Abstract
In the nematode Caenorhabditis elegans, the let-7 microRNA (miRNA) controls the timing of key developmental events and terminal differentiation in part by directly regulating lin-41. C. elegans lin-41 mutants display precocious cell cycle exit and terminal differentiation of epidermal skin cells. lin-41 orthologues are found in more complex organisms including both mice and humans, but their roles are not known. We generated Mlin41 mouse mutants to ascertain a functional role for Mlin41. Strong loss of function Mlin41 gene-trap mutants demonstrated a striking neural tube closure defect during development, and embryonic lethality. Like C. elegans lin-41, Mlin41 also appears to be regulated by the let-7 and mir-125 miRNAs. Since Mlin41 is required for neural tube closure and survival it points to human lin-41 (HLIN41/TRIM71) as a potential human development and disease gene.
Keywords: microRNA, let-7, mir-125, mlin41, lin-41, mouse, knock-out, development, cell cycle
Introduction
Tissue morphogenesis requires strict spatial and temporal cues during development. While the process of spatial patterning has been well characterized, only a small number of genes involved in temporal patterning, known as heterochronic genes, have been identified. Heterochronic genes have been shown to specifically regulate the timing of cell fate determination in C. elegans.1 For example, lin-41 controls the timing of seam cell differentiation during the C. elegans larval to adult transition.2 C. elegans lin-41 mutants display precocious expression of adult seam cell fates and are also dumpy, uncoordinated and sterile,2 showing that lin-41 is required for fertility, neural or muscle function, and seam cell development. In addition, LIN-41 is expressed in the affected tissues, i.e., seam cells, neurons, muscles and the somatic gonad,2 suggesting that it functions cell autonomously.
Recent work has shown that several members of the heterochronic pathway, including genes which encode microRNAs (miRNAs),3 are conserved by sequence in humans and mice.2,4–7 Furthermore, these genes are temporally expressed as determined by northern blot or in situ hybridization.2–10 While it is not known whether these heterochronic homologues function to regulate the timing of specific developmental events, their temporal expression patterns suggest a functional role for this pathway during early mouse development. 5,7,8,11 Specifically, we were interested in determining the biological role for lin-41 in the mouse.
Mouse (Mlin41/Trim71) and human (HLIN41/TRIM71) lin-41 homologues contain the same RBCC and C-terminal NHL domain as C. elegans lin-41.2,7,8 Not only are the coding sequences of Mlin41 and HLIN41/TRIM71 highly homologous,7,8 but they also possess a high degree of homology in their 3'UTRs.7 Since C. elegans lin-41 negatively regulated by the binding of let-7 miRNA to complementary sequences in the lin-41 3'UTR,12,13 we proposed that the mammalian homologues are also regulated by miRNAs in a 3'UTR dependent manner.7 Putative let-7 complementary sites (LCS) and miR-125 complementary sites (MCS) in both the mouse and human lin41 3'UTRs have been identified.7 Recent work has shown that HLIN41/TRIM71 and zebrafish lin41 are regulated by let-7.14 Here, we provide evidence that regulation of mammalian lin41 is also mediated by miR-125 binding to complementary sequences in the 3'UTR.
To determine the function of Mlin41 during mammalian development, we analyzed mouse lines generated from gene-trap insertions into the Mlin41 locus (BayGenomics). Loss of Mlin41 leads to embryonic lethality and neural tube closure defects. These studies show that Mlin41 is an essential gene during mammalian development and is required for proper anterior neural tube closure.
Results
Characterization of Mlin41 gene-trap lines
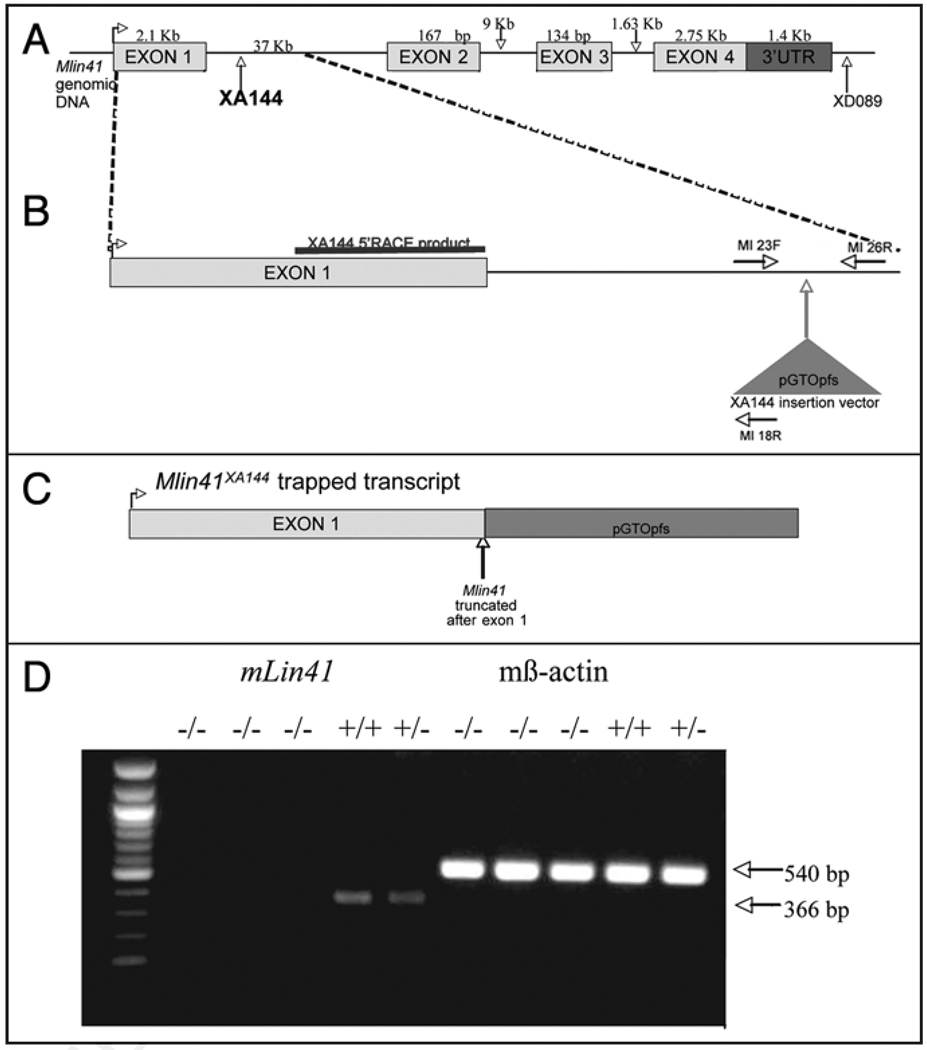
When the cDNA sequence of Mlin41,8 was mapped to the corresponding genomic sequence, we found that the Mlin41 locus consisted of 4 exons spread over 51 Kb of sequence (Fig. 1). Two gene-trap insertions in the Mlin41 locus were obtained from BayGenomics (BayGenomics, CA). We determined that the gene-trap insertion for Mlin41XA144 occurred in the first intron (37 Kb) of the Mlin41 gene (Fig. 1), located 865 bp after the end of exon 1 (see Methods). This insertion was predicted to truncate the Mlin41 protein after exon 1 leaving only 271 amino acids at the N-terminus (Fig. 1C) and removing the NHL domain and 3'UTR. The Mlin41XA144 truncation was predicted to lead to a severe loss of function and perhaps correspond to a null mutation. We generated homozygous Mlin41XA144 mice (see Methods) and designed PCR primers to genotype Mlin41XA144 animals (Fig. 1B, Suppl. Fig. 2). Consistent with being a severe Mlin41 allele, we detect substantially reduced levels of Mlin41 RNA by quantitative RT-PCR in the Mlin41XA144 embryos (Fig. 1D).
Figure 1.
Schematic of Mlin41XA144 gene-trap insertion in the Mlin41 genomic locus (not to scale). (A) The XA144 insertion in intron 1 (37 Kb). (B) Region of XA144 gene-trap insertion enlarged. The XA144 5'RACE product is highlighted over exon 1. Exon 2 is 167 bp not 1676 bp. The gene-trap insertion vector for this line is pGT0pfs. The genotyping primers are shown as MI 23F, MI 18R and MI 26R. (C) Resulting XA144 gene-trapped insertion transcript. (D) RT-PCR analysis of homozygous and heterozygous Mlin41XA144 animals. RT-PCR of B-Actin from the same animals is shown as a control.
A second gene-trap insertion in Mlin41, XD089 (Fig. 1), generated a mouse line known as Mlin41XD089, similarly to that described for Mlin41XA144. Based on inverse PCR results, this insertion occurred after the one known cleavage and poly-adenylation signal of the Mlin41 gene (Fig. 1 & Suppl. Fig. 1). However, based on RACE-PCR results (Suppl. Fig. 1), the transcript generated from the XD089 gene-trap insertion was predicted to generate a protein product twenty-four amino acids shorter than the predicted endogenous Mlin41 protein and the mRNA would not contain the native 3'UTR. We predicted that this mutation was likely to result in a less severe loss of function phenotype than the Mlin41XA144 mutation.
Mlin41 expression patterns in the gene-trap mutant line
In order to discern the expression pattern driven by the Mlin41 promoter in Mlin41XA144 and Mlin41XD089 mice, we took advantage of the presence of a lacZ reporter in the gene-trap vector, assaying embryos for beta-Galactosidase activity. As expected, all wild type embryos failed to stain blue, whereas both heterozygotes and homozygotes displayed lacZ expression. While the lacZ expression pattern in homozygotes and heterozygotes mutants was largely the same, homozygous embryos (Fig. 3A) stained more intensely than heterozygotes (Fig. 2), likely due to the presence of two copies of the gene-trap vector. The Mlin41 expression pattern analysis presented here is based on the lacZ pattern detected in heterozygous embryos, because homozygous animals possessed altered morphology, as described below.
Figure 3.
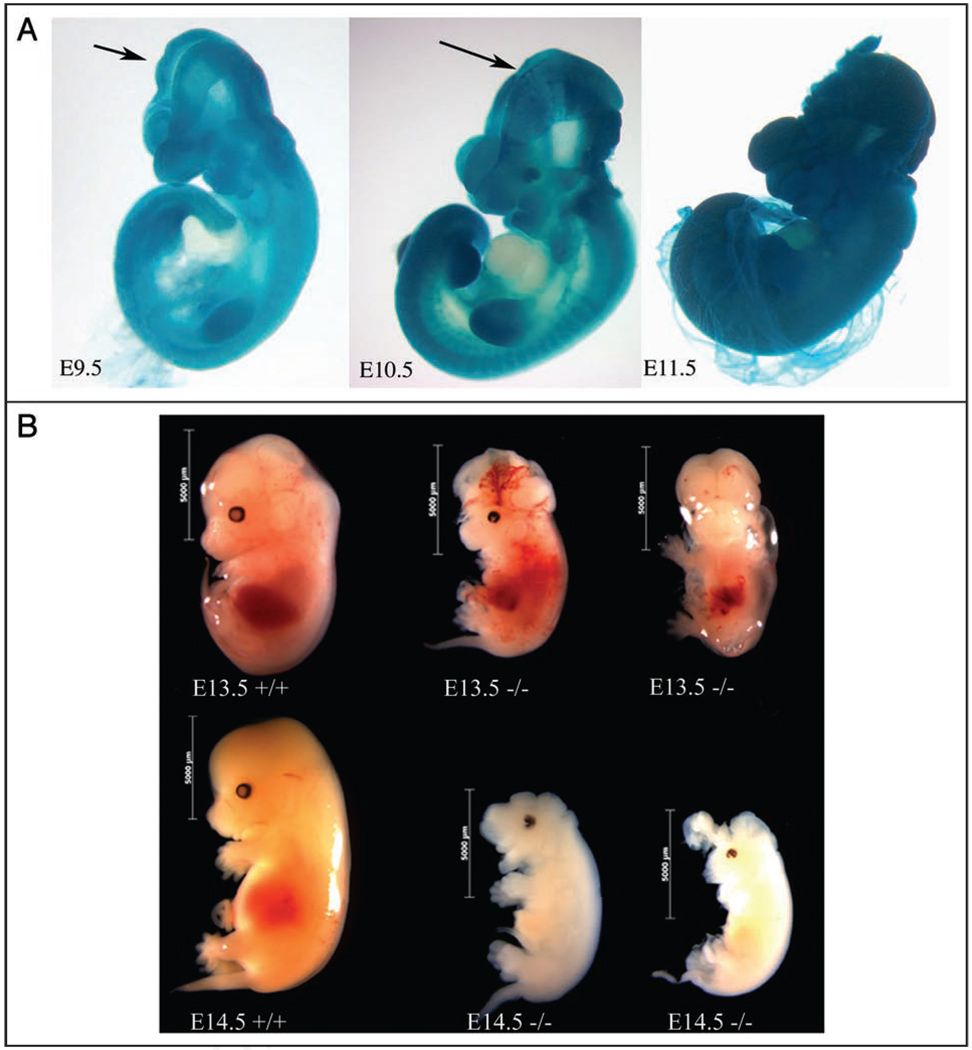
(A) Mlin41XA144 homozygous embryos. The arrows mark sites of neural tube closure defects. Embryos not to scale. (B) Mlin41XA144 homozygous embryos compared to heterozygous littermates to scale. First row is E13.5 staged embryos. Embryo 3rd from left is a ventral view. Second row shows E14.5 staged embryos.
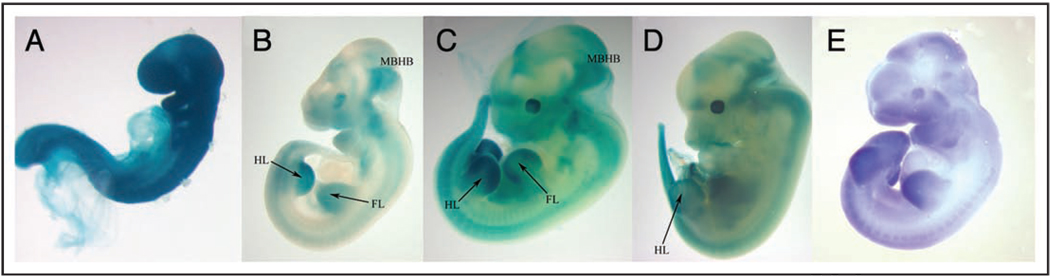
Figure 2.
The lacZ expression pattern from Mlin41XA144 heterozygotes. Nasal process (NP), branchial arches (BA), forelimb (FL), hindlimb (HL), midbrain/hindbrain border (MB/HB). Embryos not to scale. (A) E9.0. (B) E10.5. (C) E11.5. (D) E12.5. (E) Mlin41 expression pattern by in situ hybridization using probe pBRM1.7
In situ hybridization studies of Mlin41 revealed temporal expression in the neuroepithelium, branchial arches, facial prominence, tailbud, dorsal root ganglia and eyes along with dynamic expression patterns in both forelimb buds and hindlimb buds (Fig. 2E).7,8 Temporal lacZ expression was observed in Mlin41XA144 heterozygous embryos (Fig. 2). At E9.0, lacZ expression appeared intense and ubiquitous (Fig. 2A), reflecting the abundance of Mlin41 detected at this stage by northern blot and in situ hybridization previously reported.7 By E10.5, when total Mlin41 mRNA levels were found to decrease,7 a restricted lacZ expression pattern could be detected in these heterozygous embryos, with intense staining in the midbrain/hindbrain border and within the limb buds, as well as staining in the branchial arches, facial prominence, eye, tail bud and dorsal root ganglia (Fig. 2B). lacZ expression could still be detected in the brain, dorsal root ganglia, facial prominence, eyes and limbs at E11.5 and E12.5. In the forelimb bud at E11.5, expression was seen more distally than at earlier stages while expression in the hindlimb bud was found broadly but with intense expression in the distal portion (Fig. 2C). By E12.5, expression in the forelimb bud could no longer be detected and the hindlimb bud expression was localized to the distal tip (Fig. 2D). The Mlin41XA144 lacZ expression pattern in the developing limbs was dynamic during embryonic development, similar to what is observed by in situ hybridization analysis.7,8 Similar expression domains were seen with a second Mlin41 gene trap line, Mlin41XD089 (Suppl. Fig. 5). Based on the dynamic expression of Mlin41, we predicted that Mlin41 would play important roles during development in these tissues.
Mlin41XA144 mutants exhibit embryonic lethality
Heterozygous Mlin41XA144 mice were mated and resulting embryos were collected at developmental stages E8.5 through E16.5 for analysis. Genotypic analysis revealed that when embryos were sacrificed at E8.5 or earlier, Mendelian ratios were observed (n = 44 total embryos). Following E8.5, embryo genotypes did not segregate with the expected Mendelian ratios. There was a marked decrease in homozygous mutant embryos, implying an embryonic lethality occurring some time after E8.5. Specifically, only 15.6% homozygous mutant embryos were recovered between E9.0 and E9.5 (n = 135 total embryos). A Chi-square analysis to test for Mendelian ratios among collected homozygous mutant, heterozygous and wild type embryos, demonstrated that the percentages of homozygous mutant embryos were significantly less than the expected 25% (p value ≤ 0.05). The percentage of homozygous mutant embryos dramatically decreased after that, reaching 0% by E16.5 (n = 145 total embryos aged E14.5–E16.5). No visible early limb defects were detected in the few −/− animals that lived long enough to make limb buds (Fig. 3B), but the few surviving E13.5 day embryos exhibited abnormal blood pooling (Fig. 3B). Two rare E14.5 embryos looked abnormal and were both smaller and white in color when compared to wild type, presumably undergoing the process of resorption (Fig. 3B).
Mlin41XA144 mutants displayed abnormal neural tube closure
Homozygous Mlin41XA144 mutant embryos demonstrated anterior neural tube closure defects resulting in exencephaly (100%, n = 76). Open neural folds were everted, enlarged and failed to fuse15 and brain tissue was exposed (Fig. 3). However, in the posterior spinal region of the embryos neural tube closure was properly completed. Embryos collected at E14.5 had exposed brain tissue and appeared smaller than wild type littermates (Fig. 3B). Similar exencephaly defects were seen in the Mlin41XD089 allele (Suppl. Fig. 6, described below).
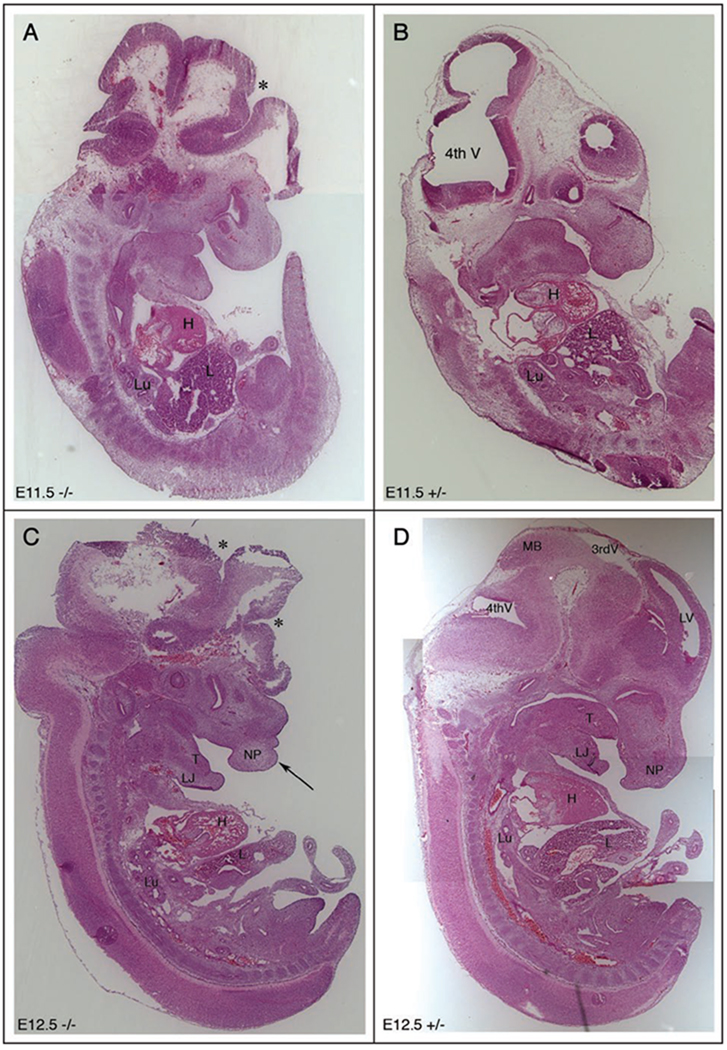
To evaluate more closely the open neural tube defect and other potential abnormalities to anatomical structures, serial sections at several stages of wild type and Mlin41XA144 heterozygote embryogenesis were collected and compared (Fig. 4). In −/− embryos, the open neural tube was clearly seen (Fig. 4 and Suppl. Fig. 3) and brain structures appeared to be altered as a result. The ventricles of the brain and other structures did not appear to be in the same locations as those of +/− embryos. At E11.5, the open neural tube (Fig. 4A) and exposed brain tissue were also clearly detected and the placement of brain ventricles was not in the correct locations as compared to the +/− littermate (Fig. 4A and B). Other anatomical structures in the trunk region, including the heart, liver, lungs and other organs, appeared similar in size, structure and placement between homozygous mutant and heterozygous littermates at E11.5. By E12.5, the open neural tube (Fig. 4C) and exposed brain tissue were still detectable and brain ventricles were also difficult to detect when compared to the heterozygous littermate (Fig. 4C and D). We analyzed other structures in the facial and trunk regions and most facial structures were detectable and appeared normal, including the tongue. However, possible clefting defects of the facial prominence and abnormalities of the nasal process, (the site of the future nose), were also observed (Fig. 4C arrow).
Figure 4.
Neural tube closure defects in Mlin41 mutants. Mlin41XA144 sagittal sections (A) E11.5 homozygous mutant (−/−). (B) E11.5 heterozygous embryo (+/−) 14 µm each, (C) E12.5−/− [arrow] demarcates abnormal nasal process. (D) E12.5+/−, [*] demarcates open neural tube (A and C), nasal process (NP), 3rd ventricle (3rd V), 4th ventricle (4th V), lateral ventricle (LV), midbrain (MB), heart (H), liver (L), lung (Lu), tongue (T), lower jaw (LJ). Embryos not to scale.
Mutant E12.5 embryos were analyzed by transverse section to more closely observe brain abnormalities (Suppl. Fig. 4A–H). Again, the open neural tube was obvious when compared to a wild type littermate, and the exposed brain tissue was splayed out around the head. Lateral ventricles (Fig. 4G LV in +/+) were not detectable in the mutants, most likely due to a failure of the surrounding tissue to close as a result of the open neural tube defect.
Mlin41XD089 homozygous embryos, similar to the Mlin41XA144 embryos, demonstrated an open neural tube defect leading to exencephaly, but at a much lower penetrance, 56% of homozygous embryos (n = 23) (Suppl. Fig. 6). LacZ expression patterns were also similar to those obtained for the Mlin41XA144 line (Suppl. Fig. 5). These results support our claim that Mlin41 functions to promote proper neural tube closure.
The Mlin41 3'UTR imparts let-7 and miR-125 mediated repression on a reporter gene
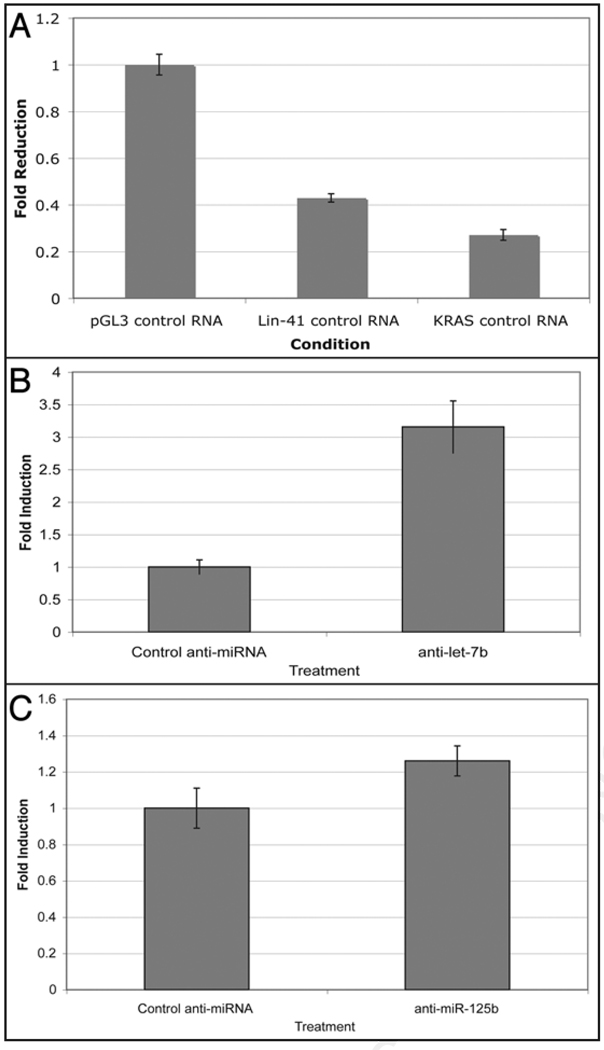
C. elegans lin-41 is regulated by binding of the let-7 and lin-4 miRNAs to its 3'UTR. The human and mouse lin-41 orthologues, HLIN41 and Mlin41, both contain multiple conserved let-7 and mir-125 (the lin-4 orthologue) complementary sites in their 3'UTRs.7 To test whether the Mlin41 3'UTR imparts regulatory information, the entire published Mlin41 3'UTR was fused behind the luciferase reporter gene in the pGL3-control vector (Promega) to generate pGL3-Mlin41. This construct was transfected into human HeLa cells, which are known to express both the let-7 and miR-125 family of miRNAs.16 Compared to the control 3'UTR, the Mlin41 3'UTR exerted 2 to 3-fold repression on the reporter gene (Fig. 5). The Mlin41 3'UTR acted to almost the same levels as the 3'UTR of an established let-7 target gene, KRAS (Fig. 5).16 This demonstrated that the Mlin41 3'UTR promotes gene repression.
Figure 5.
(A) The Mlin41 3'UTR is sufficient to repress expression of a luciferase reporter gene in HeLa cells compared to the control SV40 3'UTR in pGL3 control (n = 3 for each data point). Error bars represent standard deviations. (B) Repression exerted by the Mlin41 3'UTR depends on let-7 and mir-125. HeLa cells were transfected with anti-miRs to let-7b (B) and mir-125b (C) and the relative levels of luciferase expression were measured compared to control anti-miR. Both anti-let-7b (n = 3, p < 0.05) and anti-mir-125b (n = 6, p < 0.05) lead to a statistically significant derepression of reporter gene expression. Error bars represent standard deviations.
Based on the presence of let-7 and miR-125 sites in the 3'UTR of Mlin41,7 we tested whether binding of these miRNAs is responsible for the repressing function of the Mlin41 3'UTR. Human HeLa cells were co-transfected with the pGL3-Mlin41 plasmid and either control, let-7 or miR-125 anti-miRs. Anti-miRs are inhibitors consisting of a 2'O-methylated RNA oligonucleotide complementary to the miRNA.17 These anti-miRs have been shown to block endogenous let-7 or miR-125 function.16 The expression seen using these anti-miRs was compared to that observed with a control consisting of scrambled sequence of the same length as let-7 or miR-125. We found that luciferase expression was increased in the presence of either the let-7 or mir-125 anti-miRs (Fig. 5B and C), demonstrating that repression exerted by the Mlin41 3'UTR is dependent on let-7 and miR-125.
Discussion
Much of our knowledge of vertebrate developmental genes borrows heavily from discoveries made initially in invertebrates. By continuing to investigate the functions of vertebrate orthologues of invertebrate developmental genes, we hope to gain a better understanding of mammalian development. Based on the temporal role of lin-41 during C. elegans development, in addition to the expression of Mlin41 detected by in situ hybridization and northern blot analysis,7 we hypothesized that Mlin41 is involved in controlling events in mouse embryogenesis. In order to characterize the function of Mlin41 during mouse development, we generated two gene-trap insertion lines, Mlin41XA144 and Mlin41XD089, and analyzed their mutant offspring. The XA144 gene-trap inserts in the 5' region of intron 1 and is predicted to cause an N terminal truncation of the Mlin41 protein, while the XD089 insertion is predicted to cause a short C-terminal truncation. Both alleles caused penetrant neural tube closure defects and embryonic lethality. Although Mlin41XA144 mutants die during embryogenesis, it is unlikely that the embryonic lethality is caused by their neural tube defects. There are well characterized mutants that demonstrate neural tube closure defects leading to exencephaly, but that are able to survive late into embryogenesis and even to birth.15
We previously reported that endogenous Mlin41 expression is downregulated in the developing mouse embryo sometime around E9.5 and corresponds to an increase in expression of the let-7 and mir-125 miRNAs.7 There are several miRNA complementary sites in the Mlin41 3'UTR for let-7, miR-125,7 and perhaps other miRNAs as well. It was recently shown that when the chicken lin41 (Glin41) or the Mlin41 3'UTRs are transfected into let-7 expressing HeLa cells, luciferase reporter activity is repressed8 and we have confirmed and extended this result (Fig. 5). Further experiments performed by Kanamoto et al. making specific changes to the LCSs in Glin41 show that the repression of reporter activity required LCSs in the Glin41 3'UTR. We extend this analysis to show that let-7 and miR-125 also regulate Mlin41. Experiments using the Mlin41 3'UTR downstream of a luciferase reporter and a let-7 or miR-125 anti-miR cotransfected into HeLa cells, showed that there was increased reporter expression compared to the negative control. This demonstrates that the Mlin41 3'UTR can potentially mediate let-7 and miR-125 repression in vivo. These experiments represent the first known examples of let-7 and miR-125 directly interacting with mammalian lin-41. Thus, similar to the regulation of C. elegans lin-41 by let-7,12,13 our results demonstrate that the Mlin41 gene is regulated by the miR-125 miRNAs binding to complementary sequences in the Mlin41 3'UTR.
We detected very similar expression patterns for Mlin41 when comparing the lacZ expression pattern of the Mlin41XA144 embryos to the in situ hybridization pattern obtained using a Mlin41 probe.7 One expression area of distinction, the midbrain/hindbrain border may prove to be illustrative of Mlin41 regulation. Robust Mlin41 expression seen in the gene-trap line at midbrain/hindbrain border was barely detectable by in situ hybridization (Fig. 2). In order to understand this difference in expression, we point out that the Mlin41XA144 gene-trap insertion occurs in intron 1 of Mlin41 and truncates the gene’s mRNA (Fig. 1). Therefore, the remaining mRNA of the coding sequence and 3'UTR are missing in the Mlin41XA144 gene-trapped animals, and it thus may not be subjected to 3' negative influences on mRNA stability, for example by miRNA regulation. The Mlin41 in situ hybridization probe hybridizes to the full length Mlin41 mRNA and thus, in contrast to the gene-trap insertion line, detects where the Mlin41 mRNA is stably expressed rather than just where the promoter is active. We speculate that the lacZ reporter in Mlin41xa144 is not subject to the destabilizing effect of miRNAs on the Mlin41 mRNA, due to it’s lack of its normal 3'UTR, leading to ectopic lacZ expression in this brain region. We hypothesize that miRNA-mediated downregulation of Mlin41 through its 3'UTR is vital for proper Mlin41 expression and perhaps function.
In support of this hypothesis, it has recently been shown that miR-125 is expressed in the E9.5 mouse embryonic brain in a region just anterior to and overlapping the midbrain/hindbrain border9 and thus just anterior to and overlapping the Mlin41XA144 lacZ expression domains. The adjacent expression domains and the presence of miR-125 binding sites in the Mlin41 3'UTR and their repression of Mlin41 by miR-125 in HeLa cells, leads us to speculate that it may also downregulate Mlin41 expression in the brain. It has also been shown that let-7 can be detected in the brains of E9.5 mouse embryos by in situ hybridization in a pattern similar to that seen for miR-125.18 It is therefore possible that let-7 and miR-125 regulate Mlin41 expression through the 3'UTR and that this downregulation at the midbrain/hindbrain border is critical for proper development and perhaps neural tube closure.
The lacZ expression we observe at the midbrain/hindbrain border of the Mlin41XA144 heterozygotes combined with the open neural tube defect seen in the Mlin41XA144 homozygotes is strong evidence that Mlin41 plays a role in vertebrate neural tube closure. It should be noted that failure to close the neural tube anywhere in the midbrain/hindbrain region, leads to exencephaly.15 Mutations in Mlin41 could alter initiation or proper closure of the neural tube at the midbrain/hindbrain border, leading to exencephaly. We speculate that under normal conditions Mlin41 is broadly expressed early in the neuroepithelial region where it may block the function of genes repressing neural tube closure or the timing of neural tube closure. Then, prior to and during neural tube closure, we propose that Mlin41 is downregulated, perhaps by let-7 or miR-125, and downstream genes important for complete closure of the neural tube are able to function. However, Mlin41-driven lacZ expression can still be detected at the midbrain/hindbrain border in heterozygous embryos since it cannot be regulated through its 3'UTR, although enough wild-type Mlin41 was present for proper neural tube closure.
We show that the loss of Mlin41 leads to defects in neural tube closure, but the molecular interactions driving this are not known. Two Drosophila RBCC-NHL proteins, homologous to Mlin41, Mei-P26 and Dappled interact with Argonaute proteins and miRNAs, and specifically, Dappled is regulated by let-7.19,20 It is interesting that another mammalian heterochronic gene homologue, Ago2 (alg-2 in C. elegans), has been shown to cause embryonic lethality and neural tube closure defects in mouse embryos.5 Further research will shed light on how Mlin41 functions during vertebrate development specifically in the neural tube. This will enable a greater knowledge of neural tube closure and the role of vertebrate heterochronic gene homologues.
Material and Methods
Gene trap insertion lines
The Mlin41 sequence was used to probe the BayGenomics database (www.baygenomics.com) for potential ES clones with gene trap insertions in the lin41 locus, revealing the XA144 (pGT0pfs vector) and XD089 (pGT1Lxf vector) insertions (Suppl. Fig. 1). The Gene Trap insertion vectors contain a strong splice acceptor site and a lacZ cassette, and are usually found inserted in an intron. The affected gene is detected via 5'RACE using a vector specific primer (Fig. 1, Suppl. Fig. 1).
The ES cells lines obtained from BayGenomics were generated from 129P2 (129Ola) mice and injected in to C57BL/6J blastocysts by the Yale Animal Genomics Service and implanted into wild type CD-1 pseudo-pregnant female mice. The chimeric offspring were mated to C57BL/6J mice and F1 progeny were screened for the presence of the gene trap insertion vector to determine founders (Fig. 1).
Mlin41XA144 genotyping
The XA144 gene trap insertion resides in intron 1 of Mlin41 as determined by 5'RACE (BayGenomics database, Suppl. Fig. 1). The exact location of XA144 within intron 1 was found using a primer 66R, specific to the gene trap vector (pGT0pfs) and an intronic primer, MI 1. This was verified by sequencing and nested PCR using primers MI 13F and MI 12R. Once the precise location of the XA144 gene trap insertion was determined (Fig. 1), a multiplex PCR genotyping strategy was implemented using a primer from within the gene trap insertion vector (MI 18R) and a forward (MI 23F) and reverse (MI 26R) primer from the genomic mouse sequence on either side of the XA144 insertion. The products generated by PCR were as follows: wild type offspring—a single band of 350 bp (MI 23F and MI 26R); homozygous Mlin-41XA144—a single band of 230 bp representing the product of PCR with MI 23F and MI 18R; heterozygous Mlin-41XA144 animals—both a 350 bp and 230 bp band (Suppl. Fig. 2). Genotyping and PCR primer sequences: MI 18R (5'-GTG GAG GGT GGT GTG GGA AAG CCT TC-3'), MI 23F (5'-GCA TGT TCT GGT CGG TGA GTA CCC-3'), MI 26R (5'-GAT AAG AGG GGT TCT GCC TAG CCTC-3'), 66R (5'-CGC CAG GGT TTT CCC AGT CAC GAC-3'), MI 1 (5'-CTC GGA GAA CCA GGA TTT GAG GAC-3'), MI 13F (5'-CGA ACC CAG AGC TTA TGC TAC CC-3') and MI 12R (5'-GAA AGA CCT GAC TTG GGT CCC CGGG-3').
Mlin41XD089 genotyping
The XD089 insertion, based on 5'RACE reactions reported by BayGenomics is in or downstream of exon 4 of Mlin41 (Suppl. Fig. 1). To determine the precise location of its insertion in the Mlin41 locus, inverse PCR was used. XD089 ES cell DNA was digested with MspI, purified and ligated back onto itself. This ligation product was used as the template for PCR with primers specific to gene-trap insertion vector, pGT1Lxf, 66R and 80F. Nested PCR conferred the same product using primers 66R and 79F and this product was sequenced. This showed that the insertion occurs 2.7 Kb downstream of the end of the known Mlin41 3'UTR, in sequence that had not previously been predicted to be part of the lin-41 cDNA (Fig. 1). A multiplex PCR strategy for genotyping embryonic tissue from the XD089 gene trap line was implemented. The products generated by PCR could include a single band of 570 bp (OFSEST 24 and OFSEST 25) for wild type; a band of 605 bp representing the XD089 insertion (80R and OFSEST 24); or both bands in heterozygous animals. Genotyping and PCR primer sequences: OFSEST 24 (5'-GGC GTG CCC TTT TAG TGG GAA CTC-3'), OFSEST 25 (5'-CAT TAA AAG GGG TCT AAG ACC AAT GC-3'), 80R (5'-ATT CAG GCT GCG CAA CTG TTG GG-3'), 66R (5'-CGC CAG GGT TTT CCC AGT CAC GAC-3'), 80F (5'-CCC ACC AGT TGC GCA GCC TGAA-3'), 79F (5'-CGA TCT TCC TGA GGC CGA TACT-3').
lacZ detection
The gene trap insertions in line XA144 and XD089 contain a lacZ cassette that allows for expression analysis of the Mlin41 locus. Embryos at various developmental stages were obtained for beta-Galactosidase detection. Embryos were dissected away from their yolk sacs in PBS and fixed in Tissue Fixative (Specialty Media) for 40 to 90 minutes. Yolk sacs were processed and genotyped. Following the fix, embryos were washed 2 times in Tissue Rinse Solution A (Specialty Media) and 2 times in Tissue Rinse Solution B (Specialty Media) at room temperature. An X-gal detection solution was made by warming Tissue Stain Base Solution (Specialty Media) at 37°C and adding X-Gal Stock Solution (Speciatly Media) at 1:40 dilution. The X-gal Stain Solution was applied to embryos and placed at 37°C for 1 hour and up to overnight. Embryos staining blue were subsequently photographed.
RNA extraction and RT-PCR
Day-8.5 mice embryos were microsurgically isolated to use as the templates for the RT-PCR. Total RNA was extracted from the embryos using TRIzol reagent (Invitrogen, Carlsbad, CA), following the manufacturer’s instruction after homogenization of the sample pellets by pestle motor (Kontes). cDNA was made from the total RNA with Iscript cDNA Synthesis Kit (Bio-Red, Hercules, CA), according to the manufacturer’s protocol, and subjected to PCR analysis using primers designed from MLin41 exon 3 and 4. Primer 1 (5'-GAA GGT GGT CCA GTC TGA GGTC-3'), Primer 2 (5'-CTG ACA AAG CCA ATA GAC TTG AGG-3').
Mouse B-actin control primers were as follows: F (5'-GTG GGC CGC TCT AGG CAC CAA-3'), R (5'-CTC TTT GAT GTC ACG CAC GAT TTC-3'). PCR conditions were 95°C, 3 minutes; 94°C, 30 seconds; 60°C, 1 minute; 72°C, 1 minute for 29 cycles using GoTaq Flexi DNA Polymerase (Promega, Madison, WI ).
Embryo sectioning and H&E staining
Mouse embryos were fixed overnight in 4% paraformaldehyde at 4°C. Yolk sacs were processed and genotyped. Embryos were then stored in 0.5 M Sucrose at 4°C until being frozen. Embryos were placed in OCT media in plastic molds and flash frozen in liquid nitrogen. These blocks were stored at −80°C. Embryos were sectioned at 14 µm using a cryostat (Leica 3050S generously shared by Wiemin Zhong’s lab) onto Superfrost/Plus slides (Fisher) and stained with Hematoxylin and Eosin (Polysciences) for histological analysis.
Luciferase reporter assays
To construct the pGL3-Mlin41 plasmid, the Mlin41 3' UTR was amplified by PCR using mouse genomic DNA as a template, digested with XbaI, and ligated to pGL3-control (Promega) digested with the same enzyme. Primer sequences were: mLin41S: (5'-TCA TCT AGA TTG TGT CTT CTG-3'), mLin41AS: (5'-TCA TCT AGA AGA CAT ATA CTG TG-3').
HeLa S3 cells grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) at 37°C and 5% CO2 were co-transfected in 24-well plates with 0.25 ng/well of pGL3-Mlin41 or pGL3 and 0.1 ng/well of pRL-TK (Promega), as well as anti-miRs (Ambion) at a final concentration of 200 nM, using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s provided protocol. After a 24-hour incubation, the cells were harvested and assayed using the Dual luciferase assay kit (Promega), following the manufacturer’s directions. The values of firefly luciferase activity were normalized over those of Renilla luciferase.
Supplementary Material
Acknowledgements
Gene-trap ES cells were generously provided by BayGenomics, CA. ES cells were injected into mice by the Yale Animal Genomics Service with Dr. Tim Notolli. Frozen sections were cut on a cryostat provided by the Wiemin Zhong lab. Special thanks to Imran Babar, Michelle Boehm and Aurora Esquela-Kerscher for help in the preparation of this manuscript. This work was supported by a grant from the Yale Core Center for Musuloskeletal Disorders, and grants from NIH (GM 62594 and GM 64701).
Footnotes
Note
Supplementary materials can be found at: www.landesbioscience.com/supplement/SchulmanCC7-24-Sup.pdf
References
- 1.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 2.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 3.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 6.Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- 7.Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn. 2005;234:1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanamoto T, Terada K, Yoshikawa H, Furukawa T. Cloning and regulation of the vertebrate homologue of lin-41 that functions as a heterochronic gene in Caenorhabditis elegans. Dev Dyn. 2006;235:1142–1149. doi: 10.1002/dvdy.20712. [DOI] [PubMed] [Google Scholar]
- 9.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 10.Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 11.Lancman JJ, Caruccio NC, Harfe BD, Pasquinelli AE, Schageman JJ, Pertsemlidis A, et al. Analysis of the regulation of lin-41 during chick and mouse limb development. Dev Dyn. 2005;234:948–960. doi: 10.1002/dvdy.20591. [DOI] [PubMed] [Google Scholar]
- 12.Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA∷target interaction. Chem Biol. 2004;11:1619–1623. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Lin YC, Hsieh LC, Kuo MW, Yu J, Kuo HH, Lo WL, et al. Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Mol Biol Evol. 2007;24:2525–2534. doi: 10.1093/molbev/msm195. [DOI] [PubMed] [Google Scholar]
- 15.Copp AJ, Greene ND, Murdoch JN. The genetic basis of mammalian neurulation. Nat Rev Genet. 2003;4:784–793. doi: 10.1038/nrg1181. [DOI] [PubMed] [Google Scholar]
- 16.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. Faseb J. 2006 doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- 19.Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, et al. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–245. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Farrell F, Esfahani SS, Engstrom Y, Kylsten P. Regulation of the Drosophila lin-41 homologue dappled by let-7 reveals conservation of a regulatory mechanism within the LIN-41 subclade. Dev Dyn. 2008;237:196–208. doi: 10.1002/dvdy.21396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.