Abstract
Bacterial chemoreceptors mediate chemotaxis by recognizing specific chemicals and regulating a noncovalently associated histidine kinase. Ligand binding to the external domain of the membrane-spanning receptor generates a transmembrane signal that modulates kinase activity inside the cell. This transmembrane signaling is being investigated by novel strategies, which have revealed a remarkably subtle conformational signal carried by a signaling helix that spans the entire length of the >350-Å-long receptor. Multiple, independent lines of evidence indicate that, in the periplasmic and transmembrane domains, conformational signaling is a piston-type sliding of the signaling helix towards the cytoplasm.
Like other motile bacteria, Escherichia coli and Salmonella typhimurium respond to chemical gradients by moving towards higher concentrations of attractants and lower concentrations of repellents (reviewed in Refs 1–4). This behavior, termed chemotaxis, is mediated by a dedicated sensory system comprising transmembrane chemoreceptors, histidine and aspartate kinases, an SH3-like coupling protein, and two enzymes that mediate sensory adaptation by covalently modifying the chemoreceptors (Box 1). Homologs of these sensory components occur in virtually every motile bacterium or archaeon investigated to date, making this type of sensory pathway one of the most prevalent in nature. It is likely that the homologous components possess conserved molecular mechanisms. For example, chemoreceptors are expected to share similar mechanisms of transmembrane signaling.
Chemoreceptors are stable homodimers both in the absence and presence of ligands5. Each homodimer is an elongated helical bundle thought to be oriented normal to the membrane (Fig. 1)6–11. The periplasmic domain consists of eight helices arranged in two symmetric four-helix bundles, one per subunit (helices α1–α4, α1′–α4′). Two helices from each subunit span the bilayer, where they form a transmembrane four-helix bundle (helices TM1, TM2, TM1′, TM2′). The cytoplasmic domain is a distinct four-helix bundle, formed by association of two helical hairpins, one per subunit (helices CD1, CD2, CD1′, CD2′). One helix in each subunit extends the entire length of the structure (helix α4/TM2/linker/CD1), connecting the ligand-binding site at the membrane-distal end of the periplasmic domain with the kinase-interaction region at the opposite end of the receptor. The only major region that has not yet been shown experimentally to be helical is the conserved linker connecting the transmembrane and cytoplasmic domains12 but, regardless of its structure, the linker is stably folded and thus can communicate signals between receptor domains13.
Fig. 1.
Structure of a dimeric bacterial chemoreceptor. (Left) Atomic structural model generated by combining crystal structures of the periplasmic and cytoplasmic domains of the aspartate and serine receptors, respectively, with modeled structures of the transmembrane and linker regions11. The two symmetric subunits of the homodimer are in blue and gold, respectively. (Right) Schematic diagram showing structural and functional regions. For simplicity, helix supercoiling is omitted, and the pathway components that dock to the receptor in the assembled signaling complex are shown schematically (ellipsoids, spheres). Kinase docking, regulation and phosphotransfer events occur at the extreme cytoplasmic tip of the receptor. The adaptation enzymes interact with a conserved sequence at the C-terminus of certain receptors. Cytoplasmic sites of methylation and demethylation are shown as small ovals.
The first step in signal transduction is the binding of attractant or attractant-occupied binding protein to the periplasmic domain at one of two interfacial sites between the two symmetric four-helix bundles. Much evidence (summarized below) indicates that attractant binding sends a conformational signal within the dimer, travelling from the periplasmic domain through the transmembrane helices to the cytoplasmic domain. Ultimately, the ligand-induced signal reaches the bound histidine kinase, where it inhibits autophosphorylation. The simplest signaling mechanism would alter the average receptor conformation in such a way as to displace one or more transmembrane helices relative to one another or to the membrane. Such a displacement would trigger movements in the cytoplasmic domain. To characterize this transmembrane signal one must determine which helices move and the manner in which they move.
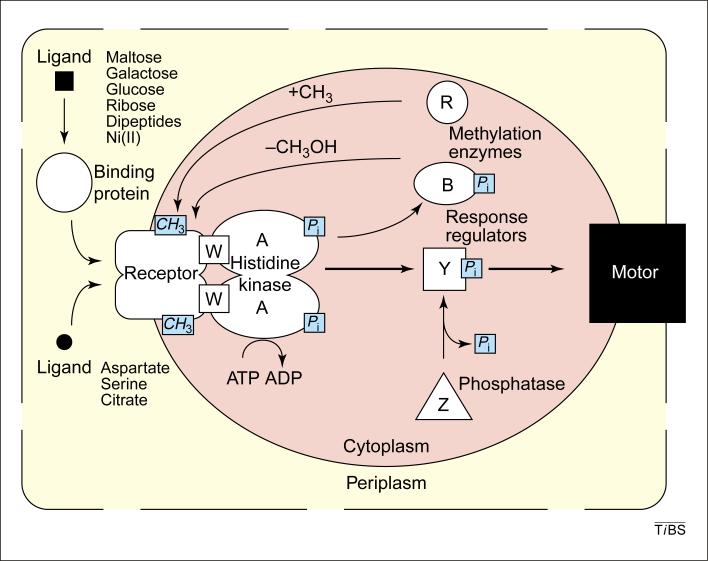
Box 1. The bacterial chemosensory system.
In Escherichia coli and Salmonella typhimurium, chemoreceptors are homodimers that form supermolecular complexes with the homodimeric histidine kinase CheA (A), the SH3-like coupling protein CheW (W), and the response regulators/aspartate kinases CheY (Y) and CheB (B). One class of receptor has a C-terminal interaction site for the enzymes of adaptational modification, the methyltransferase CheR (R) and the methylesterase CheB (B) (Fig. I) (reviewed in Refs 1–4). There are chemoreceptors for aspartate (Tar), ribose and galactose (Trg), serine (Tsr), dipeptides (Tap), citrate (Tcp) and cytoplasmic redox potential (Aer)2,5,6. Complex formation activates autophosphorylation of CheA, in which the γ-phosphate of ATP is transferred to a His residue of the kinase. In turn, that phosphate is transferred to an aspartyl residue of the response regulator CheY, activating it for interaction with the flagellar motor.
Phosphorylated CheY controls swimming behavior by binding to the flagellar rotary motor and changing its rotational state from counterclockwise to clockwise, thereby switching from forward swimming to uncoordinated tumbling that reorients the cell in a new, randomly chosen swimming direction1–4. CheZ (Z) stimulates the naturally rapid hydrolysis of phosphorylated CheY by an unknown mechanism. In the absence of a chemoeffector gradient, the steady-state level of phosphorylated CheY results in alternation between forward swimming and tumbling, creating a 3D random walk. In a gradient, chemoattractant binding to receptor inhibits CheA autophosphorylation, causing a reduction in the cellular content of the short-lived phosphorylated CheY, reducing the probability of tumbles and thus extending forward swimming.
Attractant binding activates a feedback loop of sensory adaptation. When the cellular population of receptors experiences an increase in net attractant occupancy, thereby altering its net signal output, the adaptation system restores the signal output to the basal level by increasing the proportion of receptor adaptation sites that are methyl esterified1–4. Attractant binding inhibits kinase activity; compensatory methylation increases kinase activity. Thus, the feedback loop functions to maintain kinase activity at an intermediate level that provides the cellular concentration of phosphorylated CheY needed to generate a functional swim:tumble ratio. On a molecular level, attractant occupancy both enhances the propensity for methylation of the adaptation sites on the occupied receptors and reduces the kinase activity of CheA. Lower CheA kinase activity reduces the cellular content of the active phosphorylated form of the methylesterase CheB, and thus triggers a global reduction in receptor demethylation. The net result is increased methylation of the population of occupied receptors, which increases CheA kinase activity and counters the inhibitory effects of attractant binding.
Evidence that a specific helix–helix interface carries the signal
Initial studies set out to identify the helices that carry the transmembrane signal, and to ascertain whether the signal was transmitted via the subunit interface or within individual subunits. One indication that chemoreceptor signaling involves specific intrasubunit helix–helix movements came from an early 19F nuclear magnetic resonance (NMR) study of the isolated periplasmic domain of the aspartate receptor (Tar). In this study, ligand binding perturbed 4-fluoro-Phe probes at the α1–α4 interface but not at the α1–α1′ interface that dominates subunit contacts14.
Many studies have employed site-directed Cys residues and sulfhydryl chemistry to probe the molecular details of the helix movements, an approach facilitated by the fact that chemoreceptors lack intrinsic Cys residues or contain only a few that can be replaced without functional consequences8,15. Some of the most productive strategies made use of engineered inter-helix disulfide bonds15,16. Significantly, engineered disulfides in chemoreceptors block signaling if placed across certain helical interfaces but not others. In vitro studies of the aspartate receptor found that receptors containing one or even two α1/TM1–α1′/TM1′ disulfide bonds that constrain the subunit interface retained transmembrane signaling, as assayed by ligand effects on methylation at adaptation sites15,17. An in vivo study of the ribose and galactose receptor (Trg) in intact, functional cells found that four α1/TM1–α1′/TM1′ disulfides that constrain the subunit interface each allowed normal receptor signaling, but two TM1–TM2 disulfides each eliminated cellular responses to attractant stimulation18 (Fig. 2a). Seven of nine α1/TM1–α1′/TM1′ disulfides that constrain the subunit interface of the aspartate receptor each allowed normal receptor regulation of kinase activity in vitro9 (Fig. 2b). By contrast, seven of eight disulfides across the α1/TM1–α4/TM2 interface disrupted kinase regulation19,20. Four of these α1/TM1–α4/TM2 disulfides covalently locked the receptor in opposite signaling states: two in the apo receptor state, characterized by high kinase activity and low attractant affinity, whereas the other two trapped the attractant-occupied state that has low kinase activity and high attractant affinity (Fig. 2d).
Fig. 2.
Engineered disulfides used to analyze conformational signaling. The structural (α1/TM1) and signaling (α4/TM2) helices of the ribose and galactose receptor (Trg, left) and the aspartate receptor (Tar, right) are shown as schematic wheel diagrams viewed from the periplasm [a,b and c,d (top)] and as ribbon diagrams viewed parallel to the membrane [c,d (bottom)]. (a and b) Helix-constraining disulfides that allow (black) or block (red) (a) response to attractant by intact cells18 or (b) attractant-induced kinase inhibition in vitro9,19. (c) Diagnostic Cys pairs for which rates of crosslinking are unchanged (black), increased (solid red) or decreased (dashed red) by ligand occupancy of Trg in vivo21. (d) Disulfides that allow native (≥50%) receptor-mediated kinase regulation in vitro (black) or that lock the receptor in one of its two native signaling states; either the low ligand-affinity state that activates the kinase (dashed red) or the high ligand-affinity state that inhibits the kinase (solid red)9,19,20. To conserve space, the ribbon diagrams have been compressed vertically.
An in vivo study of the effects of signaling on the formation of disulfides tested 67 transmembrane Cys pairs spanning neighboring helices of the ribose and galactose receptor. In the absence and presence of ligand the same 19 pairs exhibited disulfide crosslinking, indicating that conformational signaling did not produce large movements between transmembrane helices21. Among Cys pairs for which accurate rates of disulfide formation could be determined in vivo, ligand occupancy did not have a significant effect on any of four TM1–TM1′ intersubunit pairs but changed the rates for all four TM1–TM2 pairs, increasing two and decreasing two21 (Fig. 2c). Thus the TM1–TM1′ subunit interface, which could be immobilized without affecting signaling, exhibited no movement detectable by diagnostic crosslinking. By contrast, the TM1–TM2 interface, at which immobilization eliminated signaling, was the same interface at which signaling altered the crosslinking rate of diagnostic Cys pairs.
Genetic studies investigated whether occupancy at one interfacial ligand-binding site sends a signal through one or both receptor subunits. The two symmetric aspartate-binding sites of the aspartate receptor are each composed of distinct half-sites – one from each subunit6. By combining subunits containing different half-site mutations, it is possible to generate receptor heterodimers in which one binding site is functional and the other non-functional. Three studies combined these periplasmic or complementing mutations in the transmembrane domain with cytoplasmic mutations that inactivate or truncate only one cytoplasmic domain of the heterodimer22–24. These studies revealed that the aspartate-induced signal is generated at a specific half-site and is transmitted through only one subunit to the cytoplasmic domain. The signaling half-site includes residues at the periplasmic end of the α4/TM2 helix that interact with the amino group of aspartate2,6, thereby providing a simple mechanism by which attractant binding could displace the α4/TM2 helix and perturb the α1/TM1–α4/TM2 interface in the subunit that carries the transmembrane signal2.
Other studies examined the functional effects of point mutations at specific helix interfaces25. Cys substitutions at any one of the positions in TM1 and TM2 of chemoreceptor Trg did not eliminate receptor function, but ~40% had effects on receptor signaling in vivo, as assessed by altered methylation at the receptor adaptation sites. Substitutions that increased methylation, thus mimicking the attractant signal, clustered along the TM1–TM2 interface within a subunit. Substitutions that inhibited the attractant-induced methylation increase clustered along the TM1–TM1′ interface between subunits. Because mutational substitutions usually perturb interactions, the clustering of mutations that mimic the effects of attractant binding along the TM1–TM2 interface implies that this interface is perturbed by conformational signaling. Similarly, clustering along the TM1–TM1′ interface of substitutions that reduce attractant-induced methylation implies that signaling is optimal when native subunit interactions are maintained25.
These collective observations (Table 1) point to a common conclusion: in the periplasmic and transmembrane domains the α1/TM1–α4/TM2 interface within a receptor subunit is the locus of conformational signaling. By contrast, any movement between subunits that is crucial to signaling in these domains must be sufficiently modest to be allowed by disulfide bonds between the α1/TM1–α1′/TM1′ helices that dominate the subunit interface. The simplest conformational change consistent with these constraints is a displacement of helix α4/TM2, which we thus call the signaling helix, relative to the static subunit interface in the periplasmic and transmembrane domains.
Table 1.
Summary of evidence implicating specific helix–helix interfaces in signal transduction
| Domain assessed | Feature assessed | Receptor characteristics | Environment | Interface implicated | Refs |
|---|---|---|---|---|---|
| Periplasmic | Environments of 19F probes ± ligand by NMR | Periplasmic fragment of Tar with C36–C36′ disulfide | Solution | α1–α4 | 14 |
| Periplasmic | Distance-difference matrix of X-ray coordinates ± ligand | Periplasmic fragment of Tar with engineered C36–C36′ disulfide | Crystal | α1–α4 | 20 |
| Periplasmic | Distance-difference matrix of X-ray coordinates ± ligand | Periplasmic fragment of Tar (no disulfide) | Crystal | α1–α1′ | 26 |
| Periplasmic, transmembrane | Kinase control and adaptational methylation of disulfide-constrained receptors ± ligand | Intact Tar in complex with kinase | Isolated cell membranes | α1/TM1–α4/TM2 | 9,19 |
| Periplasmic, transmembrane | Chemotaxis mediated by heterodimers with complementary subunit mutations | Hemi-truncated Tar heterodimers | Cells | α1/TM1–α4/TM2 | 23,24 |
| Periplasmic, transmembrane | Gene expression controlled by kinase domain in chimeric construct ± ligand | Chimera of Tar and cytoplasmic kinase domain of EnvZ | Cells | α1/TM1–α4/TM2 | 22 |
| Transmembrane | Flagellar rotation controlled by disulfide-constrained receptors ± ligand | Intact Trg in complex with kinase | Cells | TM1–TM2 | 18 |
| Transmembrane | Rates of disulfide formation in Cys-containing receptors ± ligand | Intact Trg in complex with kinase | Cells | TM1–TM2 | 21 |
| Transmembrane | Patterns of adaptational methylation of Cys-containing receptors ± ligand | Intact Trg in complex with kinase | Cells | TM1–TM2 | 25 |
| Transmembrane | Distances between site-directed EPR spin labels ± ligand | Intact Tar | Reconstituted lipid vesicles | TM1–TM2 | 32 |
| Transmembrane | Distances between isotopically labeled side chains ± ligand by solid state NMR | Intact Tsr | Isolated cell membranes | TM1–TM2 | 33 |
| Cytoplasmic | Kinase control by disulfide-constrained receptors ± ligand | Intact Tar in complex with kinase | Isolated cell membranes | CD1–CD1′ and CD2–CD2′ | 10 |
| Cytoplasmic | Kinase control by receptors with interfacial substitutions | Intact Tar in complex with kinase | Isolated cell membranes | CD1–CD1′ | 38 |
Abbreviations: EPR, electron paramagnetic resonance; NMR, nuclear magnetic resonance; Tar, aspartate receptor; Trg, ribose and galactose receptor.
Evidence for specific types of helix displacement
In principle, the transmembrane signal could be carried by helix sliding, tilting or rotation, or altered helix dynamics. Several independent biophysical and biochemical approaches have been employed to investigate the structural basis of the signal. Because there is general agreement that the signal is small in amplitude, it is especially important to synthesize information provided by multiple techniques. For clarity, studies of different domains are considered separately.
Periplasmic domain
X-ray crystallography has provided 3D structures of several variants of the dimeric periplasmic domain of the aspartate receptor, both in the absence and presence of bound ligand. The initial structures were of a periplasmic domain fragment containing an engineered α1–α1′ disulfide bond (C36–C36′) between the subunits6. Subsequent structures were of the wild-type fragment that lacks the disulfide-forming Cys residue26. In all cases, the aspartate-free and aspartate-bound structures were similar, making it difficult to identify the ligand-induced conformational change. The issue is best approached by a model-independent technique such as distance difference analysis, which mathematically compares the atomic coordinates of two conformations to identify their structural differences20,26,27. However, the answer provided by distance difference analysis depends on whether the disulfide bond is present between the two subunits. In its presence, the major locus of ligand-induced change is the α1–α4 interface in the subunit interacting with the amino group of the bound aspartate. In its absence the major displacement occurs at the α1–α1′ interface between the subunits. Thus, the question becomes which form of the dimeric periplasmic fragment is a better model for the intact, membrane-bound receptor. The native receptor has no disulfide crosslink between the subunits, suggesting that the structure without a crosslink would be the better choice. However, much evidence indicates that the membrane-proximal ends of helices α1 and α1′ are in close proximity7–9,15,17. In crystals of the periplasmic domain, such α1–α1′ proximity occurs in the presence of the crosslink but not in its absence6,26. In addition, intact receptor that contains this same crosslink exhibits normal signaling in vitro and in vivo9,15,18. Thus, the crosslinked fragment appears the best model for the periplasmic domain of the intact receptor.
Excess aspartate saturates only one of the two ligand-binding sites in the crosslinked fragment because negative cooperativity significantly lowers the affinity of the unoccupied site6,14,28. Distance difference analysis of the crosslinked fragment revealed that the structure of one subunit was unaltered by attractant binding, and therefore could be used to guide the superposition of the apo and attractant-occupied structures20. That superposition provided a high-resolution view of the attractant-induced conformational change (Fig. 3), a 1.6 Å piston displacement of signaling helix α4 down its long axis towards the membrane accompanied by a 5° tilt of the same helix20. Rearrangements of the α1–α1′ subunit interface were minor except near the aspartate-binding site. Thus, analysis of the crosslinked structures provided further evidence that signaling affects primarily the interface between helices α1 and α4 (Table 1)20. The piston displacement was less than the ~2 Å upper limit for low-energy sliding of packed helices29. It occurred only in the subunit in which helix α4 contacted the amino group of the bound aspartate, the same signaling subunit identified by mutational studies22,24. By contrast, in the fragment that lacked the crosslink a different ligand-induced conformational change was observed: an inter-subunit rotation of ~4° but no sliding of α4 (Ref. 26). However, such a picture might not accurately reflect the conformational change in the intact receptor (see previous paragraph).
Fig. 3.
Piston displacement of the signaling helix. (a) Structural view of the piston displacement, generated by superposition of crystal structures for the apo and aspartate-occupied periplasmic fragment containing an inter-subunit disulfide bond6. Binding of aspartate [shown as a red Corey–Pauling–Koltun (CPK) model] displaces the signaling helix α4 (red) downwards towards the cytoplasm by ~1.6 Å (Ref. 20). (b) Schematic view of information flow through the signaling helix, beginning with ligand-induced piston displacement in the periplasmic and transmembrane domains20,21, continuing with a subtle uncharacterized conformational change in the cytoplasmic domain10, and ending with modulation of the autophosphorylation activity of the histidine kinase CheA.
Transmembrane domain
Disulfides that covalently locked the aspartate receptor in its kinase-activating and -inhibiting states suggested that the same piston displacement observed in the crystal structure of the crosslinked periplasmic fragment occurs during transmembrane signaling in the intact, membrane-bound receptor-kinase complex19,20. Modeling revealed that kinase-inhibiting disulfides drive a piston displacement of signaling helix α4/TM2 downwards towards the cytoplasm relative to structural helix α1/TM1 (Ref. 20). By contrast, kinase-activating disulfides drive a piston displacement in the opposite direction (Fig. 2d)20. The modeled displacements were in the same direction and of similar magnitude as the piston movements of α4 in the disulfide-linked crystal structure (compare Fig. 2d with Fig. 3). The modest 1–2 Å displacement is consistent with the discovery of one disulfide bond (C36–C183) that crosslinks signaling helix α4/TM2 to structural helix α1/TM1 yet allows partial ligand-induced kinase regulation (Fig. 2b), indicating that a portion of the essential displacement can occur within the constraints of a single, optimally placed disulfide linkage19,20.
The nature of the transmembrane conformational change induced by attractant binding to the ribose and galactose receptor has been deduced from the effects of attractant on rates of oxidative disulfide formation between diagnostic TM1–TM2 Cys pairs21. Figure 2c shows the positions of the two Cys pairs exhibiting increased rates and the two pairs exhibiting decreased rates. The changes cannot be accounted for by helical tilting, rotation around a long axis or movement towards or away from each other. By contrast, a modest piston sliding of signaling helix TM2 towards the cytoplasm relative to TM1 would explain all four changes. If the changed rates of crosslinking for the diagnostic Cys pairs reflect the conformational signal in the transmembrane region, then substitutions near the ligand-binding site that induce signaling30 should create the same changes. This prediction has been verified31.
Emerging spectroscopic and modeling approaches have begun to shed further light on the transmembrane conformational change. The labeling of introduced Cys pairs with nitroxide spin labels and measurement of spin–spin distances by electron paramagnetic resonance has been used to explore ligand-induced conformational changes in the purified aspartate receptor. Among four α1–α4 and three TM1–TM2 pairs, the spin–spin interactions of one periplasmic pair and all three transmembrane pairs were affected by the presence of ligand32. The estimated uncertainty for distance determinations using this method is ~2.5 Å and none of the four calculated movements exceeded 1 Å. However, the directions of the apparent changes (one decreased distance, three increases) were consistent with the piston-type sliding of signaling helix α4/TM2 described above. Solid-state NMR studies of the full-length, membrane-bound serine receptor have detected a 1 Å ligand-induced distance change between helices α4/TM2 and α1/TM1 consistent with the same piston displacement33. Finally, modeling of the interaction between maltose-binding protein and the aspartate receptor suggests a mechanism by which this docking could trigger the same asymmetric piston displacement of the signaling helix induced by aspartate binding34.
Cytoplasmic domain
The cytoplasmic domain must transmit both attractant-induced transmembrane signals and adaptational signals to the bound kinase, but the structural basis of this transmission is not known. Disulfide scanning studies that tested the functional effects of 188 intersubunit cytoplasmic disulfides identified 14 that allowed the crosslinked aspartate receptor to activate, and in some cases super-activate, the kinase10,13,35,36. Seven of these disulfides locked the receptor in the kinase-activating state (Fig. 4). This high frequency of intersubunit, activity-locking crosslinks indicates that, in contrast to the periplasmic and transmembrane domains, the dimer interface is crucial to conformational signaling in the cytoplasmic domain (Table 1). The differing involvement of the subunit interface in signaling through these regions corresponds to the changing disposition of the signaling helix relative to the subunit interface: the periplasmic and transmembrane regions of the signaling helix (α4/TM2) are distal to the largely static subunit interface, whereas the cytoplasmic region of the signaling helix (CD1) is an integral component of the subunit interface. Notably, six of the seven activity-locking disulfides were clustered near a position where many side-chain substitutions superactivate the kinase, in some cases constitutively38. This cluster lies near the surface glutamyl residues that are the sites of adaptational modification. Changing these sites from all anionic Glu residues to neutral methyl esters or Gln residues switches the receptor signaling state from one that inhibits the kinase to one that superactivates the kinase39,40. Clearly, this region of the cytoplasmic domain plays a central role in kinase regulation.
Fig. 4.
Diagnostic Cys residues and disulfides in the cytoplasmic domain. Schematic view of locations of inter-subunit disulfides in the aspartate receptor that (1) trap the kinase-activating state (red bonds) or (2) retain normal kinase regulation by attractant (black and white bonds). In addition, the adaptation sites (black circles) and positions implicated in crucial kinase-docking or dimer–dimer interactions between receptors (black squares) are shown10,11,37.
As in the periplasmic and transmembrane regions, the ligand-induced signal that passes through the cytoplasmic domain appears to be subtle. Seven intersubunit, cytoplasmic disulfides were found to allow ligand-induced kinase inhibition (Fig. 4), indicating that the conformational change within the cytoplasmic domain can be transmitted within the flexibility constraints imposed by disulfides placed between helices CD1–CD1′ and CD2–CD2′ (Ref. 10). Overall, it appears that the transmembrane signal subtly rearranges the packing of the cytoplasmic four-helix bundle, and that this alteration transmits the signal to the kinase.
Higher-order structures and conformational signaling
Chemoreceptors are clustered, and clusters are found primarily at the poles of the bacterial cell41. Clustering implies that interactions might occur between receptor dimers, and modeling studies42 have generated much interest in the possible functions of such higher-order interactions. Studies of receptor adaptation have shown that receptors possessing a C-terminal interaction site for the adaptational enzymes (CheR, CheB) facilitate the adaptation of receptors that lack the interaction site via an intermolecular mechanism43–47. A structural basis for clustering is suggested by crystals of the serine receptor cytoplasmic domain fragment in which three dimeric domains associate at their membrane-distal tips to form trimers11,42. Strong evidence for the involvement of higher-order interactions in signaling is provided by the distinct positive cooperativity observed for ligand-induced inhibition of kinase activity in vitro48,49. For the aspartate receptor, Hill analysis indicates that cooperativity depends weakly on the level of adaptational modification and arises from interactions between as few as two or three dimers49, consistent with the crystallographic trimer of dimers11. Hill analysis of the serine receptor indicates that cooperativity varies strongly with modification of the adaptational sites and corresponds to as many as 12 interacting dimers for the fully modified receptor48, consistent with models proposing higher-order interactions42,50,51. The different degrees of cooperativity observed for the aspartate and serine receptors could reflect different levels of receptor expression (the serine receptor was more highly overexpressed) or inherent differences between the receptors.
The discovery of the cooperative nature of transmembrane signaling extends our notions of chemoreceptor signaling but does not alter the significance of conformational changes within a single receptor dimer. Much of the evidence for piston-type sliding of the signaling helix has been provided by in vivo and in vitro studies using the working receptor–kinase signaling complex (conditions in which native cooperative interactions are presumed to occur). It seems likely that cooperative interactions among receptor dimers are not alternatives to helical sliding as a mechanism of transmembrane signaling, but rather arise from interactions between individual dimers, in which the signal is carried by a piston displacement of the signaling helix.
Conclusions
What conveys the informational signal from the ligand-binding site of a chemoreceptor to the associated kinase ~350 Å away on the other side of the membrane? The extended structure we call the signaling helix (helix α4/TM2/linker/CD1) appears to provide the direct, physical connection between ligand and kinase (Fig. 3). It is striking that multiple, independent lines of evidence either implicate signal-induced movement of the signaling helix, or can be explained by such a movement. By contrast, comparatively little evidence supports alternative possibilities for signaling, such as a change in receptor dynamics, side-chain displacement, or rotational displacement of receptor subunits17,26,52–54. Essentially all data indicate that the signaling movement is subtle and almost all relevant observations of signaling movement in the periplasmic and cytoplasmic domains identify or are consistent with a modest (1–2 Å) sliding of the signaling helix towards the cytoplasm in a piston displacement. The notion is satisfying because ligand binding occurs at one end of this helix and it is easy to see how binding could displace this helix towards the cytoplasm. In addition, the displacement uses the relatively rigid, incompressible long-axis of the helix to transmit information over a great distance. By contrast, a signal carried by helix tilting or twisting would be more susceptible to dissipation by helix bending or torsional flexibility. The remarkably small magnitude of the displacement is consistent with the limited energy provided by attractant binding, because such a displacement avoids the disruption of numerous polar side-chain interactions and ridges-in-grooves packing between supercoiled helices. A large piston displacement would not only disrupt these interactions, but also require the energetically unfavorable movement of charged and hydrophobic residues across the water–membrane interface. In the cytoplasmic domain, the conformational change follows the path of the signaling helix to the subunit interface, but the nature of the conformational signal sent through the cytoplasmic four-helix bundle to the associated kinase remains to be established.
A large number of transmembrane proteins, including many receptors and allosterically regulated channels and transporters, couple ligand binding on one side of the membrane to a regulatory change on the other side. Such proteins are often constructed from helical bundles. We expect that the subtle helical sliding central to transmembrane signaling in chemoreceptors will be found to be a mechanistic feature in other examples of transmembrane coupling.
Fig. I.
Components of the chemotaxis system in Escherichia coli and Salmonella typhimurium. Abbreviations: Ni(II), nickel ion; Pi, inorganic phosphate; A, B, R, W, Y and Z represent CheA, CheB, CheR, CheW, CheY and CheZ, respectively.
Acknowledgements
Support provided by NIH Grant GM40731 (to J.J.F.) and GM29963 (to G.L.H.). We thank our current and previous lab members, and many colleagues in the field for stimulating conversations. We regret that, owing to space constraints, we could not cite all relevant publications.
Contributor Information
Joseph J. Falke, Dept of Chemistry and Biochemistry, University of Colorado, Boulder, CO 80309-0215, USA. falke@colorado.edu
Gerald L. Hazelbauer, Dept of Biochemistry, University of Missouri-Columbia, Columbia, MO 65211, USA. hazelbauerg@missouri.edu
References
- 1.Blair DF. How bacteria sense and swim. Annu. Rev. Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 2.Falke JJ, et al. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djordjevic S, Stock AM. Structural analysis of bacterial chemotaxis proteins: components of a dynamic signaling system. J. Struct. Biol. 1998;124:189–200. doi: 10.1006/jsbi.1998.4034. [DOI] [PubMed] [Google Scholar]
- 4.Robinson VL, et al. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 2000;7:626–633. doi: 10.1038/77915. [DOI] [PubMed] [Google Scholar]
- 5.Milligan DL, Koshland DE. Site-directed crosslinking: establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- 6.Milburn MV, et al. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 7.Pakula AA, Simon MI. Determination of transmembrane protein-structure by disulfide crosslinking: the E. Coli Tar receptor. Proc. Natl. Acad. Sci. U. S. A. 1992;89:4144–4148. doi: 10.1073/pnas.89.9.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee GF, et al. Deducing the organization of a transmembrane domain by disulfide crosslinking: the bacterial chemoreceptor Trg. J. Biol. Chem. 1994;269:29920–29927. [PubMed] [Google Scholar]
- 9.Chervitz SA, et al. Transmembrane signaling by the aspartate receptor: engineered disulfides reveal static regions of the subunit interface. Biochemistry. 1995;34:9722–9733. doi: 10.1021/bi00030a010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bass RB, Falke JJ. The aspartate receptor cytoplasmic domain: in situ chemical analysis of structure, mechanism and dynamics. Struct. Fold. Des. 1999;7:829–840. doi: 10.1016/s0969-2126(99)80106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim KK, et al. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 12.Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 13.Butler SL, Falke JJ. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry. 1998;37:10746–10756. doi: 10.1021/bi980607g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danielson MA, et al. Attractant-induced and disulfide-induced conformational-changes in the ligand-binding domain of the chemotaxis aspartate receptor: a F-19 NMR study. Biochemistry. 1994;33:6100–6109. doi: 10.1021/bi00186a009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falke JJ, Koshland DE. Global flexibility in a sensory receptor: a site directed disulfide approach. Science. 1987;237:1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- 16.Careaga L, Falke JJ. Thermal motions of surface α-helices in the D-galactose chemosensory receptor. J. Mol. Biol. 1992;226:1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott WG, Stoddard BL. Transmembrane signaling and the aspartate receptor. Structure. 1994;2:877–887. doi: 10.1016/s0969-2126(94)00088-3. [DOI] [PubMed] [Google Scholar]
- 18.Lee GF, et al. Transmembrane signaling characterized in bacterial chemoreceptors by using sulfhydryl crosslinking in vivo. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3391–3395. doi: 10.1073/pnas.92.8.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chervitz SA, Falke JJ. Lock on/off disulfides identify the transmembrane signaling helix of the aspartate receptor. J. Biol. Chem. 1995;270:24043–24053. doi: 10.1074/jbc.270.41.24043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chervitz SA, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughson AG, Hazelbauer GL. Detecting the conformational change of transmembrane signaling in a bacterial chemoreceptor by measuring effects on disulfide crosslinking in vivo. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11546–11551. doi: 10.1073/pnas.93.21.11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, et al. Ligand-binding induces an asymmetrical transmembrane signal through a receptor dimer. J. Mol. Biol. 1993;232:493–498. doi: 10.1006/jmbi.1993.1405. [DOI] [PubMed] [Google Scholar]
- 23.Gardina PJ, Manson MD. Attractant signaling by an aspartate chemoreceptor dimer with a single cytoplasmic domain. Science. 1996;274:425–426. doi: 10.1126/science.274.5286.425. [DOI] [PubMed] [Google Scholar]
- 24.Tatsuno I, et al. Signaling by the E. coli aspartate chemoreceptor Tar with a single cytoplasmic domain per dimer. Science. 1996;274:423–425. doi: 10.1126/science.274.5286.423. [DOI] [PubMed] [Google Scholar]
- 25.Lee GF, et al. Identification of functionally important helical faces in transmembrane segments by scanning mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5416–5420. doi: 10.1073/pnas.92.12.5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chi YI, et al. Apo structure of the ligand-binding domain of aspartate receptor from Escherichia coli and its comparison with ligand-bound or pseudoligand-bound structures. FEBS Lett. 1997;414:327–332. doi: 10.1016/s0014-5793(97)01027-2. [DOI] [PubMed] [Google Scholar]
- 27.Bjorkman AM, et al. Mutations that affect ligand binding to the E. coli aspartate receptor: implications for transmembrane signaling. J. Biol. Chem. 2001;276:2808–2815. doi: 10.1074/jbc.M009593200. [DOI] [PubMed] [Google Scholar]
- 28.Biemann HP, Koshland DE. Aspartate receptors of E. coli and S. typhimurium bind ligand with negative and half-of-the-sites cooperativity. Biochemistry. 1994;33:629–634. doi: 10.1021/bi00169a002. [DOI] [PubMed] [Google Scholar]
- 29.Chothia C, Lesk AM. Helix movements in proteins. Trends Biochem. Sci. 1985;10:116–118. [Google Scholar]
- 30.Yaghmai R, Hazelbauer GL. Ligand occupancy mimicked by single residue substitutions in a receptor: transmembrane signaling induced by mutation. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7890–7894. doi: 10.1073/pnas.89.17.7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beel BD, Hazelbauer GL. Signaling substitutions in the periplasmic domain of chemoreceptor Trg induce or reduce helical sliding in the transmembrane domain. Mol. Microbiol. doi: 10.1046/j.1365-2958.2001.02446.x. in press. [DOI] [PubMed] [Google Scholar]
- 32.Ottemann KM, et al. A piston model for transmembrane signaling of the aspartate receptor. Science. 1999;285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 33.Murphy OJ, et al. Site-directed solid-state NMR measurement of a ligand-induced conformational change in the serine bacterial chemoreceptor. Biochemistry. 2001;40:1358–1366. doi: 10.1021/bi0015109. [DOI] [PubMed] [Google Scholar]
- 34.Zhang YH, et al. Model of maltose-binding protein/chemoreceptor complex supports intrasubunit signaling mechanism. Proc. Natl. Acad. Sci. U. S. A. 1999;96:939–944. doi: 10.1073/pnas.96.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danielson MA, et al. Cysteine and disulfide scanning reveals a regulatory α-helix in the cytoplasmic domain of the aspartate receptor. J. Biol. Chem. 1997;272:32878–32888. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bass RB, et al. Signaling domain of the aspartate receptor is a helical hairpin with a localized kinase docking surface: cysteine and disulfide scanning studies. Biochemistry. 1999;38:9317–9327. doi: 10.1021/bi9908179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu JD, Parkinson JS. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by E. coli. J. Bacteriol. 1991;173:4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trammell MA, Falke JJ. Identification of a site critical for kinase regulation on the central processing unit (CPU) helix of the aspartate receptor. Biochemistry. 1999;38:329–336. doi: 10.1021/bi981964u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ninfa EG, et al. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J. Biol. Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 40.Borkovich KA, et al. Attenuation of sensory receptor signaling by covalent modification. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the E. coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu TS, et al. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat. Cell Biol. 2000;2:792–796. doi: 10.1038/35041030. [DOI] [PubMed] [Google Scholar]
- 43.Wu J, et al. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 44.Li JY, et al. The serine chemoreceptor from E. coli is methylated through an inter-dimer process. Biochemistry. 1997;36:11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- 45.LeMoual H, et al. Methylation of the E. coli chemotaxis receptors: intra- and inter-dimer mechanisms. Biochemistry. 1997;36:13441–13448. doi: 10.1021/bi9713207. [DOI] [PubMed] [Google Scholar]
- 46.Barnakov AN, et al. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng X, et al. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J. Bacteriol. 1997;179:1–7. doi: 10.1128/jb.179.21.6714-6720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li GY, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of E. coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 49.Bornhorst JA, Falke JJ. Attractant regulation of the aspartate receptor-kinase complex: limited cooperative interactions between receptors and effects of receptor modification state. Biochemistry. 2000;39:9486–9493. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ames P, et al. Methylation segments are not required for chemotactic signaling by cytoplasmic fragments of Tsr, the methyl-accepting serine chemoreceptor of E. coli. Mol. Microbiol. 1996;19:737–746. doi: 10.1046/j.1365-2958.1996.408930.x. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, et al. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 1997;24:7231–7240. doi: 10.1093/emboj/16.24.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim SH. Frozen dynamic dimer model for transmembrane signaling in bacterial chemotaxis receptors. Protein Sci. 1994;3:159–165. doi: 10.1002/pro.5560030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maruyama IN, et al. A model for transmembrane signaling by the aspartate receptor-based on random-cassette mutagenesis and site-directed disulfide crosslinking. J. Mol. Biol. 1995;253:530–546. doi: 10.1006/jmbi.1995.0571. [DOI] [PubMed] [Google Scholar]
- 54.Cochran AG, Kim PS. Imitation of E. coli aspartate receptor signaling in engineered dimers of the cytoplasmic domain. Science. 1996;271:1113–1116. doi: 10.1126/science.271.5252.1113. [DOI] [PubMed] [Google Scholar]
- 1.Blair DF. How bacteria sense and swim. Annu. Rev. Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 2.Falke JJ, et al. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djordjevic S, Stock AM. Structural analysis of bacterial chemotaxis proteins: components of a dynamic signaling system. J. Struct. Biol. 1998;124:189–200. doi: 10.1006/jsbi.1998.4034. [DOI] [PubMed] [Google Scholar]
- 4.Robinson VL, et al. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 2000;7:626–633. doi: 10.1038/77915. [DOI] [PubMed] [Google Scholar]
- 5.Bibikov SI, et al. Domain organization and flavin adenine dinucleotide-binding in the aerotaxis signal transducer Aer of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5830–5835. doi: 10.1073/pnas.100118697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Repik A, et al. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol. Microbiol. 2000;36:806–816. doi: 10.1046/j.1365-2958.2000.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]