Abstract
Background & Aims:
Induction of immediate early transcription factors (ITF) represents the first transcriptional program controlling mitogen stimulated cell cycle progression in cancer. Here, we examined the transcriptional mechanisms regulating the ITF protein c-Myc and its role in pancreatic cancer growth in vitro and in vivo.
Methods:
Expression of ITF proteins were examined by RT-PCR and immunoblotting, and their implications in cell cycle progression and growth were determined by flow cytometry and [3H] thymidine incorporation. Intracellular Ca2+ concentrations, calcineurin activity and cellular NFAT distribution were analyzed. Transcription factor complex formations and promoter regulation were examined by immunoprecipitations, reporter gene assays and chromatin immunoprecipitation (ChIP). Using a combination of RNAi knockdown technology and xenograft models we analyzed the significance for pancreatic cancer tumor growth.
Results:
Serum promotes pancreatic cancer growth through induction of the proproliferative NFAT-c-Myc axis. Mechanistically, serum increases intracellular Ca2+ concentrations and activates the calcineurin/NFAT pathway to induce c-Myc transcription. NFAT binds to a serum responsive element within the proximal promoter, initiates p300-dependent histone acetylation and creates a local chromatin structure permissive for the inducible recruitment of ELK-1, a protein required for maximal activation of the c-Myc promoter. The functional significance of this novel pathway was emphasized by impaired c-Myc expression, G1-arrest and reduced tumor growth upon NFAT depletion in vitro and in vivo.
Conclusion:
Our study uncovers a novel mechanism regulating cell growth and identifies the NFAT-ELK complex as modulators of early stages of mitogen stimulated proliferation in pancreatic cancer cells.
Keywords: Pancreatic cancer, c-Myc, NFAT, transcription
Introduction
A hallmark in cell cycle re-entry and G1-progression is the rapid, sustained and protein-synthesis independent induction of immediate early transcription factors, ITF1,2. They function as mediators coupling external short-term signals to G1 transition through their ability to induce specific transcriptional programs resulting in up-regulation of D-type cyclins and their partnering kinases CDK4 and CDK63. Immediate inducible transcription factors themselves are tightly controlled by mitogenic and antiproliferative signaling pathways on the levels of promoter regulation and proteasomal degradation 4-6. Induction of ITF gene products is caused by direct promoter interaction, occurs within minutes upon stimulation and requires activation of latent signaling regulated transcription factors already present in the cell1,4,6. Both, the composition and activation level of these upstream transcriptional regulators (e.g. SRF and Smads) as well as the expression pattern of downstream ITF proteins are depending on the cell-type and the activation status of the cell at a given time 5,7-10. In cancer cells activation of oncogenic signaling and transcription pathways can override growth suppressor activities and stimulate cell cycle progression, either through rapid induction of ITF subsets or via stimulation of single master ITF proteins. Not surprisingly, induction and transcriptional activation of ITF factors has frequently been associated with fast growing malignancies 11-13. Therefore, understanding the signaling and transcriptional mechanisms underlying mitogen induced ITF expression and activation not only helps to better understand cancer growth promotion, but also can provide a platform for the development of novel therapeutic options in cancer therapy. This might be particularly true for pancreatic cancer, which is characterized by a rapid disease progression, an aggressively invasive tumor phenotype and a general resistance to conventional chemotherapy 14,15.
With the final goal to develop novel innovative therapeutic strategies in pancreatic cancer, we conducted the presented study and examined the expression, transcriptional regulation and function of the most prominent ITF members in pancreatic cancer. We identified the c-Myc proto-oncogene as a master ITF in G1-S cell cycle progression and growth of pancreatic cancer. Serum induced c-Myc expression and cancer growth is mediated by fast and sustained stimulation of the Ca2+ /calcineurin/NFAT signalling and transcription pathway. Mechanistically, serum rapidly enhances intracellular Ca2+ concentrations, leading to calcineurin activation and subsequent nuclear translocation and c-Myc promoter binding of NFAT factors. NFAT binding to the serum responsive −84/−63 promoter region enhances the net histone acetylase activity associated with the c-Myc promoter and subsequently allows recruitment of the co-factor ELK-1. Depletion of NFAT in pancreatic cancer cells prevents mitogen induced c-Myc expression and blocks tumor cell growth in vitro and in a xenograft mouse model, confirming a key role of the NFAT-Myc axis in pancreatic cancer growth. Together, these findings define a novel insight into the networks of event controlling early stages of cell cycle progression and provide a platform to develop novel therapeutics approaches for pancreatic cancer.
Methods
Cell lines, plasmids, RT-PCR analysis and siRNA oligonucleotides
Detailed descriptions on cell lines, plasmid generation, siRNA oligonucleotides and transfection protocols used in this study are available under Supplementary Information online.
Immunohistochemistry and fluorescence microscopy
8988t cells grown on chambered coverslips were left untreated or treated with 10% FCS for 60 min. Cells were washed, fixed, blocked and probed with anti-NFATc2 antibody (1:150; Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A) as previously described16. NFATc2 was detected with a fluorochrome-conjugated secondary antibody and nuclei were counterstained with DAPI. Coverslips were mounted on glass slides and cells were evaluated with a fluorescence microscope (Carl Zeiss, Inc., Oberkochen, Germany). Immunohistochemical analysis of tumors explanted from the nude-mice was performed as previously described16. Briefly, paraffin sections were stained with anti-NFATc1 (1:150; Abcam, Cambridge, UK) or anti c-Myc antibody (1:250; Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A). Antibody binding was visualized using a biotinylated secondary antibody, avidine-conjugated peroxidase (ABC method; Vector Laboratories, Burlingame, CA), and 3,3′diaminobenzidine tetrachloride (DAB) as a substrate, and hematoxylin as counter stain. The sections were counterstained with hematoxylin and slides were evaluated by standard light microscopy.
Subcellular fractionation, co-immunoprecipitation and immunoblotting
Subcellular fractionation, co-immunoprecipitation and western blotting were performed as described previously16,17. Detailed description of these technical procedures are available under Supplementary Information online. For immunoblotting, membranes were probed with antibodies against NFATc1 (Abcam), NFATc2, c-Myc, c-Jun, c-Fos, p300, and ELK-1 (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A). CdK4, CdK6, cyclin D1, cyclin D3, cyclin E, EGR-1, anti-HA, and lamin a/c were purchased from Cell Signaling (Danvers, MA), or ß-actin (Sigma-Aldrich, Saint Louis, MI).
Chromatin immunoprecipitation (ChIP) assays
The ChIP was done in PaTu8988t and Panc-1 cells treated with serum or untreated for indicated time periods. Cells were cross-linked with 1% formaldehyde for 10min at 37°C, harvested in SDS lysis buffer (Upstate Biotechnology), and DNA was shredded to fragments of 500bp by sonication. Antibodies against NFATc1, NFATc2, AcH3 (Milipore) or ELK-1 were added and pre-cleared chromatin was incubated over night. Protein G agarose beads were added and incubated for 1.5h at 4°C. After reversing the cross-links, DNA was isolated and used for PCR reactions. Specific primer pairs were designed with the PrimerExpresss 3.0 (Applied Biosystems) as followed: 5′ gagggatcgcgctgagtat 3′ and 5′ tctaactcgctgtagtaattccagc 3′ for qualitative or quantitative PCR amplifying the −84/−63 element of the c-Myc promoter.
[3H]-Thymidine assay and Flow cytometry
Cell growth was measured by [3H]-thymidine incorporation and flow cytometry as described previously16. A detailed protocol is available in the Supplementary Information section online.
Reporter gene assays
Luciferase reporter gene assays were performed as described previously18. Cells were seeded in 24-well tissue culture dishes and transfected with the indicated constructs before treatment. Luciferase activity was measured using the Lumat LB 9501 (Berthold Technologies, Mannheim, Germany) luminometer and the Dual-Luciferase®-Reporter Assay System (Promega, Madison, WS) according to the manufacturer's instructions. Firefly luciferase values were normalized to Renilla luciferase activity and were expressed as mean “fold induction”. Mean values are displayed ± standard deviations.
Generation of aequorin expressing cells and measurement of intracellular Ca2+ concentrations
For calcium measurements a GFP-aequorin (G5A) fusion protein was expressed using a lentiviral expression system19. Detailed descriptions are available in Supplementary Information. Ca2+ concentrations were measured as described previously20. In brief, PaTu8988t cells stably expressing the Ca2+ sensitive photoprotein (GFP-) aequorin were incubated for 30min in culture medium containing 5μM coelenterazine (Biaffin, Kassel, Germany), which constitutes the chromophore of apoaequorin. The cells were then washed twice with HBS buffer (140mM NaCl, 5mM KCl, 1mM MgSO4, 2mM CaCl2, 20mM KH2PO4, 20mM HEPES, 5.5mM glucose, pH 7.4). Afterwards, cells were stimulated as indicated and the luminescence signal was detected in a PolarSTAR plate reader from BMG (Offenburg, Germany).
Calcineurin activity assay
Cellular calcineurin phosphatase activity was measured with the Biomol GreenTM Cellular Calcineurin Assay Kit Plus according to the manufacturers instructions (Enzo Life Sciences GmbH, Lörrach, Germany). Cells were left untreated or treated with 10% FCS for indicated time periods before harvesting. Mean values are displayed ± standard deviations.
Tumor xenografts
Panc-1 cells were transfected with the pSilencer3.1-H1 Hygro vector (Ambion) bearing a hairpin-structured shRNA cassette for NFATc1 silencing (5′ ggactccaaggtcattttctt 3′) or control shRNA. Stable clones were selected in Hygromycin containing media, and the efficacy of NFATc1 silencing was validated by western blot. We injected 2 ×106 Panc-1 cells subcutaneously into the hind leg of a male athymic nude mice and measured the tumor weekly21. We estimated tumor volume (V) from the length (l) and width (w) of the tumor using the formula: V = (π/6) × [(l + w)/2]3. Animal experiments were carried out using protocols approved by the Institutional Animal Care and Use Committee at the University of Ulm.
Results
Time-dependent cell cycle propagation and growth in pancreatic cancer cells stimulated with serum
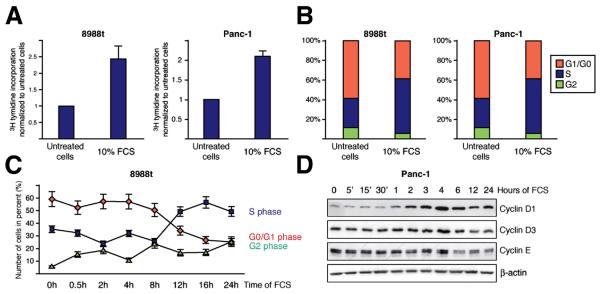
The effect of serum on pancreatic cancer cell growth was determined in PaTu8988t and Panc-1 cell lines by thymidine assays and flow cytometry analysis. Pancreatic cancer cells were arrested by serum deprivation and growth was stimulated through application of 10% FCS. Serum stimulation caused a significant increase in cell proliferation (Figure 1A) resulting from accelerated cell cycle propagation (Figure 1B). Flow cytometry analysis revealed a significant shift from G1 to S phase after 8 to 16h upon treatment and this effect was associated with induction of D-type cyclins (Figure 1C and 1D). Thus, serum induction of cell growth involves the expression of G1-promoting cyclins, cell cycle re-entry of arrested cells and subsequently G1-S phase propagation in pancreatic cancer cells.
Figure 1. Serum promotes pancreatic cancer growth through accelerated cell cycle transition.
(A) Cell proliferation was examined after treatment with 10% FCS for 24h. Thymidine incorporation was measured and calculated relative to untreated control, which was arbitrarily set to 1 for each experiment. Data are representative of triplicate experiments and displayed ± SD. (B) Cell cycle analysis of PaTu8988t (left) and Panc-1 (right) cells after treatment with 10% FCS or without FCS for 24h were performed by propidium iodide staining and following flow cytometry. Data are shown from one representative experiment (n=4). (C) Time course analyses of serum induced cell cycle progression in PaTu8988t cells. Cells were treated with 10% FCS for indicated time periods and analyzed by flow cytometry. Data are representative of triplicate experiments and displayed ± SD. (D) Western blot analysis to determine expression of cyclins following serum stimulation over time.
Expression and induction of immediate early transcription factors (ITF) in pancreatic cancer cells
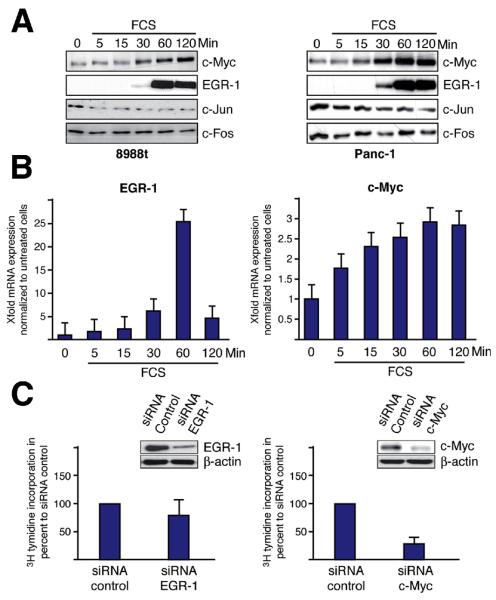
We next investigated the expression and serum-responsiveness of ITFs in pancreatic cancer cells. Serum starved pancreatic cancer cells were treated with 10% FCS over time and the expression was determined for the most prominent members of this group of transcription factors: EGR-1, c-Myc, c-Jun and c-Fos. Figure 2A displays moderate to high expression levels of c-Jun and c-Fos in serum starved pancreatic cancer cells and these levels remained unaffected by serum stimulation. In contrast, EGR-1 and c-Myc were modestly expressed in growth-arrested cells, but significantly induced upon mitogen stimulation (Figure 2A and B). To investigate the functional implication of c-Myc and EGR-1 in serum driven cell growth, we performed [3H]-thymidine assays following genetic silencing of either factor. Successful knockdown of EGR-1 by RNA interference caused only a moderate inhibition of cell proliferation, while depletion of c-Myc resulted in a strong reduction of pancreatic cancer growth (Figure 2C). Together, these experiments identify c-Myc as the early mediator of serum induced cell cycle progression in pancreatic cancer cells.
Figure 2. Serum induced cell proliferation depends on c-Myc induction.
(A) PaTu8988t (left) and Panc-1 (right) cells were treated with FCS as indicated and the expression levels of c-Jun, c-Fos, c-Myc and EGR-1 were examined by immunoblotting. (B) PaTu8988t cells were treated with FCS to determine EGR-1 (left) and c-Myc (right) mRNA expression by quantitative RT-PCR. (C) Specific siRNA oligonucleotides were transfected to either silence EGR-1 (left panel) or c-Myc (right panel) and [3H] thymidine incorporation was measured in Panc-1 cells. Successful gene silencing was confirmed by immunoblotting.
Serum stimulates c-Myc promoter activation through activation of the Ca2+/calcineurin/NFAT signaling and transcription pathway
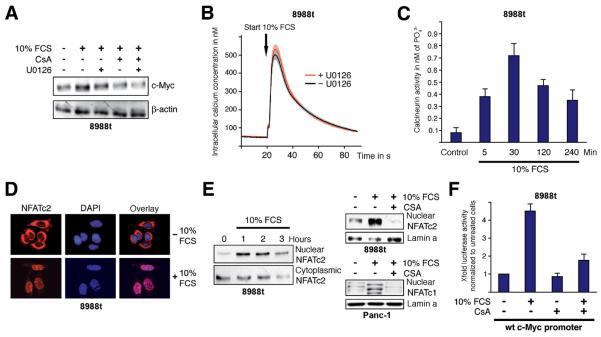
Depending on the cell-type, mitogen stimulation can utilize different intracellular signaling cascades to activate the ITF response. Here, we focused on two major mitogenic pathways, the MAP-kinase network and Ca2+ dependent signaling. Both pathways exert oncogenic potential with known implications in cell proliferation and progression of pancreatic cancer16,22. Serum starved cells were pretreated with either cyclosporine A (CsA) to block Ca2+/calcineurin signaling, the ERK-MAPK inhibitor U0126 or a combination of both agents. Cells were then stimulated and c-Myc expression was determined. Again, serum treatment enhanced the level of c-Myc expression in 8988t cells (Figure 3A). Surprisingly, pharmacological inactivation of ERK-MAPK only marginally interfered with serum inducibility on both mRNA (data not shown) and protein level (Figure 3A). In contrast, CsA prevented c-Myc up-regulation, indicating a role of the Ca2+/calcineurin cascade in c-Myc induction by serum. Consequently, we tested whether serum affects intracellular Ca2+ concentrations in cancer cells and generated 8988t cells stably expressing aequorin, a photoprotein from jellyfish that allows monitoring of intracellular Ca2+ fluctuations by luminescence technology. Serum treatment of growth arrested cancer cells caused a rapid increase in intracellular Ca2+ concentrations (Figure 3B), and this effect was independent from ERK-MAPK activation but resulted in a tremendous induction of the downstream phosphatase calcineurin (CaN) (Figure 3C). Transcription downstream of calcineurin is in large part funnelled through the transcription factor nuclear factor of activated T cells (NFAT), a heavily phosphorylated protein that is cytoplasmic in resting cells, but enters the nucleus when dephosphorylated by calcineurin. We have most recently demonstrated high expression levels of NFATc1 and NFATc2 in pancreatic cancer cells16. Interestingly, all tested cell lines displayed high expression of either NFATc1 or NFATc2 but none showed significant expression of both factors. In growth arrested 8988t cells, NFATc2 is primarily localized in the cytoplasm, suggesting inactivation of the protein (Figure 3D). Serum stimulation promoted rapid and sustained NFATc2 translocation into the nucleus for several hours, as revealed by immunofluorescence microscopy and subcellular fractionation assays (Figure 3D and 3E). Similar effects were observed in Panc-1 cells with accumulation of nuclear NFATc1 upon serum application (Figure 3E). Moreover, nuclear translocation of both factors was antagonized following CsA treatment, indicating that serum stimulates nuclear shuttling of NFAT through activation of the Ca2+/calcineurin cascade. We then analyzed whether calcineurin/NFAT signaling mediates serum induction on the level of promoter regulation and performed reporter gene assays using a 2.8 Kb spanning c-Myc promoter reporter construct. Serum treatment caused a 4-fold induction of the wild-type c-Myc promoter and consistent with our expression data this effect was diminished upon application of CsA (Figure 3F).
Figure 3. Serum utilizes the calcium/calcineurin/NFAT pathway to induce c-Myc.
(A) Western blot analysis to illustrate c-Myc expression in 8988t cells depending on treatment with either 1μM CsA, 10μM U0126 or a combination of both agents before application of 10% FCS. ß-actin was used for loading control. (B) 8988t cells stably expressing the calcium sensor protein aequorin were loaded with coelenterazine. Luminescence was continuously recorded following application of FCS +− UO126 that was given 30 min before stimulation (gray line). Data represent means of three independent experiments. The light and dark gray areas indicate S.E.M. (C) Calcineurin phosphatase activity was determined in 8988t cells by measurement of phosphate released from the RII phosphopeptide following treatment with FCS for the indicated time periods. Data are mean values ± SD. (D) Immunocytological detection of NFATc2 localization in 8988t cells. Cells were serum deprived and then treated with FCS for 60 min. Serum stimulation led to a rapid translocation of NFATc2 (red signals) from the cytosol to the nucleus. Nuclei were visualized by DAPI staining (blue signals). (E) 8988t and Panc-1 cells were either pretreated with CsA to block calcineurin or directly stimulated with 10% FCS as indicated. Following subcellular fractionation, nuclear and cytoplasmic fractions were analyzed separately for the presence of NFATc2/NFATc1 proteins by immunoblotting. (F) The luciferase activity of the human wild-type c-Myc promoter construct was measured upon treatment of 8988t cells with FCS for 24h in the presence or absence of 1μM CsA. Reporter gene activities are expressed as ‘fold activation’ relative to untreated controls.
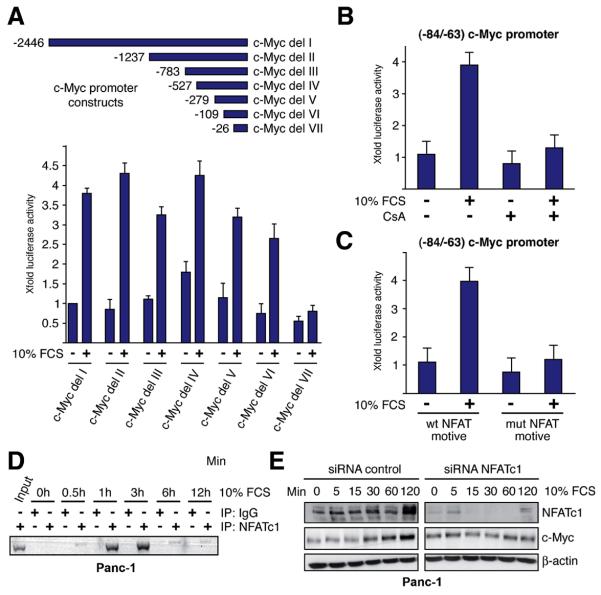
To identify the serum responsive element of the human c-Myc promoter, we performed reporter assays using sequential deletion fragments fused to luciferase. Analysis of the promoter fragments revealed that sequences up to −109bp did not significantly decrease serum inducibility, while further deletion up to position −26 (relative to the P2 transcription initiation site) rendered the promoter insensitive to serum (Figure 4A). The −109 to −26pb region of the human c-Myc promoter encompasses a short region that is known as a key convergence node for multiple signalling and transcription cascades (located between −84 and −63 bp) 23. Similar to the full-length c-Myc promoter, the −84/−63 promoter region was responsive to serum treatment and showed an approximate 4-fold induction (Figure 4B). Furthermore, inactivation of the calcineurin/NFAT pathway through application of CsA or mutational disruption of the NFAT binding sequence abolished serum responsiveness, indicating that NFAT binding is required for c-Myc promoter induction by serum (Figure 4B and 4C). Support for this assumption came from chromatin immunoprecipitation assays demonstrating inducible NFAT binding to the proximal c-Myc promoter upon serum stimulation (Figure 4D). Thus, NFAT binds to the serum responsive element to induce transcription from the c-Myc promoter. Consistently, depletion of NFAT impaired serum-induced expression of c-Myc in pancreatic cancer cells (Figure 4E). These data strongly indicated that serum utilizes the Ca2+/calcineurin/NFAT pathway to induce c-Myc expression in cancer cells.
Figure 4. Serum promotes NFAT promoter binding to induce c-Myc.
(A) PaTu8988t cells transfected with the full-length c-Myc promoter construct (c-Myc del-I) or various deletion constructs were treated with FCS and luciferase activities were assayed after 24h. Reporter gene activities are expressed as ‘fold activation’ relative to the activity of the untreated full-length promoter. Promoter activities are expressed as mean values ± SD. (B) The (−84/−63) c-Myc promoter fragment was transfected into 8988t cells and serum responsiveness was measured in the presence or absence of 1μM CsA. Reporter gene activities are expressed as ‘fold induction’ relative to the untreated control. (C) 8988t cells were transfected with the wild-type (−84/−63 wt) promoter or a mutant that lacks the NFAT-binding site before treatment with FCS. Firefly luciferase reporter activities were normalized to Renilla luciferase activity and expressed as fold induction relative to the activity of untreated −84/−63 wt c-Myc promoter. (D) Chromatin immunoprecipitations were performed with specific NFATc1 antibodies and in vivo binding to the c-Myc promoter was determined by PCR using primers specific for the c-myc promoter region harboring the −84/−63 region. ChIP assay demonstrates time-dependent binding of NFATc1 to the c-Myc −84/−63 element upon serum. (E) Cells were transfected with siRNA against NFATc1 or control siRNA and western blot analysis were performed to detect c-Myc expression after stimulation with 10% FCS over time.
NFAT recruits ELK-1 to induce maximal activation of the c-Myc promoter
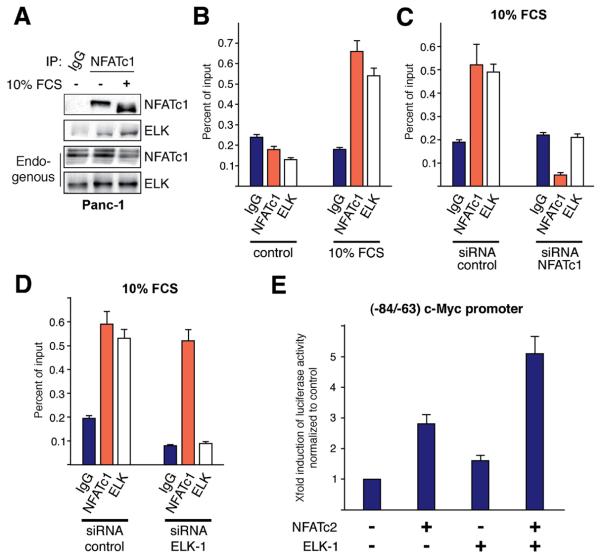
Sufficient NFAT promoter induction often requires recruitment of other DNA binding proteins24. The NFAT binding region (−75/−71) of the c-Myc promoter encompasses the core GGAA/T consensus sequence for ETS-domain transcription factors. We have tested whether NFAT and ELK-1 cooperate in c-Myc transcription and precipitated endogenous NFAT proteins to analyze inducible complex formation with ELK-1. Figure 5A illustrates our findings and shows the activation of NFATc1 by serum application (lane 2 and 3), as indicated by the shift of the protein from a slow to a fast migrating form, representing dephosphorylation and activation of the factor. NFAT and ELK modestly precipitated in unstimulated cells (lane 2) but this interaction was strongly enhanced by serum treatment (lane 3). Moreover, NFAT/ELK-1 complex formation was associated with increased binding of both transcription factors to the −84/−63 promoter region, as revealed by ChIP experiments (Figure 5B). Further analysis demonstrated that ELK-1 functions as a co-factor to ensure maximal transcription from the c-Myc promoter by NFAT. In fact, while Elk-1 alone barely induced c-Myc promoter activation, it strongly enhanced NFAT driven transcription. Moreover, recruitment of ELK-1 to the c-Myc promoter is strictly dependent on the presence and promoter binding of NFAT (Figure 5C and 5E), as evidenced by the loss of Elk-1 promoter occupancy following NFAT depletion. Together, these studies indicate that activated NFAT factors recruit ELK-1 to induce maximal activation of the c-Myc promoter in pancreatic cancer cells.
Figure 5. NFAT recruits ELK-1 for cooperative c-Myc promoter activation.
(A) Endogenous NFATc1/ELK-1 interaction was examined by coimmunoprecipitation in Panc-1 cells. Cells were treated with 10% FCS before NFATc1 was isolated using specific antibodies. NFATc1 bound ELK-1 was identified by the use of a specific anti-ELK-1 antibody. (B) Chromatin immunoprecipitations were performed with specific NFATc1 and ELK-1 antibodies to determine inducible promoter binding. PCR was done using primers specific for the c-myc promoter region harboring the serum responsive region. (C+D) ChIP analysis to demonstrate NFAT and ELK-1 binding to the −84/−63 c-Myc promoter region following depletion of either NFATc1 (C) or ELK-1 (D) in Panc-1 cells. (E) The (−84/−63) c-Myc promoter reporter was co-transfected with either NFATc2, ELK-1 or both factors into Panc-1 cells and luciferase activity was expressed as fold induction relative to basal (−84/−63) c-Myc promoter activity.
NFAT-ELK-1 transcriptional complex formation requires the histone acetyltransferase p300 to activate the c-Myc promoter
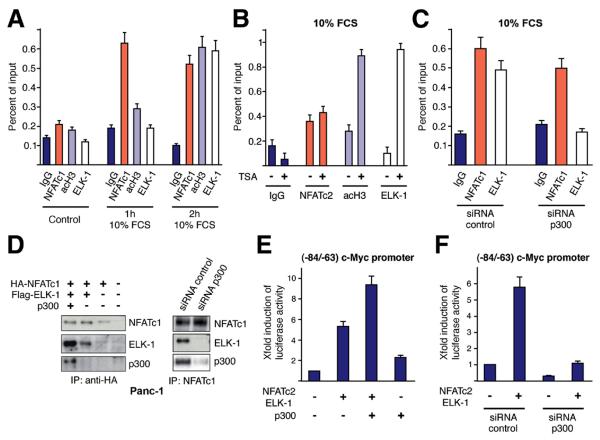
p300 is commonly recruited to transcriptional complexes to bridge DNA-binding partners and enhance gene transcription through creation of a hyperacetylated chromatin state25. ChIP assays indeed displayed a sequential occupancy of the proximal c-Myc promoter that is initiated by NFAT factors and followed by consecutive recruitment of acetylated histones and ELK-1 (Figure 6A). In detail, serum rapidly induced NFAT binding within 1 hour, followed by enhanced local promoter acetylation and ELK-1 binding. NFAT-promoter binding precedes histone acetylation and thus remains unaffected by increased global acetylation through TSA treatment (Figure 6B). In contrast, NFAT/ELK-1 complex formation and ELK-1's recruitment to the c-Myc promoter require the p300 histone acetyltransferase (Figure 6C and D). Hence, depletion of p300severely compromised both transcription factor complex formation and promoter binding, while introduction of p300 stimulates binding to NFAT and enhances its ability to activate the c-Myc promoter (Figure 6C and E). Together, NFAT binding to the serum responsive element is the initial event in c-Myc promoter activation that stimulates HAT-induced histone acetylation at the c-myc promoter, and consequently allows accessibility of ELK-1.
Figure 6. NFAT binding to the c-Myc promoter induces sequential histone acetylation and ELK-1 recruitment.
(A) Time dependent binding of NFATc1, ELK-1 and acetylated histone 3 (acH3) to the −84/−63 c-Myc promoter was tested by ChIP analysis. Quantitative PCR was performed using primers specific for the c-myc promoter region harboring the −84/−63 (B) ChIP analysis to test NFAT/ELK-1 and acH3 occupancy of the c-Myc promoter in response to TSA treatment. (C) ChIP analysis to study NFAT and ELK-1 binding to the −84/−63 c-Myc promoter region in Panc-1 cells following siRNA mediated depletion of the HAT p300. (D) p300 dependent interaction between NFATc1 and ELK-1 was examined by co-immunoprecipitation following overexpression (left panel) or genetic depletion (right panel) of p300. (E) The (−84/−63) c-Myc promoter was co-transfected with NFAT and ELK-1 alone or together with p300. Luciferase activity was expressed as fold induction relative to basal (−84/−63) c-Myc promoter activity. (F) Cells were transfected with the (−84/−63) c-Myc promoter construct along with NFATc2/ELK-1 and either siRNA against p300 or control siRNA. Luciferase activity was expressed as fold induction relative to basal (−84/−63) c-Myc promoter activity
NFAT mediates mitogen induced pancreatic cancer growth in vitro and in vivo
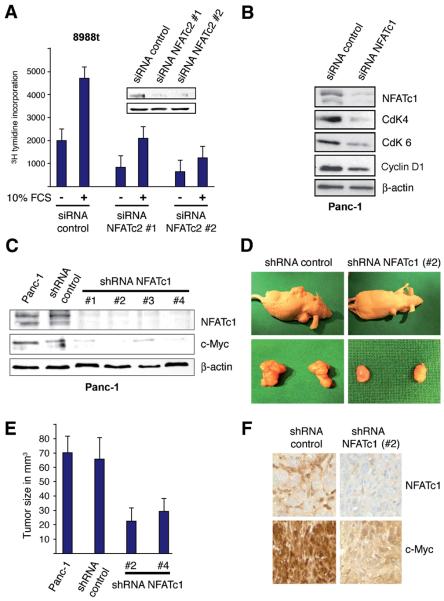
Finally, we analysed the relevance of the NFAT-c-Myc axis in serum induced pancreatic cancer growth. To this end, we arrested cells by serum deprivation and allowed cells to re-enter the cell cycle through application of serum. These experiments, performed in control and NFAT depleted Panc-1 and 8988t cells, revealed a critical function of NFAT proteins in serum driven pancreatic cancer growth. Consistent with the data shown in figure 1, serum significantly stimulates proliferation in cancer cells (Figure 7A). However, depletion of NFAT proteins rendered cells refractory to serum and prevented pancreatic cancer growth through induction of a G1 arrest, as evidenced by a significant down-regulation of D-type cyclins and their partner kinases (Figure 7B). The biological relevance of NFAT proteins in c-Myc expression and pancreatic cancer growth was examined in a xenograft mouse model using Panc-1 cells stably transfected with shRNA against NFATc1 or control shRNA. Similar to our results using siRNA against NFAT, stable shRNA mediated knockdown of NFATc1 was associated with reduced levels of c-Myc (Figure 7C). We then injected untransfected Panc-1 control cells, control shRNA Panc-1 cells, NFATc1 shRNA clone #2 and NFATc1 shRNA clone #4 cells in the flanks of athymic nude mice and determined the effect of NFAT silencing on tumor growth. After five weeks, the animals were sacrificed and the tumors extracted. The results, shown in Figure 7D-F, demonstrate that genetic silencing of NFATc1 caused a significant reduction in tumor mass and this effect was closely associated with reduced c-Myc expression (Figure 7E and F). A representative example of the tumor mass is shown in Figure 7D. Together, these experiments suggest an important role of the NFAT-c-Myc axis in mitogen driven cell cycle progression and tumor growth in the pancreas.
Figure 7. NFAT depletion reduces c-Myc expression and pancreatic cancer growth in vitro and in vivo.
(A) Proliferation of PaTu8988t cells after treatment with siRNA against NFATc2 or control siRNA and in the presence or absence of 10% FCS for 24h was analyzed by [3H] thymidine incorporation. Data are representative of triplicate experiments and displayed ± SD. Successful NFATc2 knockdown is demonstrated by western blot analysis. (B) Immunoblotting to determine the expression of cyclin D1 and its related kinases CdK4 and CdK6 upon depletion of NFATc1. (C) Western blot analyses to verify successful stable NFATc1 knockdown in Panc-1 cells. (D) A representative example of tumor mass is shown for the Panc-1 cell line transfected with either shRNA against NFATc1 or control shRNA. (E) Panc-1 cells were transfected with the indicated constructs and injected in the flank of male athymic nude mice (n = 6 for each experimental condition). Weekly measurements were taken from the tumors, and the mean tumor volume was determined after 5 weeks. (F) Expression of NFATc1 and c-Myc was determined by immunohistochemical analysis of Panc-1 tumors
Discussion
In this study we examined the expression, transcriptional regulation and function of the most prominent ITF members in pancreatic cancer and identified the c-Myc protooncogene as the predominant factor in serum stimulated G1 cell cycle transition and growth. While other ITF proteins have proven to be either resistant to mitogen stimulation (c-jun and c-fos) or dispensable (egr-1) for pancreatic cancer cell proliferation, only c-Myc fulfilled both criteria. c-Myc induction by serum, on the other hand, requires prior activation of the Ca2+/calcineurin/NFAT signaling and transcription cascade. In fact, serum rapidly enhances intracellular Ca2+ concentrations in pancreatic cancer cells, and as a consequence of this, causes activation of the calcium responsive phosphatase calcineurin. Calcineurin activation in turn promotes nuclear translocation of pre-existing cytoplasmic NFAT proteins and allows their subsequent transcriptional activation. The NFAT family of transcription factors comprises a group of five proteins related to the Rel family that play a critical role in the function of many vertebrate tissues 26. Besides their well-documented role in T-cell activation, NFAT proteins are now being increasingly recognized for their implications in tumorigenesis27-29. This is particularly true for NFATc1 and NFATc2, which are frequently deregulated in cancer. Depending on the cell type and organ NFAT proteins can have similar or opposing functions during tumor development and progression 16,29-30. In NIH-3T3 fibroblasts, for instance, NFATc1 acts as a positive regulator of cell proliferation, inhibits cell death and induces cell transformation, while NFATc2 blocks cell cycle progression and subverts oncogenic transformation 29. We could most recently confirm a pro-proliferative function of NFATc1 in pancreatic cancer. In fact, high levels of NFATc1 were found in pancreatic intraepithelial precursor lesions (PanIN lesions) and with a high frequency in advanced pancreatic cancers 16. We have also shown that NFATc1 is highly active in pancreatic cancer cells and induces anchorage-independent growth and cell proliferation through activation of the c-Myc oncogene. In this study, we extent our knowledge on the role of the calcineurin/NFATc1/c2-cMyc axis in the regulation of pancreatic carcinogenesis and identified this signaling-modulated transcriptional mechanism as a central module in the mitogen driven pancreatic cancer growth in vitro and in vivo. Accordingly, inactivation of this pathway, either through application of cyclosporine A or through RNAi mediated silencing of NFAT factors, prevented c-Myc up-regulation by serum and arrested pancreatic cancer cells at G1 cell cycle phase. The relevance of the NFAT-c-Myc axis in pancreatic cancer growth was also emphasized in xenograft mice models confirming loss of c-Myc expression and significantly reduced tumor growth of subcutaneously injected pancreatic cancer cells upon stable silencing of NFAT. Therefore, disruption of the NFAT-c-Myc interplay through specific inactivation of the calcineurin/NFAT pathway in tumor cells might be an attractive target for future approaches in pancreatic cancer therapy.
To better understand how NFAT confers serum induction of c-Myc, we performed extensive c-Myc promoter studies and identified a proximal region that is occupied by NFAT and responsible for serum responsiveness. Upon mitogenic activation of the Ca2+/calcineurin pathway NFAT factors rapidly bind to and induce transcription from a consensus promoter sequence localized within the serum responsive element.
Mechanistically, NFAT binding to this promoter region enhances the net histone acetylase activity associated with the c-Myc promoter and subsequently allows recruitment of ELK-1, an ETS-like factor required for maximal activation of the promoter. Detailed sequential chromatin immunoprecipitation analysis revealed that NFAT binding is the initial event in c-Myc promoter activation that stimulates the histone acetyltransferase (HAT) p300 to create a hyperacetylated chromatin state at the proximal promoter that allows accessibility of ELK-1. Consistently, depletion of the HAT p300 severely compromised local histone acetylation, prevented NFAT/ELK-1 complex formation and abolished inducible ELK-1 promoter occupancy, resulting in inappropriate c-Myc promoter activation. Together, these studies extent our understanding of how NFAT proteins regulate gene expression through interaction with partnering transcription factors. Based on their weak DNA-binding affinities, NFAT proteins frequently interact with site-specific transcriptional regulators for sufficient promoter transactivation. Here, we show for the first time that this protein-protein interaction (NFAT-ELK-1) involves local chromatin re-organization and requires p300 mediated histone acetylation. Future approaches will analyze whether this mechanism is c-Myc promoter specific or holds true for other NFAT regulated target promoters.
Taken together, this study uncovered a master role of the NFAT-p300-ELK axis in mitogen induced pancreatic cancer growth in vitro and in vivo . Based on our findings we propose a model in which NFAT binding to target promoters can trigger a HAT relay switch and thus allows recruitment of additional transcription factors through alteration of the local chromatin structure.
Supplementary Material
Acknowledgments
Grant support: V.E. is supported by the Deutsche Forschungsgemeinschaft (DFG, SFB-TR17) and the Max Eder program of the German Cancer Research Foundation (Deutsche Krebshilfe, 70-3022-El I). M.E.F.Z. is supported by the Miles and Shirley Fiterman Center for Digestive Diseases, Division of Gastroenterology and Hepatology, Division of Oncology Research, Mayo Clinic Cancer Center, Mayo Clinic Pancreatic SPORE P50 CA102701 and CA136526.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have nothing to disclose.
References
- 1.Almendral JM, Sommer D, Macdonald-Bravo H, et al. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol. 1988;8:2140–2148. doi: 10.1128/mcb.8.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bravo R. Growth factor-responsive genes in fibroblasts. Cell Growth Differ. 1990;1:305–309. [PubMed] [Google Scholar]
- 3.Lee MH, Yang HY. Regulators of G1 cyclin-dependent kinases and cancers. Cancer Metastasis Rev. 2003;22:435–49. doi: 10.1023/a:1023785332315. [DOI] [PubMed] [Google Scholar]
- 4.Murphy LO, MacKeigan JP, Blenis J. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol Cell Biol. 2004;24:144–153. doi: 10.1128/MCB.24.1.144-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CR, Kang Y, Siegel PM, et al. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 6.Sng JC, Taniura H, Yoneda Y. A tale of early response genes. Biol Pharm Bull. 2004;27:606–12. doi: 10.1248/bpb.27.606. [DOI] [PubMed] [Google Scholar]
- 7.Yordy JS, Muise-Helmericks RC. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]
- 8.Herold S, Herkert B, Eilers M. Facilitating replication under stress: an oncogenic function of MYC? Nat Rev Cancer. 2009;9:441–444. doi: 10.1038/nrc2640. [DOI] [PubMed] [Google Scholar]
- 9.Murphy LO, Blenis J. MAPK signal specificity: the right place at the right time. Trends Biochem Sci. 2006;31:268–275. doi: 10.1016/j.tibs.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Hsu T, Trojanowska M, Watson DK. Ets proteins in biological control and cancer. J Cell Biochem. 2004;91:896–903. doi: 10.1002/jcb.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- 12.Jochum W, Passegue E, Wagner EF. AP-1 in mouse development and tumorigenesis. Oncogene. 2002;20:2401–2412. doi: 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- 13.Eilers M. Eisenman RN Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghaneh P, Costello E, Neoptolemos JP. Biology and management of pancreatic cancer. Postgrad Med J. 2008;84:478–497. doi: 10.1136/gut.2006.103333. [DOI] [PubMed] [Google Scholar]
- 15.Schneider G, Siveke JT, Eckel F, et al. Pancreatic cancer: basic and clinical aspects. Gastroenterology. 2005;128:1606–1625. doi: 10.1053/j.gastro.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Buchholz M, Schatz A, Wagner M, et al. Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway. EMBO J. 2006;25:3714–3724. doi: 10.1038/sj.emboj.7601246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buck A, Buchholz M, Wagner M, et al. The tumor suppressor KLF11 mediates a novel mechanism in transforming growth factor beta-induced growth inhibition that is inactivated in pancreatic cancer. Mol Cancer Res. 2006;4:861–872. doi: 10.1158/1541-7786.MCR-06-0081. [DOI] [PubMed] [Google Scholar]
- 18.Ellenrieder V, Zhang JS, Kaczynski J, et al. Signaling disrupts mSin3A binding to the Mad1-like Sin3-interacting domain of TIEG2, an Sp1-like repressor. EMBO J. 2002;21:2451–2460. doi: 10.1093/emboj/21.10.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baubet V, Le Mouellic H, Campbell AK, et al. Chimeric green fluorescent protein-aequorin as bioluminescent Ca2+ reporters at the single-cell level. PNAS. 2000;97:7260–7265. doi: 10.1073/pnas.97.13.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones K, Hibbert F, Keenan M. Glowing jellyfish, luminescence and a molecule called coelenterazine. Trends Biotechnol. 1999;17:477–481. doi: 10.1016/s0167-7799(99)01379-7. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Zapico ME, Gonzalez-Paz NC, Weiss E, et al. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell. 2005;7:39–49. doi: 10.1016/j.ccr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 22.Dunn KL, Espino PS, Drobic B, et al. Ras-MAPK signal transduction pathway, cancer and chromatin remodeling. Biochem Cell Biol. 2005;83:1–14. doi: 10.1139/o04-121. [DOI] [PubMed] [Google Scholar]
- 23.Yagi K, Furuhashi M, Aoki H, et al. c-myc is a downstream target of the Smad pathway. J Biol Chem. 2002;277:854–861. doi: 10.1074/jbc.M104170200. [DOI] [PubMed] [Google Scholar]
- 24.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Tang Y, Cole PA, et al. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr Opin Struct Biol. 2008;18:741–747. doi: 10.1016/j.sbi.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109:67–79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 27.Neal JW, Clipstone NA. A constitutively active NFATc1 mutant induces a transformed phenotype in 3T3-L1 fibroblasts. J Biol Chem. 2003;278:17246–17254. doi: 10.1074/jbc.M300528200. [DOI] [PubMed] [Google Scholar]
- 28.Buchholz M, Ellenrieder V. An emerging role for Ca2+/calcineurin/NFAT signaling in cancerogenesis. Cell Cycle. 2007;6:16–19. doi: 10.4161/cc.6.1.3650. [DOI] [PubMed] [Google Scholar]
- 29.Robbs BK, Cruz AL, Werneck MB, et al. Dual roles for NFAT transcription factor genes as oncogenes and tumor suppressors. Mol Cell Biol. 2008;28:7168–7181. doi: 10.1128/MCB.00256-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jauliac S, López-Rodriguez C, Shaw LM, et al. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol. 2002;4:540–544. doi: 10.1038/ncb816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.