Abstract
Dynamic histone lysine methylation involves the activities of modifying enzymes (writers), enzymes removing modifications (erasers) and readers of the histone code. One common feature of these activities is the recognition of lysines in methylated and unmethylated states, whether they are substrates, reaction products or binding partners. We applied the concept of adding a lysine mimic to an established inhibitor (BIX-01294) of histone H3 lysine 9 methyltransferases G9a and G9a-like protein (GLP) by including a 5-aminopentyloxy moiety, which is inserted into the target lysine-binding channel and becomes methylated by GLP, albeit slowly. The compound enhanced its potency in vitro and reduced cell toxicity in vivo. We suggest that adding a lysine or methyllysine mimic should be considered in the design of small molecule inhibitors for other methyl-lysine writers, erasers and readers.
Keywords: Epigenetics, Histone lysine methylation, enzymatic inhibition, lysine mimics
Chromatin, rather than being a passive platform to store genetic information, can regulate transcriptional processes based on modifications of both DNA and histones. Histones are subject to an array of posttranslational modifications, including methylation of lysines. These marks are generated by a host of histone methyltransferases, removed by histone demethylases and recognized by reader domains in the methylated and unmodified states. Importantly, such enzymes are novel targets for therapeutics 1; 2.
BIX-01294 (a diazepin-quinazolin-amine derivative) inhibits activities of G9a and G9a-like protein (GLP) lysine methyltransferase (IC50 in low μM range) 3; 4; 5 and reduces the methylation levels of histone H3 lysine 9 (H3K9) at several G9a target genes 3; 6; 7. BIX-01294 (BIX) consists of a central quinazoline ring linked to a seven-membered diazepane ring and a benzylated six-membered piperidine (Fig. 1a). Structural comparison of GLP of BIX-bound with that of substrate peptide-bound 8 suggested that BIX resembles the bound conformation of histone H3 Lys4 to Arg8, residues N-terminal to the target lysine, but leaves the target lysine binding channel unoccupied 4. Herein we report the use of the co-crystal structure of GLP-BIX complex 4 and molecular modeling to guide the design, synthesis, and validation of new BIX derivatives with moieties mimicking lysine and methyllysine.
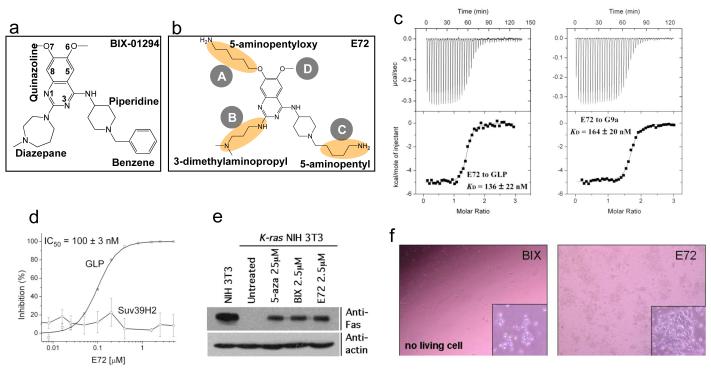
Figure 1.
Chemical structures for (a) BIX-01294, (b) E72, highlighted with changes. (c) The binding (KD values) of E72 compound to GLP (left panel) and G9a (right panel) are measured by isothermal titration calorimetry with a VP-ITC instrument (MicroCal) at 25°C. The titrations were conducted in buffer containing 20 mM Tris pH 8.0, 150 mM NaCl, 1.3-1.45% DMSO and 50 μM AdoMet. The sample chamber and syringe were filled with 20-30 μM GLP (or 21 μM G9a) and 260-350 μM (or 286 μM) compound, respectively. The data were processed using Origin 7.0 software. (d) The inhibition (IC50 values) of E72 against GLP or Suv39H2 is plotted against various concentrations of compound, under the conditions of 0.075 μM GLP or Suv39H2, 10 μM H3 peptide (residues 1-15), 100 μM AdoMet in the buffer of 20 mM Tris pH 8.5, 5 mM dithiothreitol (DTT), and 2% DMSO. The reaction mixture was incubated for 5 min (GLP) or 15 min (Suv39H2) at 30 °C, and subjected to mass-spectrometry-based inhibition assay as described 4. (e) Ras-mediated epigenetic silencing of Fas is derepressed with E72, BIX and 5-aza treatments, as described 4. (f) Mouse embryonic stem (ES) cells (TT2) 18 were treated for 24 h with each compound at 10 μM concentration, BIX-01294 (left panel), E72 (right) (see also Supplementary Table 1 and Fig. 3b).
Adding lysine and methyllysine mimics
We replaced the O7-methoxy group with a 5-aminopentyloxy substituent at site A (Fig. 1b). The length of the aliphatic chain and the presence of a terminal amino group for the 5-aminopentyloxy moiety were expected to extend into the active site of GLP and mimic the side chain of a substrate lysine. In addition, we replaced the diazepane ring and the benzyl with a 3-dimethylaminopropyl and a 5-aminopentyl group at sites B and C, respectively. These modifications generated compound E72 (Fig. 1b). The 3-dimethylaminopropyl unit contains a three-carbon aliphatic chain and a di-methylated amino moiety that was expected to form a favorable electrostatic interaction with Asp1131 of GLP 4. The benzyl functionality is not well defined in the structure of GLP-BIX complex 4, because the branched benzene moiety has little direct contact with the enzyme. Taken together, these modifications resulted in the dissociation constant (KD) of approximately 136 nM (Fig. 1c, left panel) and half-maximal inhibitory concentration (IC50) of 100 nM (Fig. 1d) for E72 against GLP under linear reaction conditions by mass spectrometry-based inhibition assay 4. The cumulative effect of these changes in E72 is to decrease IC50 by a factor of approximately 7 in comparison to that of BIX measured by the same assay 4.
E72 retains the binding affinity towards G9a (Fig. 1c, right panel), selectivity over the related H3K9 methyltransferase Suv39H2 (Fig. 1d; Supplementary Fig. S1) and the ability to reactivate K-ras–mediated epigenetic silencing of the proapoptotic Fas gene in NIH 3T3 cell (Fig. 1e). More importantly, E72 has much reduced cell cytotoxicity (Fig. 1f). In all three cell types treated (Supplementary Table 1), very little toxicity was observed for E72 at 10 μM concentration, while BIX killed almost all cells at 10 μM and approximately 50% at 1 μM.
Structures of GLP bound with E72
We solved the ternary structure of GLP in complex with E72, in the presence of S-adenosyl-L-homocysteine (AdoHcy), at resolution of 2.19 Å (Supplementary Table 2). The E72 compound was co-crystallized with GLP-AdoHcy in the space group P21, at 16°C by mixing equal volumes of protein (at 18-20 mg/ml) with solution containing 2 mM compound in 0.1 M Hepes pH 7.5, 14% polyethylene glycol 4000, 9% isopropanol, and 12% dimethyl sulfoxide (DMSO). There are four molecules per crystallographic asymmetric unit. The structures of the protein component are highly similar (with root-mean-square-deviation of ~0.2-0.6Å of 1020 pairs of main chain atoms between the monomers).
E72 is bound to the acidic surface of the histone H3 peptide-binding groove (Fig. 2a), surrounded by many acidic residues including four aspartates, Asp1131, Asp1135, Asp1140, and Asp1145. Two of them, Asp1140 and Asp1145, maintain the same sets of hydrogen bonding interactions described previously for BIX 4 with the linker NH group between the quinazoline and piperidine rings and the N1 ring nitrogen atom of the quinazoline, respectively (Fig. 2b). Additionally, Asp1131 forms a salt bridge with the dimethylamino group as expected (Fig. 2a-b). The three-carbon aliphatic chain may be optimal for the formation of this charge-charge interaction, as reducing or increasing the chain length by one carbon resulted in less inhibition under a single inhibitor concentration tested (Supplementary Fig. S2).
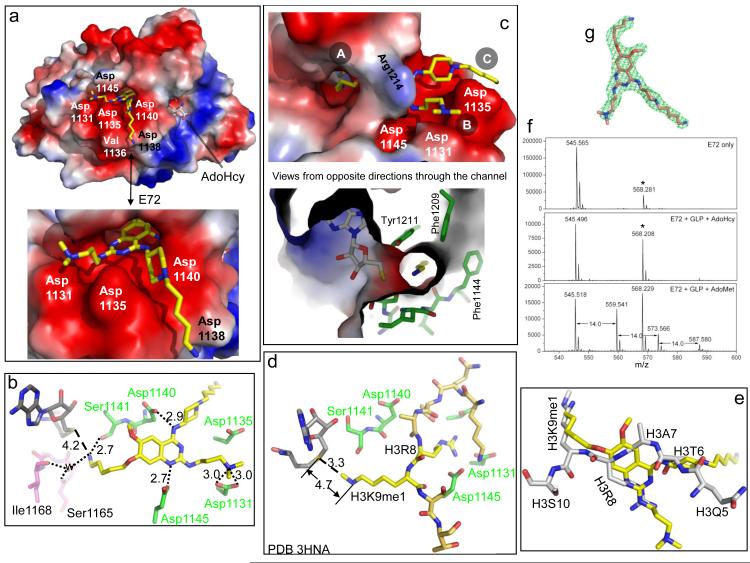
Figure 2. Structure of GLP-E72-AdoHcy complex.
(a) Surface representation of the GLP catalytic domain with AdoHcy and E72 bind in two distinctive pockets. The surface charge at neutral pH is displayed as red for negative, blue for positive and white for neutral. (b) Network of hydrogen bonds centered on E72. (c) The 5-aminopentyloxy moiety occupies the target lysine-binding channel, views from opposite directions. (d) The structure of GLP-AdoHcy-histone H3 peptide containing H3K9me1 (PDB 3HNA) 8. (e) Superimposition of histone H3 peptide (taken from PDB 3HNA) and E72 (yellow). (f) E72 is methylated by GLP under the reaction conditions of 1.5 μM GLP, 1.6 μM E72, 100 μM AdoMet in the buffer of 20 mM Tris pH 8.5, 5 mM DTT and 2% DMSO. The mixture was incubated for 3 h at 30 °C and then overnight (~16 h) at room temperature (~21 °C). The mass peak at 568 Da (indicated by an asterisk) is from the matrix used in the mass spectra measurement (Supplementary Fig. S3b). (g) Omit electron density, Fo-Fc (green mesh), contoured at 4σ above the mean, is shown for E72.
In E72, the O7 5-aminopentyloxy moiety at site A is extended into the target lysine binding channel (Fig. 2c). Aromatic residues, Phe1144, Phe1209, and Tyr1211, wrap around the aliphatic chain (Fig. 2c), while the terminal amino group forms a hydrogen bond with the main-chain carbonyl oxygen of Ser1141 (Fig. 2b). The terminal amino group is only approximately 4.2Å from the AdoHcy sulfur atom (Fig. 2b), where the transferable methyl group would be attached on S-adenosyl-L-methionine (AdoMet). In comparison, in the ternary structure of GLP in complex with AdoHcy and histone H3 peptide containing monometylated lysine 9 (H3K9me1) 8, the distances between the AdoHcy sulfur atom and the methyl group or the target nitrogen are approximately 3.3 Å (S…CH3) or 4.7 Å (S…N) (Fig. 2d), respectively. Superimposition of the E72 complex and the H3K9me1 peptide complex structures reveals that the position of the 5-aminopentyloxy moiety coincides with that of lysine 9 of histone H3 (Fig. 2e). This observation suggested that E72 might potentially be a substrate for methylation by GLP. Indeed, after long incubations (overnight) in nearly equimolar ratio of enzyme to compound, E72 is mono, or even di, and tri-methylated by GLP (Fig. 2f).
The 5-aminopentyl moiety attached to the piperidine ring at site C sits in an acidic surface groove that is formed by Asp1135 and Val1136 on one side, Asp1140 and Asp1138 on the other and the main-chain atoms of Arg1137 on the floor (Fig. 2a). The terminal amino group makes no specific protein interactions as it points out into the solvent. In the structures of GLP in complex with BIX 4 as well as E67 (the compound retains the benzyl moiety on the piperidine; Fig. 3a), the density for the benzyl moiety is not well defined (Supplementary Fig. S3a). This result suggests that, with respect to the 5-aminopentyl moiety at site C, the enhanced potency of E72 by approximately a factor of 3 (comparing the IC50 value to that of E67; Fig. 3a) is likely due to van der Waals contacts between the groove and the aliphatic chain of this moiety.
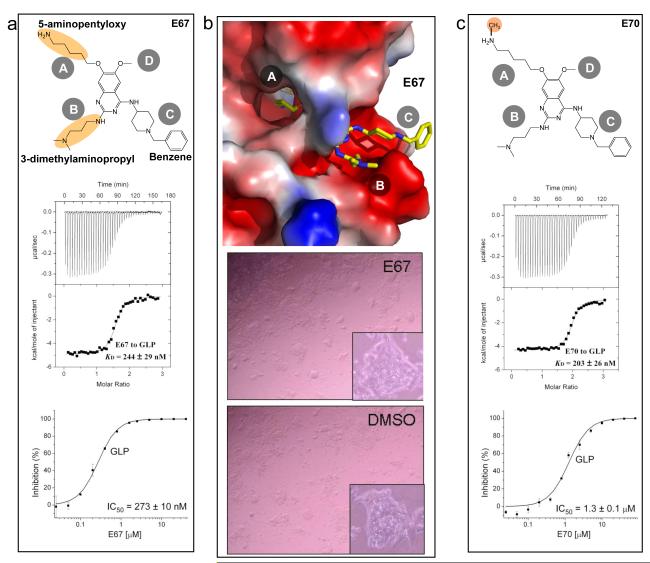
Figure 3.
Comparison of compound E67 and E70 (mono-methylated form of E67). (a) Top panel shows the chemical structure of E67, the middle and bottom panels show the measurements of KD and IC50 against GLP. (b) Top panel shows the structure of GLP-AdoHcy-E67 complex (Supplementary Table 2). The 5-aminopentyloxy moiety occupies the target lysine-binding channel. Bottom panels show the reduced cell toxicity of E67. Mouse embryonic stem (ES) cells (TT2) were treated for 24 h with E67 compound at 10 μM concentration. The experiments were performed together with BIX-01294 and E72 (Fig. 1f), and control DMSO (see Supplementary Table 1). (c) Top panel shows the chemical structure of E70, the middle and bottom panels show the measurements of KD and IC50 against GLP.
Compound E67 and its mono-methylated form (E70)
The 5-aminopentyloxy moiety at site A that is inserted into the target lysine binding channel in both E72 (Fig. 2) and E67 (Fig. 3b) appears to be optimal for inhibition because compounds containing one fewer methylene unit are weaker inhibitors of the enzyme (4C compounds in Supplementary Fig. S4). Interestingly, the compounds containing one additional methylene (6C compounds) showed comparable inhibition to that of compounds with 5 methylene units (5C in Supplementary Fig. S4). This is probably because the length of the 6C aliphatic chain mimics the methylated 5-aminopentyl group. The mono-methylated E67 (compound E70) has KD value similar to that of unmethylated form, but with reduced inhibitory effect by a factor of approximately 4 (Fig. 3c). Perhaps E70-GLP complex represents a methylated product resulted in a faster off rate of the ligand. Recently, Liu et al. synthesized a BIX analog (UNC0224)with a 3-(dimethylamino)propoxy replacing the O7-methoxy group at site A 5. While the central quinazoline ring overlaps well with that of E72 and E67, the 3-(dimethylamino)propoxy side chain is two-carbons too short to mimic a di-methylated lysine side chain in the active sites of G9a and GLP (Supplementary Fig. S5 and Supplementary Table 3).
DISCUSSION
By including a moiety to mimic the lysine side chain at site A, we discovered that such modification can result in more effective inhibition of GLP (a H3K9 methyl writer) via a slow methylation reaction. The observations provide avenues for designing small molecule inhibitors for other methyl-lysine writers, erasers and readers by including a lysine or methyllysine mimic. For example, the 3-dimethylaminopropyl unit at site B containing a di-methylated amino group could be targeted by a di-methyl-lysine specific Jumnoji demethyase. The 5-aminopentyl group at site C might be targeted by a different SET domain protein. The modification of the O6 methoxy at site D, which interferes with the G9a/GLP-ligand interaction (data not shown), might provide an anchor for binding with other SET- or Jumonji-domain containing proteins. Once the side chain mimic at a particular branch is identified for a particular target, lysine or methyllysine mimics at other sites can conceivably be eliminated or replaced to improve the selectivity, and the length of the aliphatic chain can be optimized to improve potency. In conclusion, iterative cycles of crystallography, synthesis and bioassay will aid successful design of epigenetic inhibitors of histone lysine methyltransferases as well as provide knowledge for future therapeutics that might be directly applicable to patients who are receiving epigenetic-based therapies (5-aza-2′-deoxycytidine and HDAC inhibitors) 9.
Interestingly, the KD and IC50 values for compounds E67 and E72 are approximately the same, whereas the KD values for compounds BIX and E11 (Supplementary Fig. S2) is much lower than that of the corresponding IC50 value (Supplementary Table 3). It was somewhat puzzling that E72 and E67 compounds have lower IC50 than BIX and E11 yet have similar or even higher KD for GLP. This may be explained by the different modes of binding by these two groups of inhibitors. BIX and E11 only occupies part of the substrate peptide groove of GLP while leaving the target lysine channel open, so that it could be competed away by the substrate peptides relatively easily (indicated by the higher IC50 value than KD). E72 and E67, however, not only occupy the surface of the peptide binding groove, but intercalates into the lysine binding channel so that each binding event leads to effective inhibition, i.e., the KD and IC50 values are in agreement.
Molecular Modeling
Many of the compounds prepared as background to this study were modeled by Glide docking of candidate structures into the GLP-BIX complex 4 followed by MM-GBSA rescoring. Since this procedure essentially provides superposition of BIX-01294 with the crystal structure, it was used throughout in the search for improved BIX analogs. We illustrate the prediction for compound E72 in Supplementary Fig. S6. The GLP-BIX-AdoHcy complex X-ray structure (PDB 3FPD) was relieved of the BIX ligand in Maestro 8.5.111 (Schrodinger; http://www.schrodinger.com/) and subjected to precision flexible Glide docking 10; 11 with each new ligand structure. Based on the Glide scoring function 20 poses were saved and then rescored with MM-GBSA 12; 13. The resulting pose with the best-calculated binding affinity was selected as the optimal docking solution.
Chemical synthesis
Compound 1 was prepared as described 14 and reduced with tin (II) chloride 15 (Supplementary Fig. S7). The intermediate anthranilic acid was subsequently combined with sodium cyanate, followed by POCl3, to provide 2 16; 17. Treatment of the latter with 4-amino-1-benzylpiperidine in the presence of triethylamine delivered 3, which was hydrogenated to produce a 3:1 mixture of 4 and 5. These building blocks were individually coupled with 5-bromopentenitrile in the presence of potassium carbonate to produce 6 and 8. Finally, these compounds were treated with N,N-dimethylaminopropylamine at high temperature to provide precursors which were reduced by lithium aluminum hydride to furnish 7 (E67) and 9 (E72). Liquid chromatography-mass spectrometry and NMR were used to analyze the homogeneity of the synthesized compounds (Supplementary Fig. S8).
Supplementary Material
Acknowledgements
We thank Paul R. Thompson and Corey Causey for critical comments, Anup K. Upadhyay for discussion and advice on use of mass spectrometry, Dennis Liotta for encouragement. The Biochemistry Department of Emory University School of Medicine supported the use of SER-CAT beamlines and mass spectrometry. This work was supported by grants R01GM068680 (to XC and XZ) and R56DK082678 (to XC and YS), and U54HG003918 from the National Institutes of Health and the Welch Foundation Grant G-1495 (to MTB). X.C. is a Georgia Research Alliance Eminent Scholar.
Abbreviations
- GLP
G9a-like protein
Footnotes
PDB accession numbers The coordinates and structure factor for human GLP catalytic domain bound with E72, E67, and E11, respectively, and AdoHcy have been deposited with accession numbers3MO5, 3MO2, and 3MO0.
Additional information Supplementary information is available (see attached)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cole PA. Chemical probes for histone-modifying enzymes. Nat Chem Biol. 2008;4:590–7. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8:724–32. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 3.Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–81. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, Liu J, Snyder JP, Bedford MT, Cheng X. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol. 2009;16:312–7. doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wasney GA, Dong A, Barsyte D, Kozieradzki I, Senisterra G, Chau I, Siarheyeva A, Kireev DB, Jadhav A, Herold JM, Frye SV, Arrowsmith CH, Brown PJ, Simeonov A, Vedadi M, Jin J. Discovery of a 2,4-Diamino-7-aminoalkoxyquinazoline as a Potent and Selective Inhibitor of Histone Lysine Methyltransferase G9a. J Med Chem. 2009 doi: 10.1021/jm901543m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, Reinberg D. Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins. J Biol Chem. 2009;284:8395–405. doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–6. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, Allali-Hassani A, Campagna-Slater V, Vedadi M, Arrowsmith CH, Plotnikov AN, Schapira M. Structural biology of human H3K9 methyltransferases. PLoS One. 2010;5:e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karberg S. Switching on epigenetic therapy. Cell. 2009;139:1029–31. doi: 10.1016/j.cell.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 10.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 11.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–9. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 12.Guimaraes CR, Cardozo M. MM-GB/SA rescoring of docking poses in structure-based lead optimization. J Chem Inf Model. 2008;48:958–70. doi: 10.1021/ci800004w. [DOI] [PubMed] [Google Scholar]
- 13.Lyne PD, Lamb ML, Saeh JC. Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J Med Chem. 2006;49:4805–8. doi: 10.1021/jm060522a. [DOI] [PubMed] [Google Scholar]
- 14.Thurston DE, Murty VS, Langley DR, Jones GB. O-Debenzylation of a Pyrrolo[2,1-c][1,4]benzodiazepine in the Presence of a Carbinolamine Functionality: Synthesis of DC-81. Synthesis. 1990;1:81–84. [Google Scholar]
- 15.Hu WP, Wang JJ, Lin FL, Lin YC, Lin SR, Hsu MH. An efficient synthesis of pyrrolo[2,1-c][1,4]benzodiazepine. Synthesis of the antibiotic DC-81. J Org Chem. 2001;66:2881–3. doi: 10.1021/jo010043d. [DOI] [PubMed] [Google Scholar]
- 16.Andrus MB, Mettath SN, Song C. A modified synthesis of iodoazidoaryl prazosin. J Org Chem. 2002;67:8284–6. doi: 10.1021/jo026217o. [DOI] [PubMed] [Google Scholar]
- 17.Smits RA, de Esch IJ, Zuiderveld OP, Broeker J, Sansuk K, Guaita E, Coruzzi G, Adami M, Haaksma E, Leurs R. Discovery of quinazolines as histamine H4 receptor inverse agonists using a scaffold hopping approach. J Med Chem. 2008;51:7855–65. doi: 10.1021/jm800876b. [DOI] [PubMed] [Google Scholar]
- 18.Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda N, Ikawa Y, Aizawa S. A novel ES cell line, TT2, with high germline-differentiating potency. Anal Biochem. 1993;214:70–6. doi: 10.1006/abio.1993.1458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.