Abstract
Mimivirus, the prototypic member of the new family of Mimiviridae, is the largest virus known to date. Progress has been made recently in determining the three-dimensional structure of the 0.75-μm diameter virion using cryo-electron microscopy and atomic force microscopy. These showed that the virus is composed of an outer layer of dense fibers surrounding an icosahedrally shaped capsid and an internal membrane sac enveloping the genomic material of the virus. Additionally, a unique starfish-like structure at one of the fivefold vertices, required by the virus for infecting its host, has been defined in more detail.
Key Words: Atomic force microscopy; Cryo-electron microscopy; Mimivirus, three-dimensional structure; Nucleocytoplasmic large DNA viruses
Introduction
Mimivirus (Acanthamoeba polyphaga Mimivirus), the largest known virus, was discovered serendipitously in 1992 during an investigation of an outbreak of pneumonia in a hospital in Bradford, England [1]. It was mistakenly identified as a bacterium because its fibrous surface could be Gram-positively stained. However, a preliminary study of the genome showed that it lacked a number of genes required by independently living organisms [2]. The same study also showed that Mimivirus is closely related to Phycodnaviridae (e.g. PBCV-1), Iridoviridae (e.g. CIV) and Poxviridae; all members of the group of nucleocytoplasmic large DNA viruses (NCLDV). On the other hand, it was shown that Mimivirus is distinct enough from other NCLDVs, and therefore represents the prototype of the newly defined Mimiviridae family. The complete genome sequence (1.2 Mbp) [3] codes for 911 genes. Some of these genes, such as those for tRNAs or proteins involved in amino acid or lipid metabolism, have never previously been seen in a virus.
An early electron micrograph suggested that Mimivirus might have an icosahedral shape [2,4]. A subsequent cryo-electron microscopic (cryo-EM) study [4] confirmed that the Mimivirions had at least approximate icosahedral symmetry and showed that the virus had a vertex-to-vertex diameter of about 5,000 Å with 1,200-Å-long fibers on the surface. The surface fibers create a dense forest covering almost the whole virus capsid (fig. 1). Furthermore, a starfish-like structure was identified [5,6] at one of the fivefold vertices on all mature virions. The arms of the starfish-shaped structure are inserted between each of the five triangular faces associated with the special vertex. This insertion causes the shape of the virion to significantly deviate from ideal icosahedral symmetry, resulting in virions that retain only one accurate fivefold symmetry axis passing through the special vertex. Thus, it was necessary to abandon icosahedral symmetry in the interpretation of cryo-EM micrographs in order to obtain a 65-Å resolution cryo-EM map [5]. This map, as well as atomic force microscopy (AFM) of defibered particles, showed surface depressions separated by 140 Å in a hexagonal array [5]. There are two additional electron-dense layers, presumably membranes, immediately below the outer capsid layer, a feature that can also be seen in some other members of the NCLDVs such as African Swine Fever Virus [7]. Within these two layers there is a sac surrounding the genomic material. This sac is separated by a roughly constant distance of about 300 Å from the outer capsid, except in the vicinity of the special vertex where the sac has a large depression. The depression creates a cavity between the outer capsid and the sac underneath the unique pentameric vertex, which might be filled with enzymes necessary for infection [4] (fig. 2).
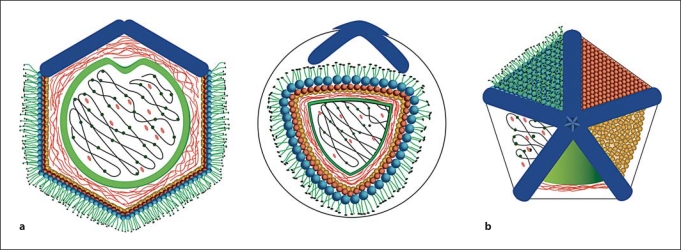
Fig. 1.
Schematic drawing of a model for Mimivirus, based principally on AFM images. a View perpendicular to the unique fivefold axis of the particle. b The stargate assembly. In a the model is presented in cross-section, and in b clockwise from exterior to interior as successive layers of structure are revealed. Most exterior (black) are the head proteins of the surface fibers that are attached to the surface fibers themselves (green). The surface fibers are attached to the anchor protein layer (blue spheres) that covers the icosahedral capsid (red spheres). Beneath the capsid is an additional layer of apparently disorganized protein (or possibly a lipid membrane studded with a variety of proteins; shown as orange spheres). The space between the inner surface of the capsid and the lipid bilayer sac (green) are the 7-nm fibers of indeterminate length (orange). Inside the membrane is the genomic DNA (black) with associated proteins (green). There are many other proteins within the membrane sac that are not directly bound to the nucleic acid (shown as pink). Reproduced from [23].
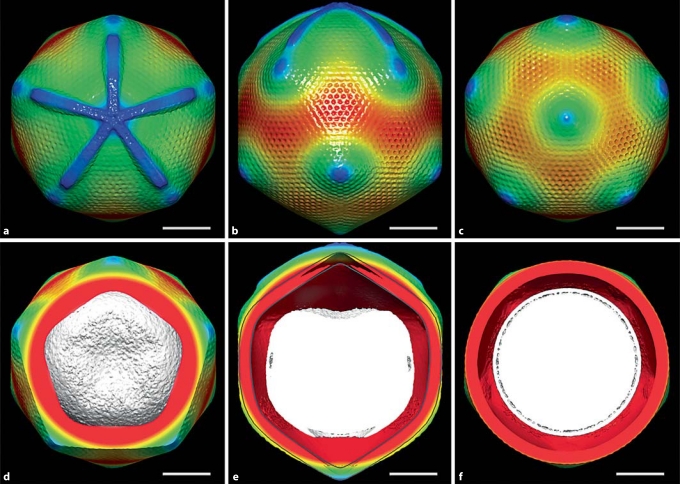
Fig. 2.
CryoEM reconstruction of Mimivirus applying only fivefold symmetry averaging. a–c Surface-shaded rendering of cryo-EM reconstruction of Mimivirus. a View onto the starfish-shaped feature associated vertex. b, c Views from opposite sides of the starfish-associated vertex. d The starfish-associated vertex removed to show the internal nucleocapsid with its concave surface facing the special vertex. e Central slice of the reconstruction viewed from the side of the particle showing the concave face of the nucleocapsid and the low-density space beneath the starfish-associated vertex. A perfectly icosahedral particle is outlined in gray to show the extension of the unique vertex. f Central slice of the reconstruction viewed along the fivefold axis from the starfish-shaped feature showing the enveloped nucleocapsid surrounded by a lower density space. The coloring is based on radial distance from the center of the virus. Gray = 0–1,800 Å; red = 1,800–2,100 Å; rainbow coloring from red to blue = 2,100–2,500 Å. Scale bars = 1,000 Å. Reproduced from [5].
Capsomer Surface Lattice
There are only a few structural motifs used in the assembly of viral capsids. One of the most common of these motifs is the ‘jelly-roll’ fold found in both RNA and DNA viruses [8,9]. It consists of two opposing β-sheets, each consisting of four anti-parallel β-strands creating a wedge-shaped structure. When the strands are sequentially named A through I along the polypeptide chain, then the order of β-strands in each sheet will be given by BIDG and CHEF. The capsomers in many viruses consist of six jelly rolls with their wedge-shaped ends pointing towards their sixfold axis. In many viruses, as in the RNA picornaviruses [9], the capsomers consist of two different proteins, each folded into a jelly-roll which occur alternatively in the capsomer. In yet other viruses, the two jelly-roll folds are joined into a single polypeptide by a short linker as in some NCLDVs [10,11] including adenovirus [12]. The double jelly-roll motif also occurs in some bacteriophages, such as PRD1 [13] or PM2 [14]. Presumably, at some point in the evolution of these viruses the jelly-roll motif had been duplicated to have two consecutive jelly-roll folds within the same polypeptide. In NCLDVs the capsomers are arranged into a close-packed hexagonal array with local p3 plane group symmetry on each icosahedral face (fig. 3). In addition to these hexameric capsomers, most NCLDVs have pentameric capsomers at each of the 12 vertices consisting of five monomeric jelly-rolls.
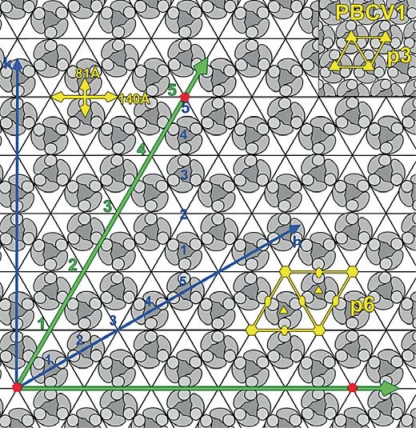
Fig. 3.
Capsomer arrangement of Mimivirus. Diagram showing the p6 plane group lattice of capsomers. Each monomer of the major capsid protein consists of two consecutive jelly-roll folds indicated by a kidney-shaped outline with the circle at one end representing the inserted ‘tower’ loop. Three monomers constitute one capsomer. The depressions along one edge are shown in green. Three fivefold vertices at the corners of a face are shown with red pentagons. As an example the fivefold vertices are placed five depressions apart. This would correspond to h = 5 and k = 5 in terms of the Caspar-Klug p3 plane group hexagonal array of capsomers (see blue axes). For Mimivirus, there are 19 depressions along an edge. One unit cell of the p6 (Mimivirus) and p3 (PBCV1) are outlined in yellow. The distance between depressions (140 Å) and between capsomers (81 Å) is shown in yellow. Reproduced from [5].
The major capsid protein of Mimivirus is homologous with that of PBCV-1 [5] which has a double jelly-roll fold [10], suggesting that Mimivirus, like other NCLDVs, has hexameric capsomers consisting of double jelly-rolls. Both AFM and cryo-EM reconstructions showed that Mimivirus has a hexagonal array of depressions separated by 140 Å on its surface [5]. Each depression is surrounded by six capsomers arranged with p6 rather than p3 symmetry as in other NCLDVs. The trimeric capsomers have alternate orientations differing by 60° surrounding each of the depressions (fig. 3). A similar p6 plane group organization had been observed by cryo-EM for infectious bursal disease virus [15], but the crystal structure of the virus showed that the cause for this is an unusual domain organization in the capsid protein of the virus [16]. Like other NCLDVs, the atomic structure of the infectious bursal disease virus major capsid protein [17] has a double jelly-roll structure.
The T Number of Mimivirus
Crick and Watson [18] suggested, as was later verified, that many viruses might have icosahedral symmetry in order to maintain the same environment for each of the 60 capsid proteins. Caspar and Klug [19] extended this concept to include quasi-equivalence for more than 60 subunits in the capsid. They suggested that icosahedral capsids of various sizes could be built by placing pentameric capsomers at certain positions in arrays of hexagonal capsomers, thereby introducing curvature. The triangulation number (T) is dependent on the coordinates (h, k) that define the relationship from one pentamer to the closest neighboring pentamer in the hexagonal array and is given by T = h2 + hk + k2[19]. For those viruses using a double jelly-roll fold and a p3 lattice as their building block for capsid assembly, the T number equals the number of jelly-roll folds in one icosahedral asymmetric unit.
The hexagonal lattice on the surface of Mimivirus is not sufficiently resolved at the current resolution of 65 Å to identify the h, k vectors and determine the T number with complete certainty. Therefore, the T number of the virus can only be estimated to be one of nine possibilities in the range of 972 ≤ T ≤ 1,200 (h = 19 ± 1 and k = 19 ± 1). As Mimivirus has a p6 plane group organization, it is missing a capsomer at each depression which implies that the number of jelly-rolls will be only 2T/3 [5].
The Starfish and Its Function
The starfish structure of Mimivirus is associated with one icosahedral fivefold vertex on each virus. The starfish arms have a width of about 500 Å, a thickness of about 400 Å and a length of about 2,000 Å, almost reaching the neighboring fivefold vertex. The arms are inserted between two neighboring triangular surfaces causing a deviation of about five degrees from the geometry of an ideal icosahedral particle for these five faces. Because there are no hexagonal arrays of depressions on the arms of the starfish it is reasonable to assume that the arms are assembled from protein(s) different from the major capsid protein [5].
Several features associated with the starfish-shaped structure suggest that it plays a role in the genome delivery of Mimivirus. EM studies of particles infecting amoeba cells showed the internal contents of the virus moving into the cell through an opening between the faces that were previously held together by the arms of the starfish [5,6,20]. Thus, the starfish is referred to as a ‘stargate’ by Zauberman et al. [6]. Furthermore, as mentioned above, the space between the starfish-shaped feature and the genomic sac might be filled with enzymes required to digest structural components of the host during the initial stages of infection, which would be comparable to the action of bacteriophages.
Surface Fibers
The dense layer of long fibers that cover the Mimivirus surface was one of the reasons why the virus was initially identified as a bacterium [2]. The surface fibers are resistant to proteases unless first treated with lysozyme, suggesting the fibers are protected by peptidoglycans. This hypothesis is supported by AFM images that showed murky material surrounding the fibers which might be peptidoglycans cross-linking neighboring fibers [5]. Additionally, the existence of peptidoglycans could account for the fact that Mimivirus can be stained Gram-positively. A possible biological role for the heavily glycosylated fibers could be that they act as a decoy for attracting amoeba [21]. Because the genome of Mimivirus contains several collagen-related genes [3], it was suggested that collagen-like proteins could build the fibers. However, even after pretreatment of virus particles with lysozyme, the fibers are resistant to collagenases, suggesting the surface fibers are not collagen-related.
AFM studies of fibers showed that most of the fibers released by protease treatment were not shortened but, instead, are cleaved at their base. In addition, groups of three or four fibers were found to be attached to a single disc-shaped feature [5], whereas the distal ends of the fibers were associated with a globular density of smaller size than the disc.
Analysis of the assembly pathway of Mimivirus in amoeba showed that fibers are attached to the capsomer late during assembly [20]. Because the depressions on the surface of the virus can be seen in the cryo-EM reconstruction of fibered particles [5], it is unlikely that the fibers are attached to the depressions in the surface hexagonal lattice.
Nucleocapsid and Internal Fibers
The 300-Å gap between the genomic sac and the capsid as well as the consistent position of the sac with respect to the special vertex, as seen in the cryo-EM reconstruction (fig. 2), shows that the nucleocapsid sac is held within the virus in a rigid and consistent manner. Therefore, some kind of support must be present in the gap to hold the nucleocapsid in place. Long internal fibers were observed by AFM after applying mechanical force to break the outer capsid layers of Mimivirus. These internal fibers have a diameter of about 60 Å with repeating units at intervals of about 70 Å [5]. The nucleocapsid might be supported by these fibers but, at this time, there is no further evidence for this suggestion.
In summary, the size and complexity of Mimivirus is greater than what is usually anticipated for a virus, but might be expected for a small cell [22]. Although structural investigations have been started with AFM and cryo-EM, these techniques clearly have limitations. AFM has possibly reached its resolution limits and cryo-EM is limited by the sample heterogeneity and a lack of accurate icosahedral symmetry. Alternative techniques, such as cryo-tomography of the whole virus or of thin sections, might be helpful. Crystallographic determinations of individual structural proteins and their assembled complexes, together with other techniques, could provide further information about the structure of Mimivirus and its interaction with amoeba.
Acknowledgements
We are grateful to Didier Raoult and Avi Minsky for helpful discussions. This work was supported by NIH grant AI11219 to M.G.R.
References
- 1.Raoult D, La Scola B, Birtles R. The discovery and characterization of Mimivirus, the largest known virus and putative pneumonia agent. Clini Infect Dis. 2007;45:95–102. doi: 10.1086/518608. [DOI] [PubMed] [Google Scholar]
- 2.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. A giant virus in amoebae. Science. 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 3.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. The 1.2-megabase genome sequence of mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 4.Xiao C, Chipman PR, Battisti AJ, Bowman VD, Renesto P, Raoult D, Rossmann MG. Cryo-electron microscopy of the giant Mimivirus. J Mol Biol. 2005;353:493–496. doi: 10.1016/j.jmb.2005.08.060. [DOI] [PubMed] [Google Scholar]
- 5.Xiao C, Kuznetsov YG, Sun S, Hafenstein SL, Kostyuchenko VA, Chipman PR, Suzan-Monti M, Raoult D, McPherson A, Rossmann MG. Structural studies of the giant Mimivirus. PLoS Biol. 2009;7:e92. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zauberman N, Mutsafi Y, Halevy DB, Shimoni E, Klein E, Xiao C, Sun S, Minsky A. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga Mimivirus. PLoS Biol. 2008;6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouiller I, Brookes SM, Hyatt AD, Windsor M, Wileman T. African swine fever virus is wrapped by the endoplasmic reticulum. J Virol. 1998;72:2373–2387. doi: 10.1128/jvi.72.3.2373-2387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Rossmann MG, Johnson JE. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 10.Nandhagopal N, Simpson AA, Gurnon JR, Yan X, Baker TS, Graves MV, Van Etten JL, Rossmann MG. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson AA, Nandhagopal N, Van Etten JL, Rossmann MG. Structural analyses of Phycodnaviridae and Iridoviridae. Acta Crystallogr D Biol Crystallogr. 2003;59:2053–2059. doi: 10.1107/s090744490302225x. [DOI] [PubMed] [Google Scholar]
- 12.Rux J, Kuser P, Burnett R. Structural and phylogenetic analysis of Adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J Virol. 2003;77:9553–9566. doi: 10.1128/JVI.77.17.9553-9566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benson SD, Bamford JK, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 14.Huiskonen JT, Kivelä HM, Bamford DH, Butcher SJ. The PM2 virion has a novel organization with an internal membrane and pentameric receptor binding spikes. Nat Struct Mol Biol. 2004;11:850–856. doi: 10.1038/nsmb807. [DOI] [PubMed] [Google Scholar]
- 15.Bottcher B, Kiselev NA, Stel'Mashchuk VY, Perevozchikova NA, Borisov AV, Crowther RA. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J Virol. 1997;71:325–330. doi: 10.1128/jvi.71.1.325-330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coulibaly F, Chevalier C, Gutsche I, Pous J, Navaza J, Bressanelli S, Delmas B, Rey F. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell. 2005;120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Luque D, Saugar I, Rodríguez JF, Verdaguer N, Garriga D, Martín CS, Velázquez-Muriel JA, Trus BL, Carrascosa JL, Castón JR. Infectious bursal disease virus capsid assembly and maturation by structural rearrangements of a transient molecular switch. J Virol. 2007;81:6869–6878. doi: 10.1128/JVI.00077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crick FH, Watson JD. Structure of small viruses. Nature. 1956;177:473–475. doi: 10.1038/177473a0. [DOI] [PubMed] [Google Scholar]
- 19.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 20.Suzan-Monti M, La Scola B, Barrassi L, Espinosa L, Raoult D. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS One. 2007;2:e328. doi: 10.1371/journal.pone.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claverie J, Ogata H, Audic S, Abergel C, Suhre K, Fournier P. Mimivirus and the emerging concept of ‘giant’ virus. Virus Res. 2006;117:133–144. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Raoult D, Forterre P. Redefining viruses: Lessons from Mimivirus. Nat Rev Microbiol. 2008;6:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- 23.Kuznetsov YG, Xiao C, Sun S, Raoult D, Rossmann MG, McPherson A: Atomic force microscopy investigation of the giant Mimivirus. Virology; in press. [DOI] [PubMed]