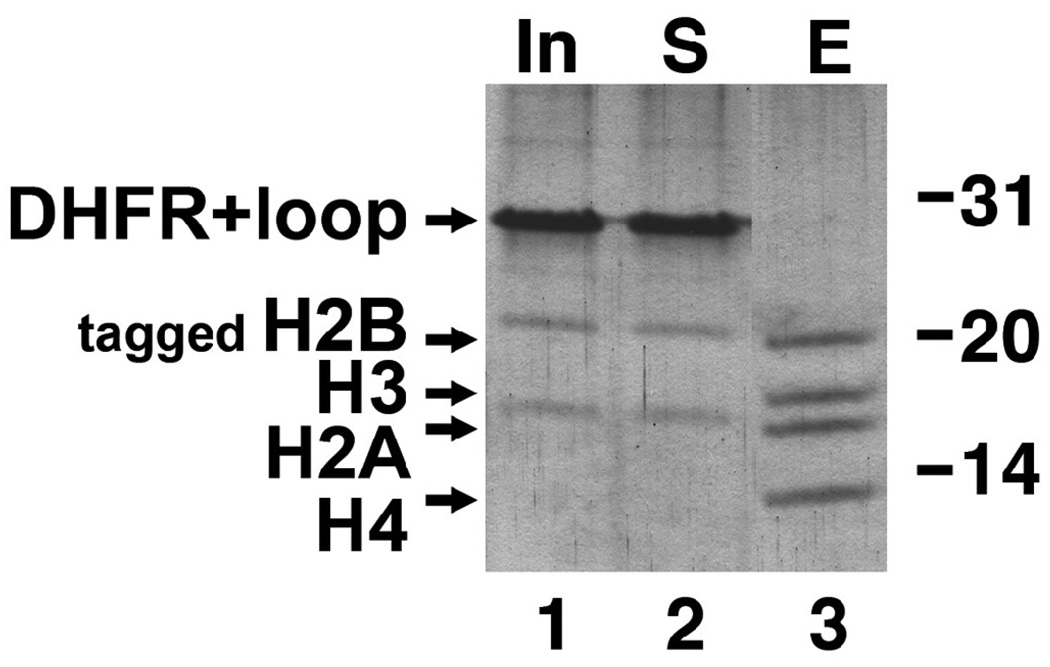
The RCC1 point mutations we studied suggest that the RCC1 switchback loop may be required for binding to the nucleosome. We were also interested to determine if the RCC1 switchback loop might be sufficient to bind to the nucleosome. Since it was not clear that the switchback loop would adopt its native conformation as an isolated peptide, we decided to graft the switchback loop (human RCC1 residues 197–223) onto a loop region of a different protein. We selected DHFR (dihydrofolate reductase) as this carrier protein because it is a well behaved, well studied monomeric protein and because we have utilized it in other unrelated protein expression studies 29. Since the ends of the RCC1 switchback loop (residues 197 and 223) are located close to each other (5.7 Å) in the human RCC1 crystal structure, we inserted these residues at the end of a β-hairpin away from the enzyme active site, i.e. between residues 145 and 146. This hybrid DHFR-switchback loop protein expressed in E. coli was found to be mostly monodisperse by dynamic light scattering (polydisperity of 31.8%) and eluted from a gel filtration column consistent with its expected molecular weight, suggesting that the hybrid protein was not misfolded (data not shown). We then employed the DHFR-switchback loop hybrid protein in our immobilized nucleosome core particle pulldown assay. Fig. 6 shows that unlike the wildtype RCC1 protein which bound to the immobilized nucleosome core particle, the DHFR-switchback loop hybrid protein remained in the unbound (supernatant, lane 2) fraction. Thus, the switchback loop does not appear to be sufficient to bind to the nucleosome when the loop is grafted onto an unrelated protein.
Figure 6.
The RCC1 switchback loop is not sufficient to confer nucleosome binding activity on DHFR. The same immobilized nucleosome pulldown assay in Fig. 5 was used to assay for nucleosome binding by a DHFR-RCC1 switchback loop hybrid protein. Lane 1 shows the input (In) DHFR-RCC1 switchback loop hybrid protein, lane 2 shows proteins that did not bind (supernatant S) to the immobilized nucleosome core particle and lane 3 shows proteins that remained bound (B) to the beads after washing.
Analysis of the RCC1 switchback loop
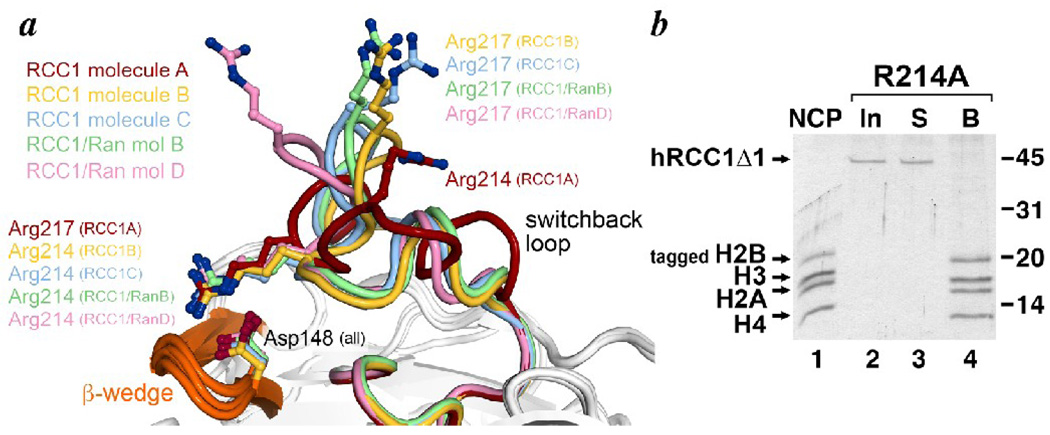
The RCC1 molecule has been crystallized on its own as well as in a complex with the Ran protein 7; 17. The three molecules of RCC1 in the asymmetric unit of the RCC1 only crystal and the two complexes of RCC1/Ran in the RCC1/Ran crystal afford five separate views of RCC1. For the most part, the structures of these five RCC1 molecules are very similar with an average rms deviation of backbone atoms of 0.71 Å (0.54 Å if the switchback loop region is not included). However, as mentioned earlier, the switchback loop is conformationally variable with one RCC1 molecule in the RCC1 only crystal adopting a significantly different backbone path than the others (RCC1 molecule A in Fig. 7a). Particularly noteworthy are the positions of Arg217, which we identified as critical for RCC1’s interaction with the nucleosome, and Arg214. In four of the five views of the RCC1 molecule, Arg217 is positioned pointing away from the β-propeller disk structure with highly similar orientations of Arg217 in three of those views (Fig. 7a). In the fifth view of RCC1 (molecule A of the RCC1 only structure), there is also an arginine residue positioned in the vicinity. However, that arginine residue is not Arg217, but is instead Arg214 (Fig. 7a). In the four RCC1 molecules where Arg217 points into solution away from the β-propeller, Arg214 interacts with the β-wedge residue Asp148 via charged hydrogen bonds. It is possible that this Arg214-Asp148 interaction stabilizes a switchback loop conformation that positions Arg217 for interactions with the nucleosome. This notion is bolstered by the observation that Arg217 of the fifth RCC1 molecule essentially switches positions with Arg214 in the other four RCC1 molecules to make the same charged hydrogen bonds that Arg214 makes with Asp148 (Fig. 7a). This remarkable conformational switch forces an alternate conformation of the switchback loop where Arg217’s interaction with Asp148 renders it unavailable for other interactions, and where Arg214 is moved significantly away from Arg217’s position in the other RCC1 structures (the Cα atom of Arg214 in the fifth RCC1 molecule is 6.9 Å from the Cα atom of Arg217 in the aligned RCC1 molecule closest to it).
Figure 7.
Conformational diversity of the RCC1 switchback loop and stabilization of a switchback loop conformation by the RCC1 β-wedge. (a) The diverse conformations of the switchback loop are illustrated in this alignment of the 5 views of the RCC1 molecule (3 molecules in the asymmetric unit of the RCC1 only crystal and 2 complexes in the asymmetric unit of the Ran-RCC1 crystal). The RCC1 A molecule in the RCC1 only crystal structure (PDB id 1A12) adopts a significantly different conformation from the other four RCC1 molecules. Whereas Arg214 in the other RCC1 molecules interacts with the β-wedge residue Asp148, Arg217 in the RCC1 A molecule makes the otherwise identical interaction with Asp148, forcing the RCC1 A molecule’s switchback loop and the Arg214 residue into a different conformation. (b) The RCC1 Arg214 residue is necessary for nucleosome binding. The immobilized nucleosome pulldown assay shows that human RCC1Δ1 containing the Arg214->Ala point mutation (input, lane 2) does not bind to nucleosomes (supernatant S, lane 3) and is therefore not found bound (B) to the immobilized nucleosomes. Input nucleosomes used in this assay are shown in lane 1, and the positions of molecular weight markers are provided on the right.
If the interactions between Arg214 and Asp148 do stabilize a conformation of the switchback loop productive for nucleosome binding, we would expect that disrupting this interaction by mutating Arg214 would affect RCC1 binding to nucleosomes. We tested this by preparing RCC1Δ1 with the R214A point mutation and examining the nucleosome binding activity of this mutant in the immobilized nucleosome pulldown assay. Our results show that RCC1Δ1 containing the R214A mutation is unable to interact with nucleosome (Fig. 7b). This finding establishes a critical role for Arg214 in RCC1/nucleosome binding although it remains to be seen if Arg214 is needed to stabilize the switchback loop for binding to nucleosomes or if Arg214 makes direct interactions with the nucleosome.
Discussion
We have analyzed how the RCC1 chromatin factor binds to nucleosomes. Our initial model for this interaction followed the conventional view that RCC1 would use opposite faces of its β-propeller domain to bind to Ran GTPase and to the nucleosome. However, mutagenesis of residues along the RCC1 face we originally thought would be responsible did not affect binding of RCC1 to the nucleosome. Instead, our second model for the RCC1/nucleosome complex and our subsequent biochemical studies have highlighted a previously unidentified region, the switchback loop, as being critical for the RCC1 β-propeller domain to bind to the nucleosome.
Our results also affirm a role for the RCC1 N-terminal tail to bind to nucleosomes. We confirm that the β-propeller domain of RCC1 is sufficient to interact with nucleosomes, but we also find that an RCC1 mutation which eliminates binding by the β-propeller domain can be compensated by including the N-terminal tail previously proposed to bind to DNA. This suppressive effect observed in the presence of the N-terminal tail can be reversed by increasing the ionic strength in the binding experiment, consistent with presumed ionic protein-DNA interactions by the tail. These results suggest that RCC1 may bind to nuclesomes via both RCC1-histone interactions using its β-propeller domain as well as RCC1-DNA interactions using its N-terminal tail. Since we employed recombinant RCC1 expressed in E. coli, the nucleosome interactions made by the N-terminal tail do not require post-translational modifications but post-translational modifications could certainly modulate such interactions in the cell.
We find that Arg217 and Arg214 in the RCC1 switchback loop are critical for the RCC1 β-propeller domain to interact with the nucleosome because removing the side chain of either by mutation to alanine renders RCC1Δ1 unable to bind nucleosomes. Based on our experiments, we are unable to determine if Arg217 and Arg214 act indirectly to stabilize an RCC1 conformation that binds to the nucleosomes or if these residues make direct contact with the nucleosome. Arg214 interacts with Asp148 from the β-wedge in four of the five views of RCC1 in the RCC1 only and RCC1-Ran complex crystal structures, and it is possible that this interaction stabilizes a switchback loop conformation appropriate for nucleosome interaction. However, while the Asp148 residue is widely conserved among RCC1 homologs, the Arg214 residue appears to be conserved only among vertebrates and other deuterostome animals, suggesting such a stabilization mechanism would not be conserved across all species. We suspect, though, that the other arginine residue, Arg217, does make direct contact with the nucleosome because this residue is nearly perfectly conserved among all RCC1-containing species and because its side chain appears to be available for intermolecular interactions in at least three of the five crystal structure views of RCC1. It is worth noting that the five views of the RCC1 switchback loop result from five different crystal packing arrangements, suggesting that the common conformation of the loop in at least three views reflects more than fortuitous crystal packing interactions and may reveal a propensity for that conformation. It is also tempting to speculate that different conformational states of the switchback loop may be used to regulate the binding of RCC1 to the nucleosome.
While our results suggest that the switchback loop may be necessary for nucleosome interaction by the RCC1 β-propeller domain, our experiment grafting this loop onto DHFR indicates that the switchback loop is not sufficient to confer nucleosome binding activity. There are at least two possible nonexclusive interpretations of this result. Firstly, the switchback loop might be insufficient because other regions of the RCC1 β-propeller domain are additionally required to directly contact the nucleosome. Alternatively, the switchback loop does interact with the nucleosome but requires stabilization by other regions of the RCC1 β-propeller domain. The possible stabilization of Arg214 in the switchback loop by Asp148 in the adjacent β-wedge might already provide an example of such a mechanism. Further examination of the switchback loop shows hydrogen bonds with other regions of the RCC1 β-propeller domain at multiple positions along the switchback loop, consistent with this hypothesis. In retrospect, it is perhaps not surprising that the switchback loop alone did not confer upon DHFR the ability to bind to nucleosomes.
One of the benefits of our crude models for how RCC1 binds to the nucleosome is that they allow us to evaluate the implications of these models for Ran recruitment. Besides the lack of experimental data to corroborate our binding model 1 (binding via RCC1 β-propeller face), one particularly discouraging aspect of that model was that it did not shed any insight into how binding of the nucleosome could increase RCC1’s guanine exchange factor (GEF) activity on Ran as observed previously. In our binding model 1, the nucleosome and Ran bind on opposite faces of the RCC1 β-propeller and it is not obvious how the information that the nucleosome has bound RCC1 would be transmitted to the nucleotide binding site in Ran. There does not appear to be precedence for other examples of conformational changes transmitted through the bodies of other β-propeller structures. The relative uniformity of the body of the RCC1 β-propeller in the five crystal structure views also argues against such a conformational change.
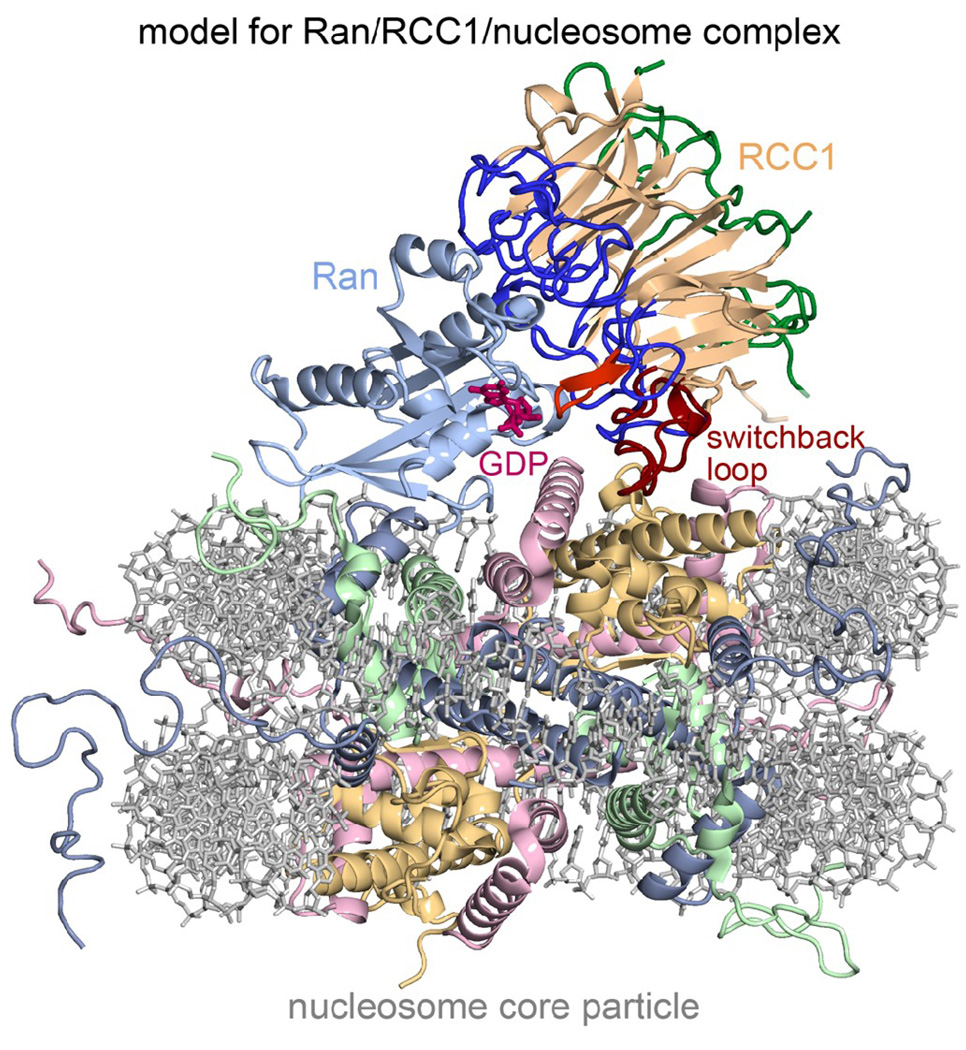
In contrast, our binding model 2 (binding via switchback loop along RCC1 β-propeller edge) suggests a model for the Ran/RCC1/nucleosome ternary complex which can explain existing results. Superposition of the Ran/RCC1 binary complex crystal structure with our RCC1/nucleosome binding model 2 via the common RCC1 subunit shows that Ran would then be positioned for direct interactions with the nucleosome (Fig. 8). This would resolve the previous dilemma of two apparently different modes by which Ran was recruited to nucleosome, one RCC1-dependent and the other RCC1-independent. This question was raised when it was found that Ran could unexpectedly bind to nucleosomes on its own, albeit with a lower binding affinity than in the presence of RCC1 30. Ran was also observed to bind to chromatin in Xenopus egg extracts without RCC1 present 31. We would like to suggest that the RCC1-dependent and RCC1-independent binding of Ran to the nucleosome reflect two different facets of the same Ran/RCC1/nucleosome complex as opposed to two different modes of binding.
Figure 8.
A model for the Ran/RCC1/nucleosome complex. This theoretical model was generated by superimposing the Ran/RCC1 crystal structure on our RCC1/nucleosome model 2 shown in Fig. 4a. The position of the GDP nucleotide was modeled by superimposing the Ran/NTF2/GDP crystal structure (PDB id 1A2K) on the Ran component. The model shows how Ran could interact directly with both RCC1 and the nucleosome in the complex, and also suggests a mechanistic explanation for how the nucleosome could further enhance Ran’s guanine exchange activity via modest conformational changes in RCC1 and Ran loop regions in the vicinity of the nucleotide binding site.
Our binding model 2 can also explain how RCC1 binding to the nucleosome enhances its ability to promote Ran’s guanine nucleotide exchange activity. In this model, the guanine nucleotide binding site on Ran is located in the middle of a triangle created by Ran, RCC1 and the nucleosome. It may be significant that the same RCC1 β-wedge that interacts with Ran and this nucleotide binding site is adjacent to the RCC1 switchback loop that makes critical interactions with the nucleosome. In particular, the RCC1 β-wedge makes direct interaction with the Ran P-loop, switch II and helix alpha3 that contribute to nucleotide binding. We suggest that binding of the nucleosome to RCC1 might induce a modest conformation change in the switchback loop, and that this conformational change is transmitted through the RCC1 β-wedge to the Ran nucleotide binding site.
In summary, our mutagenesis studies have led us to a novel model for how RCC1 binds to the nucleosome. This model is useful because it offers potential insight into how not only RCC1 interacts with the nucleosome but also how RCC1 recruits the Ran GTPase to the nucleosome to set up the RanGTP gradient around the chromosomes. We recognize, however, the limitations of our model building, and we also recognize that other binding models consistent with the available binding data are possible. Resolving this issue may require structural methods to determine precisely how RCC1 interacts with the nucleosome.
Materials and Methods
Proteins and Nucleosomes
The human RCC1 gene was cloned from HeLa cDNA by PCR amplification, and the β-propeller domain (hRCC1Δ1 corresponding to residues 21–421) expressed in E. coli as either a GST-hexahistidine tagged or a nonfusion protein using the pST50Tr T7-promoter based expression plasmid 32. The GST-hexahistidine tagged hRCC1Δ1 protein was expressed in BL21(DE3)pLysS E. coli cells at 37°C using 0.2 mM IPTG to induce expression, and purified by Talon metal affinity chromatography and SourceQ anion exchange HPLC. The nonfusion hRCC1Δ1 protein was also expressed in BL21(DE3)pLysS cells at 37°C using 0.2 mM IPTG, and purified by SP-Sepharose cation-exchange chromatography. Point mutations were introduced in hRCC1Δ1 using QuikChange mutagenesis, and the resulting mutant hRCC1Δ1 proteins expressed and purified as for the wild-type proteins. The coding regions for all expression constructs were verified by DNA sequencing. The aggregation state of selected hRCC1Δ1 proteins were analyzed using a Protein Solutions DynaPro-MS800 dynamic light scattering device.
Nucleosome core particles were prepared essentially as described previously 33. Briefly, core Xenopus histones H2A, H2B, H3 and H4 were expressed individually in E. coli and purified from inclusion bodies under denaturing conditions separately. The histone octamer was refolded from the four core histones by dialyzing equimolar mixtures of the histones in guanidine hydrochloride into a solution containing 2 M NaCl, and then purified by size exclusion chromatography over a Superdex 200 16/60 column. The purified histone octamer was then combined with recombinant Widom 601 DNA positioning sequence 34; 35 in a solution containing 2 M KCl, and the nucleosome core particle reconstituted by gradient dialysis into a solution containing 250 mM KCl. The reconstituted core particle was then purified by anion-exchange SourceQ HPLC.
Oligonucleotides containing the coding sequence of Kaposi’s sarcoma herpesvirus LANA peptide residues 1–23 were subcloned into a T7 promoter based pST50Tr E. coli expression vector to express the peptide and the mutant variant LRS->AAA as a C-terminal fusion to the hexahistidine-thioredoxin-TEV protease site (HisTrxN) 32; 36 combination tag. The wild type peptide was also expressed as a C-terminal fusion to a combination tag containing hexahistidine and two Protein A IgG binding domains. The three fusion proteins were expressed in BL21(DE3)pLysS cells at 37°C using 0.2 mM IPTG and purified from the soluble extract by Talon metal affinity chromatography and Source Q anion exchange HPLC. TEV protease and subsequent Talon metal affinity purification was used to remove the hexahistidine tag from the Protein A-LANA fusion protein.
The DHFR-switchback loop hybrid protein was created by first introducing a BamHI restriction site between DHFR residues 145 and 146 by QuikChange mutagenesis and then subcloning human RCC1 residues 197–223 PCR amplified to include 5’ BamHI and 3’ BglII restriction sites. The sequenced verified expression vector was used to express the hybrid protein in BL21(DE3)pLysS cells at 37°C using 0.2 mM IPTG. The DHFR-switchback loop hybrid protein was partially purified from the soluble extract by successive anion-exchange Q-Sepharose and SourceQ chromatography and Superdex HR200 size-exclusion chromatography.
LANA competition pulldown assay
100 µl of 8 µM GST-hexahistidine tagged hRCC1Δ1 was incubated with 20 µl of GSH-Sepharose resin (GE Healthcare) in GST binding buffer (20 mM Tris-Cl pH 8.0, 100 mM NaCl, 10 mM DTT, 200 µg/ml BSA, 0.1% NP40). After washing the resin twice with 150 µl GST binding buffer and equilibration with T50Ac buffer (5 mM Tris-Cl pH 8.0, 50 mM NaAc, 10 mM DTT, 200 µg/ml BSA, 0.1% NP40), 35 µl of 6.5 µM recombinant nucleosome core particles were added and allowed to bind to the immobilized hRCC1Δ1 protein. Unbound nucleosome core particles were removed by washing with T50Ac buffer and the bound nucleosome core particles then challenged by addition of 6.5 or 13 µM HisTrxNLANA or HisTrxNLANA(LRS->AAA) proteins in a final volume of 35 µl in the same buffer. The immobilized hRCC1Δ1 protein and remaining nucleosome core particles were then unbound from the resin using 35 µl T50Ac buffer with an additional 100 mM glutathione. Samples were fractionated on an 18% acrylamide SDS-PAGE gel and visualized by Coomassie Blue staining. Similar procedures were employed for experiments using the Protein A-LANA fusion protein.
Immobilized nucleosome pulldown assay
150 µl of 1 µM recombinant nucleosome core particles tagged with the N-terminal Strept II-hexahistidine-TEV protease site (STRHISN) combination tag 29; 32 on histone H2B were immobilized on 18 µl of Talon metal affinity resin equilibrated in H50 buffer (20 mM HEPES pH 7.5, 50 mM NaCl, 10 mM 2-mercaptoethanol, 100 µg/ml BSA and 12 mM imidazole). After washing the resin twice with 150 µl of H50 buffer, 150 µl of 1 µM hRCC1 or DHFR-switchback loop hybrid protein was incubated with the resin for 30 minutes at room temperature. Unbound protein was removed using two successive washes of 150 µl H50 buffer, and the nucleosome core particles and bound hRCC1Δ1 eluted using 30 µl of H50 buffer containing 200 mM imidazole. Samples were analyzed as described for the LANA competition pulldown assay.
Molecular graphics
The potential interactions between RCC1 and the nucleosome core particle were examined using MidasPlus (UCSF) 37 and PyMol molecular graphics software 38 and manual modeling. All molecular graphics in this manuscript were prepared using PyMol.
Acknowledgements
We thank Todd Stukenberg for advice and encouragement at the initiation of this project, and Jonathan Widom for sending the 601 nucleosome DNA positioning sequence. We also thank Andrew Fleischman for technical assistance as well as helpful input during the development of binding model 2. We gratefully acknowledge the technical assistance of Kristen Wiley, Dylan Schlaich, Michael Porzio, Allan Minns and Bryan Thurston. We also thank the Tan laboratory and the Penn State Center for Eukaryotic Gene Regulation for stimulating discussion throughout this project. This work is supported by NIH grant GM088236 to S.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Joseph J. Ran at a glance. J Cell Sci. 2006;119:3481–3484. doi: 10.1242/jcs.03071. [DOI] [PubMed] [Google Scholar]
- 2.Kuersten S, Ohno M, Mattaj IW. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 2001;11:497–503. doi: 10.1016/s0962-8924(01)02144-4. [DOI] [PubMed] [Google Scholar]
- 3.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 5.Sorokin AV, Kim ER, Ovchinnikov LP. Nucleocytoplasmic transport of proteins. Biochemistry (Mosc) 2007;72:1439–1457. doi: 10.1134/s0006297907130032. [DOI] [PubMed] [Google Scholar]
- 6.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 7.Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- 8.Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 9.Davis TL, Bonacci TM, Sprang SR, Smrcka AV. Structural and molecular characterization of a preferred protein interaction surface on G protein beta gamma subunits. Biochemistry. 2005;44:10593–10604. doi: 10.1021/bi050655i. [DOI] [PubMed] [Google Scholar]
- 10.Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gbetagamma. Science. 2003;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- 11.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 12.Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J Biol Chem. 2008;283:32158–32161. doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- 13.Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol. 2006;13:704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuetz A, Allali-Hassani A, Martin F, Loppnau P, Vedadi M, Bochkarev A, Plotnikov AN, Arrowsmith CH, Min J. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 2006;25:4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem. 2008;283:35258–35264. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJ. Snapshot of activated G proteins at the membrane: the Galphaq-GRK2-Gbetagamma complex. Science. 2005;310:1686–1690. doi: 10.1126/science.1118890. [DOI] [PubMed] [Google Scholar]
- 17.Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1) Cell. 2001;105:245–255. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- 18.Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF, Macara IG. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat Cell Biol. 2007;9:596–603. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seino H, Hisamoto N, Uzawa S, Sekiguchi T, Nishimoto T. DNA-binding domain of RCC1 protein is not essential for coupling mitosis with DNA replication. J Cell Sci. 1992;102(Pt 3):393–400. doi: 10.1242/jcs.102.3.393. [DOI] [PubMed] [Google Scholar]
- 21.Nemergut ME, Mizzen CA, Stukenberg T, Allis CD, Macara IG. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 2001;292:1540–1543. doi: 10.1126/science.292.5521.1540. [DOI] [PubMed] [Google Scholar]
- 22.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–272. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 23.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 25.Keppler BR, Archer TK. Chromatin-modifying enzymes as therapeutic targets--Part 1. Expert Opin Ther Targets. 2008;12:1301–1312. doi: 10.1517/14728222.12.10.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lall S. Primers on chromatin. Nat Struct Mol Biol. 2007;14:1110–1115. doi: 10.1038/nsmb1107-1110. [DOI] [PubMed] [Google Scholar]
- 27.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science. 2006;311:856–861. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 28.Hao Y, Macara IG. Regulation of chromatin binding by a conformational switch in the tail of the Ran exchange factor RCC1. J Cell Biol. 2008;182:827–836. doi: 10.1083/jcb.200803110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lichty JJ, Malecki JL, Agnew HD, Michelson-Horowitz DJ, Tan S. Comparison of affinity tags for protein purification. Protein Expr Purif. 2005;41:98–105. doi: 10.1016/j.pep.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 30.Bilbao-Cortes D, Hetzer M, Langst G, Becker PB, Mattaj IW. Ran binds to chromatin by two distinct mechanisms. Curr Biol. 2002;12:1151–1156. doi: 10.1016/s0960-9822(02)00927-2. [DOI] [PubMed] [Google Scholar]
- 31.Zhang C, Goldberg MW, Moore WJ, Allen TD, Clarke PR. Concentration of Ran on chromatin induces decondensation, nuclear envelope formation and nuclear pore complex assembly. Eur J Cell Biol. 2002;81:623–633. doi: 10.1078/0171-9335-00288. [DOI] [PubMed] [Google Scholar]
- 32.Tan S, Kern RC, Selleck W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr Purif. 2005;40:385–395. doi: 10.1016/j.pep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 34.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 35.Thastrom A, Lowary PT, Widlund HR, Cao H, Kubista M, Widom J. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 36.Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- 37.Ferrin TE, Huang CC, Jarvis LE, Langridge R. The MIDAS display system. Journal of Molecular Graphics. 1988;6:13–27. [Google Scholar]
- 38.Delano WL. The PyMOL molecular graphics system. Palo Alto, California, USA: DeLano Scientific LLC; http://www.pymol.org. [Google Scholar]