Abstract
Reverse genetics is a valuable tool to study the replication of many viruses in general. This tool has been particularly useful when studying viruses because of the ease of genetic manipulation, due to their relatively small genomes, and their short and quick replication rates leading to rapid phenotypic expression. For RNA viruses, reverse genetics involves the generation of a cDNA clone, which encompasses the complete viral genome, and can be genetically manipulated in many ways. For example, point mutations, insertions, or deletions can be done to a particular gene to study the function of the altered protein during the virus replication cycle or interactions with the host cell. In addition, foreign genes, e.g. reporter genes or different virus genes, can be added to the viral genomic cDNA clone and can be expressed from infected cells through virus replication. Vaccines against pathogenic viruses can also be made using this method by expressing a surface protein of a pathogenic virus from a recombinant clone of a non-pathogenic virus.
This protocol discusses the generation of an infectious, recombinant human parainfluenza virus type 3 (rHPIV-3) that expresses the enhanced green fluorescent protein (EGFP), termed rHPIV3-EGFP. The following protocol is adapted from our published work in Antiviral Res. 82:12–21 in 2009. The green fluorescence emitted from cells infected with rHPIV3-EGFP can be detected and quantitated for use as an infection tracer or direct measure of virus replication.
To study the effects of gene mutation or foreign gene expression of an RNA virus, infectious, recombinant virus needs to be rescued from the viral cDNA clone. The most important aspect of successfully rescuing a negative-sense RNA virus relies on the generation of a full-length viral antigenomic RNA from the viral cDNA clone. The viral nucleocapsid proteins (NP), phosphoprotein (P), and large protein (L) proteins only recognize and interact with viral RNA; therefore it is necessary to convert the viral cDNA clone to viral antigenomic RNA of proper length and composition. In this protocol, the transcription of the viral antigenomic RNA is driven by a T7 promoter that is strategically placed immediately upstream of the first nucleotide of the 5’ end of the viral antigenome. Previous research has shown that infectious, recombinant negative-sense RNA viruses that were rescued from positive-sense, antigenomic cDNA clones resulted in higher titers than the same viruses rescued from negative-sense, genomic cDNA clones, even though the viral RNA genome was itself negative-sense (Durbin et al., 1997a; Kato et al., 1996). In addition, the rescue of infectious, recombinant virus is enhanced when the T7 promoter and the first nucleotide of the viral antigenome are separated by two guanosine residues (Durbin et al., 1997a). The forward primer used to amplify the 5.3 kb genomic cDNA segment includes the T7 promoter adjacent to the 5’ end of the HPIV-3 antigenome separated by two guanosines. On the 3’ end of the antigenome, an antigenomic hepatitis delta virus ribozyme is positioned immediately following the last nucleotide of the viral antigenome (Perrotta and Been, 1991). The ribozyme will self-cleave itself from the viral RNA genome leaving the full-length virus RNA genome intact and at proper length. The Rib polylinker encodes the hepatitis delta ribozyme adjacent to the 3’ end of the antigenome.
To successfully express the EGFP gene from the viral genome it must mimic a viral gene by encoding the necessary viral mRNA regulation sequences. The HPIV-3 genome consists of six distinct transcriptional units, which encodes for one or more genes. Each transcriptional unit is separated by a gene end (5’-AAAUAAGAAAAA-3’), intercistronic (5’-CUU-3’), and gene start sequences (5’-AGGAUUAAAG-3’). Therefore, when inserting the EGFP gene as a seventh transcriptional unit, the insertion must contain the gene end, intercistronic, and gene start sequences to be effectively expressed through viral mRNA transcription. The reverse primer used to amplify the EGFP gene includes these regulation sequences, which will be ultimately located between the EGFP gene and the HPIV-3 nucleocapsid gene. In addition, during both virus rescue and normal infection, virus replication is most efficient when the length of the complete viral genome is a factor of six (Durbin et al., 1997b). The primers designed to amplify the EGFP gene and subsequent digestion result in an insertion of 852 nucleotides, which is a factor of six. Furthermore, a bipartite replication promoter is necessary, which consists of three equally-spaced guanosine residues at viral genome location 79, 85, and 91, and coincides with the EGFP gene transcription unit insertion site. This location represents one turn of the nucleocapsid helical structure and may co-regulate viral replication perhaps through the assembly and binding of the L–P complex with the encapsidated RNA genome (Tapparel et al., 1998). The addition of the promoter sequence to the forward primer used to amplify the EGFP gene restores the bipartite replication promoter and enhances the rescue of rHPIV3-EGFP.
The ability to rescue negative-sense RNA viruses is problematic because the viruses replicate in the cytoplasm and, thus, do not have access to the host cell’s transcriptional machinery in the nucleus (Friedman et al., 1981). In addition, their genomic viral RNA, which is negative polarity, lacks the signals necessary to initiate eukaryotic protein translation while in the cytoplasm. Therefore, the viral proteins necessary for viral RNA synthesis are packaged into virions in active transcriptase-replicase complexes for immediate replication upon infection (Storey et al., 1984). This means that during the rescue of a recombinant negative-sense virus, all necessary components for viral replication need to be present. These include the full-length viral antigenomic RNA and the proteins NP, P, and L.
To express the NP, P, and L proteins, their corresponding genes are cloned into a T7 expression vector, which will be transcribed by T7 RNA polymerase and translated by the host cell’s ribosomes. The T7 RNA polymerase, used to transcribe the viral antigenomic RNA and NP, P, and L transcripts, is supplied from a recombinant vaccinia virus, vTF7-3, which is used to infect the host cell’s during the rescue procedure (Fuerst et al., 1986). To select for the rescued rHPIV3-EGFP virus and inhibit the replication of vTF7-3, which may contaminate the infectious, recombinant virus, the antiviral compound cytosine β-D-arabinofuranoside (Ara-C) is added to the medium protecting the infected cells (Kato et al., 1996). With all these factors in place, a successful rescue should induce virus replication and infectious, recombinant virus progeny expressing EGFP should be released from the infected cells.
CAUTION: HPIV-3 and vTF7-3 are human pathogens which may cause severe illness in children, elderly, and immunocompromised individuals, but they may cause slight illness in healthy individuals. Therefore these viruses should be used in a BSL-2 laboratory.
BASIC PROTOCOL 1
CONSTRUCTION OF A FULL-LENGTH RECOMBINANT HPIV-3 CDNA CLONE CONTAINING THE EGFP GENE
The following protocol describes the procedure to amplify and assemble three viral genomic cDNA segments encompassing the entire HPIV-3 genome. It also describes the insertion of the EGFP gene into the HPIV-3 genome as a distinct transcription unit. The DrdI restriction site was chosen as the site to insert the EGFP gene because of its prime location upstream of the first gene’s start codon. To circumvent the additional DrdI sites located in the pUC19 parent vector, the pACYC177 plasmid was used as the backbone for the insertion of the EGFP gene into the HPIV-3 genome.
This protocol also describes the insertion of a customized polylinker, which contains the necessary restriction sites, the final 28 nucleotides of the HPIV-3 genome, a hepatitis delta ribozyme, and a T7 transcription termination signal, into the parent vector to facilitate the assembly of the complete genome. The first two viral genomic cDNA segments, 5.3 kb and 6.1 kb, can be added to the polylinker/parent plasmid in any order. However the genomic 4.2 kb cDNA segment needs to be added to the polylinker/parent plasmid very last because it also cuts with the PacI enzyme used to clone the 5.3 kb segment and will interfere with proper alignment of the genomic segments.
Materials
HPIV-3 virus (e.g. Strain 14702)
MA-104 cells (American Type Culture Collection, ATCC)
QIAamp Viral RNA Mini Kit (Qiagen)
-
Enzymes
ProSTAR First-Strand RT-PCR Kit (Stratagene)
PfuTurbo Hotstart DNA polymerase (Stratagene)
T4 DNA ligase (New England Biolabs, NEB)
T4 DNA polymerase (NEB)
Sequenase Version 2.0 DNA Polymerase (USB)
Calf Intestine Alkaline Phosphatase (CIP, NEB)
Thermal cycler (e.g., GENEMate)
0.1 ml thin-walled PCR tubes
QIAEX II Gel Extraction Kit (Qiagen)
QIAquick PCR Purification Kit (Qiagen)
-
Plasmids
pUC19 (NEB)
pEGFP (BD Biosciences Clontech)
pACYC177 (NEB)
-
Restriciton enzymes (NEB)
SmaI
AatII
BstEII
DrdI
KpnI
DraIII
SphI
PacI
37°C, 60°C, and 65°C water baths
Electrocomp GeneHogs E.coli (Invitrogen)
Electorporation Apparatus (e.g., Gene Pulser, Bio-rad)
imMedia Amp Blue (Invitrogen)
imMedia Amp Liquid (Invitrogen)
37°C Incubators (rotating and non-rotating)
QIAprep Spin Miniprep Kit (Qiagen)
-
Primers (See Table 1 for sequence details)
5.3kb-forward
5.3kb-reverse
6.1kb-forward
6.1kb-reverse
4.2kb-forward
4.2kb-reverse
M13/pUC Sequencing Primer (−40) (NEB)
M13/pUC Reverse Sequencing Primer (−48) (NEB)
6.1Mut-forward
6.1Mut-reverse
EGFP-forward
EGFP-reverse
Term-forward
Term-reverse
Rib-forward
Rib-reverse
QuikChange XL Site-Directed Mutagenesis (Stratagene)
Subcloning Efficiency DH5α Chemically Competent E.coli (Invitrogen)
TE buffer (e.g., Cat# BP2474, Thermo Fisher Scientific)
imMedia Amp Agar (Invitrogen)
EndoFree Plasmid Maxi Kit (Qiagen)
Additional reagents and equipment for performing agarose gel electrophoresis (Voytas, 2000).
Table 1.
Primers Used in the Cloning of the rHPIV-EGFP cDNA Clone
| Event/Primer | Sequence (5' to 3') | Highlights |
|---|---|---|
| Viral Genomic cDNA Synthesis | ||
| 5.3kb-forward | CCGACGTCTTAATTAATACGACTCACT ATAGGACCAAACAAGAGAAGAAACTT |
Bolded: AatII and PacI restrictions sites Underlined: T7 promoter sequence |
| 5.3kb-reverse | GGTCACCACAAGAGTTAGA | Bolded: natural BstEII restrictions site |
| 6.1kb-forward | TCTAACTCTTGTGGTGACC | Bolded: natural BstEII restrictions site |
| 6.1kb-reverse | ATTCATCCCAAGGGCAATA | |
| 4.2kb-forward | AGAATGGTTATTCACCTGTTC | |
| 4.2kb-reverse | GAGAAGCACTCTGTGTGGTAT | Bolded: mutated DraIII restrictions site Mutations underlined: A to C and T to G |
|
Site-Directed Mutagenesis |
||
| 6.1Mut-forward | CTTAGGAGCAAAGCGTGCTCAG AAAATGGACACTG |
Bolded: A-to-G mutation |
| 6.1Mut-reverse | CAGTGTCCATTTTCTGAGCACGC TTTGCTCCTAAG |
Reverse complement of 6.1Mut-forward |
|
EGFP Gene Amplification |
||
| EGFP-forward | TTGACTAGAAGGTCAAGAACC TGCAGGTCGACTCTAGAGGAT |
Bolded: DrdI restriction site |
| EGFP-reverse | TTGACCTTCTAGTCAATGT CTTTAATCCTAAGTTTTTCTTATTT ATTAACCGGCGCTCAGTTGGAAT |
Bolded: DrdI restriction site Underlined: HPIV-3 gene transcriptional end, intercistronic, and gene transcriptional start signals |
| Customized Polylinkers | ||
| Term-forward |
TTTTTGTGCGCCCAATACGCAAACCGCC TCTCCCCGCGCGTTGGCCGTTAATTAA GAGGGTGACCCTGCACAGAGTGCC |
Italicized: Vaccinia virus termination sequence Bolded: PacI, BstEII, and DraIII restriction sites |
| Term-reverse |
TTTTTGTAAAAAACCCCTCAAGACCCGTTT AGAGGCCCCAAGGGGTTATGCTAGTTA GGTACCCGGGCACTCTGTGCAG |
Italicized: Vaccinia virus termination sequence Underlined: T7 termination sequence Bolded: KpnI, SmaI, and DraIII restriction sites |
| Rib-forward |
ACCACACAGAGTGCTTCTCTTGTTTGGT GGGTCGGCATGGCATCTCCACCTCC TCGCGGTCCGACCT |
Bolded: DraIII restriciton site Italicized: Final 28 nucleotides of the HPIV-3 genome Underlined: Antigenomic hepatitis delta virus ribozyme sequence |
| Rib-reverse | GGCCGGTACCTCCCTTAGCCATCCGAGTG GACGACGTCCTCCTTCGGATGCCCAGG TCGGACCGCGA |
Bolded: KpnI restriction site Underlined: Antigenomic hepatitis delta virus ribozyme sequence |
| SphI Adapter | ||
| Adapter-forward | GTGACCGCGCATGCCCACAGA | Bolded: SphI restricton site Underlined: BstEII and DraIII restriction sites |
| Adapter-reverse | GTGGGCATGCGCG | Bolded: SphI restricton site Underlined: BstEII and DraIII restriction sites |
RT-PCR amplify HPIV-3 genomic segments
-
1.Purify viral RNA from the clarified supernatant of HPIV-3 infected MA-104 cells using the QIAamp Viral RNA Mini Kit.For this protocol, HPIV-3 strain 14702 was used as a source of viral RNA for cDNA synthesis, although other HPIV-3 strains may be substituted.
-
2.Synthesize three HPIV-3 genomic cDNA segments, 5.3, 6.1, and 4.2 kb, using ProSTAR First-Strand RT-PCR Kit, 300 ng of each of the forward primers (Table 1) and purified HPIV-3 viral RNA in 0.1 ml thin-walled PCR tubes in a thermal cycler according to the manufacturer’s protocol.These primers were designed from a consensus of genomic sequences of three other published HPIV-3 strains, JS, 47885, and GPv (Galinski, 1991; Ohsawa et al., 1998; Stokes et al., 1992). Other HPIV-3 specific primers may be used to incorporate other promoters and/or restriction sites. Even though this protocol uses the ProSTAR First-Strand RT-PCR Kit, other first-strand RT-PCR kits may be used.
-
3.Amplify each genomic cDNA segment using PfuTurbo Hotstart DNA Polymerase and 120 ng of both forward and reverse primers (Table 1) in 0.1 ml thin-walled PCR tubes in a thermal cycler following the manufacturer’s protocol with the following exceptions:
Annealing temperatures 50°C 5.3 kb and 6.1 kb segments 48°C 4.2 kb segment Extension time 6 min for all reactions Number of Cycles 30 During this step it is crucial to use a high fidelity proofreading DNA polymerase to reduce the number of mutations, which may be lethal to the recombinant virus. Even though this protocol uses the PfuTurbo Hotstart DNA Polymerase, other high fidelity proofreading DNA polymerases may be used. The use of a proofreading DNA polymerase during amplification will generate blunt ends that will allow cloning of PCR products into any restriction site cut with a restriction endonuclease generating blunt ends. -
4.
Check for the presence and correct length of each genomic cDNA segment by gel electrophoresis.
-
5.
If multiple bands exist in any reaction, excise and purify the bands of correct length with the QIAEX II Gel Extraction Kit. Otherwise, purify the PCR product with the QIAquick PCR Purification Kit if only one band in seen during gel electrophoresis.
Cloning of genomic segments
-
6.
Linearize pUC19 with SmaI at room temperature according to manufacturer’s protocol.
-
7.
Purify the pUC19 digestion with the QIAquick PCR Purification Kit.
-
8.
Ligate each purified genomic cDNA PCR product into the digested pUC19 vector using T4 DNA ligase according to manufacturer’s protocol.
-
9.Heat inactivate the T4 DNA ligase at 65°C for 15 min in a water bath.Alternatively, the ligated DNA may be purified with the QIAEX II Gel Extraction Kit for optimal transformation efficiency. Heat inactivation of the ligase enzyme results in increased transformation efficiencies for ligated DNA compared to untreated, ligated DNA but lower transformation efficiencies for ligated DNA compared to purified, ligated DNA.
-
10.Electroporate the ligated DNA into Electrocomp GeneHogs E.coli at 1.6 kV, 25 µF, and 200 Ω according to the manufacturer’s protocol using an electroporation apparatus.High efficiency transformation was achieved with electroporation; however other transformation methods could be used.
-
11.
Spread transformants onto imMedia Amp Blue agar plates and incubate overnight in a 37°C incubator.
-
12.Select several white bacterial colonies, inoculate into 3 ml imMedia Amp Liquid cultures, and incubate overnight in a 37°C rotating incubator.Invitrogen’s imMedia was used for reliability and convenience, although traditional LB media may also be used. Bacterial stocks can also be made by adding sterile glycerol, 17% final volume, and freezing the mixture at −80°C.
-
13.
Isolate DNA plasmids from each culture using the QIAprep Spin Miniprep Kit.
-
14.
Screen the DNA of several clones for the presence of each genomic cDNA insert by restriction digestion and gel electrophoresis.
-
15.Sequence positive clones starting with the M13/pUC Sequencing Primer (−40) and M13/pUC Reverse Sequencing Primer (−48) and continue sequencing the complete cDNA insert with virus-specific primers in both directions.The resulting positive clones were named 5.3 kb, 6.1 kb, and 4.2 kb, which represents the length of each PCR product. The final order of each genomic cDNA segment in the full-length clone is 5.3 kb–6.1 kb-4.2 kb. The complete genomic sequence for HPIV-3 strain 14702 can be found in Genbank, accession no. EU424062.
-
16.Mutate A-to-G, located at viral nucleotide position 8635, in the genomic 6.1 kb cDNA segment to eliminate a second SphI restriction site using the QuikChange XL Site-Directed Mutagenesis, following the manufacturer’s protocol.The native genomic 6.1 kb cDNA segment of HPIV-3, strain 14702 has two SphI restriction sites. Therefore, the SphI restriction site located in the middle of the 6.1 kb segment needs to be eliminated to avoid interference with further subsequent cloning. Other HPIV-3 strains may not have this undesirable SphI restriction site.
PCR Amplify EGFP ORF
-
17.PCR amplify the open reading frame of EGFP in 0.1 ml thin-walled PCR tubes in a thermal cycler using the PfuTurbo Hotstart DNA polymerase enzyme following the manufacturer’s protocol.
Components: Template 1ng pEGFP plasmid Primers (Table 1) 20uM EGFP-Forward 20uM EGFP-Reverse Cycling conditions exceptions: Annealing temperature 58°C Extension time 1 min Number of cycles 30 To abide by the “Rule of Six”, the primers used to amplify the EGFP ORF were designed to generate a PCR product that results in an 852bp band, a factor of six, when digested with DrdI in later steps. In addition, three equally spaced G nucleotides were added to the forward primer at positions 11, 17, and 23 to restore a natural bipartite replication promoter on the 3’ end of the viral genome. -
18.
Repeat steps 4–15 to clone the resulting 868 bp band, representing the PCR amplified EGFP ORF into a naive pUC19 vector.
Clone EGFP into the 5.3kb genomic cDNA segment
-
19.
Digest the pACYC177 plasmid and the plasmid containing the genomic 5.3 kb cDNA segment with AatII and BstEII restriction enzymes, sequentially, in 37°C and 60°C water baths, respectively, according to the manufacturer’s protocol.
-
20.
Separate both digestions, individually, by gel electrophoresis and purify the approximate 5.0 kb band, representing the genomic 5.3 kb cDNA segment, and the approximate 4.0 kb band, representing the pACYC177 plasmid, using the QIAEX II Gel Extraction Kit.
-
21.Ligate the purified genomic 5.3 kb cDNA segment into the purified pACYC177 vector using T4 DNA ligase according to manufacturer’s protocol.The addition of the EGFP gene into the genomic 5.3kb cDNA segment uses the DrdI restriction site. The parent plasmid pUC19 cuts twice with DrdI, so it is necessary to transfer the 5.3kb cDNA segment into a second plasmid that does not contain additional DrdI sites. The pACYC177 contains one DrdI restriction site located on a small 284 bp segment between AatII and BstEII restriction sites, which is eliminated during the purification of the larger 4.0kb segment from the smaller 284 bp segment resulting from the AatII/BstEII digestion. Other vectors, which do not contain DrdI sites, may also be used.
-
22.
Heat inactivate the T4 DNA ligase at 65°C for 15 min in a water bath.
-
23.
Electroporate the ligated DNA into Electrocomp GeneHogs E.coli at 1.6 kV, 25 µF, and 200 Ω according to the manufacturer’s protocol using an electroporation apparatus.
-
24.
Spread transformants onto imMedia Amp Agar plates and incubate overnight in a 37°C incubator.
-
25.Select several bacterial colonies, inoculate into 5 ml imMedia Amp Liquid, and incubate cultures overnight in a 37°C rotating incubator.The pACYC177 plasmid is a low-copy number vector; therefore little to no DNA may be obtained using traditional methods. Sufficient DNA can purified for cloning purposes by performing DNA isolation on the whole 5 ml culture in three separate 1.5 ml preparations.
-
26.Isolate DNA plasmids from each culture using the QIAprep Spin Miniprep Kit.To concentrate and purify more vector DNA, the supernatants from three bacterial lysates were applied to one column by allowing each supernatant to flow through the column first before loading the remaining supernatants.
-
27.
Screen the DNA of several clones for the presence of the genomic 5.3 kb cDNA insert in the pACYC177 backbone by restriction digestion, gel electrophoresis, and DNA sequencing.
-
28.
Digest the plasmids containing the PCR amplified EGFP ORF and the pACYC177/5.3 kb plasmid, with DrdI in a 37°C water bath according to the manufacturer’s protocol.
-
29.
Dephosphorylate the ends of the pACYC177/5.3 kb plasmid with CIP in a 37°C water bath according to the manufacturer’s protocol.
-
30.
Separate the PCR amplified EGFP ORF digestion by gel electrophoresis and purify the 852 bp band, representing the EGFP ORF, using the QIAEX II Gel Extraction Kit.
-
31.Repeat steps 21–27 to clone the EGFP ORF into the genomic 5.3 kb cDNA segment.The DrdI restriction site is non-palindromic; therefore directional cloning should occur using only one restriction enzyme.
Eliminate KpnI site from parent vector
-
32.Linearize the raw pUC19, containing no insert, with KpnI in a 37°C water bath according to the manufacturer’s protocol.The native KpnI restriction site located in the pUC19 multiple cloning site needs to be eliminated because a KpnI restriction site is reintroduced and used in later cloning steps. The pUC19 parent vector was selected and used because of the lack of certain restriction sites that are used in downstream applications. Other cloning vectors are commercially available and could also be used.
-
33.
Blunt the 3’ overhang ends, generated by KpnI cleavage, using T4 DNA polymerase following manufacturer’s protocol.
-
34.
Purify the reaction with the QIAquick PCR Purification Kit.
-
35.
Recircularize the plasmid with T4 DNA Ligase according the manufacturer’s protocol.
-
36.
Transform the ligated DNA into Subcloning Efficiency DH5α Chemically Competent E.coli following manufactures protocol.
-
37.
Spread transformants onto imMedia Amp Blue agar plates and incubate overnight in a 37°C incubator.
-
38.
Select several white bacterial colonies, inoculate into 3 ml imMedia Amp Liquid cultures, and incubate overnight in a 37°C rotating incubator.
-
39.
Isolate DNA plasmids from each culture using the QIAprep Spin Miniprep Kit.
-
40.Screen the DNA several clones for the presence of the SmaI restriction site by restriction digestion and gel electrophoresis.The KpnI and SmaI restriction sites overlap each other in the pUC19 multiple cloning site. Clones that have the typical four nucleotide deletions also eliminate the SmaI site. On the other hand, clones that have five deletions may leave the SmaI restriction site intact.
-
41.Confirm the elimination of the KpnI restriction site and presence of the SmaI site by DNA sequencing.The resulting plasmid was named pUC19-T
Add customized polylinkers to parent vector
-
42.Separately heat 1 µg of the forward and reverse oligonucleotides for Term and Rib (Table 1) to 70°C for 10 min in TE buffer.The forward and reverse primers for both Term and Rib contain a 14 nucleotide overlap on their 3’ ends so they can anneal to each other.
-
43.
Slowly cool the mixture to 37°C.
-
44.Extend each oligonucleotide with Sequenase Version 2.0 DNA Polymerase (USB) according to the manufacturer’s protocol.As long as the primers are annealed properly, secondary structure is of no concern. The Sequenase enzyme ignores secondary structure during elongation. Elongation should occur at each 3’ end using the opposite oligonucleotide as template.
-
45.
Purify the small double-stranded DNA products with QIAEX II Gel Extraction Kit.
-
46.
Blunt the ends of the small double-stranded DNA products using T4 DNA polymerase following manufacturer’s protocol.
-
47.
Purify the products a second time with QIAEX II Gel Extraction Kit.
-
48.
Digest pUC19-T with SmaI at room temperature according to the manufacturer’s protocol.
-
49.
Ligate the purified small double-stranded DNA products Term and Rib into the digested pUC19-T, separately, using T4 DNA Ligase according the manufacturer’s protocol.
-
50.
Transform the ligated DNA into Subcloning Efficiency DH5α Chemically Competent E.coli following manufactures protocol.
-
51.
Spread transformants onto imMedia Amp Agar plates and incubate overnight in a 37°C incubator.
-
52.
Select several bacterial colonies, inoculate into 3 ml imMedia Amp Liquid cultures, and incubate overnight in a 37°C rotating incubator.
-
53.
Isolate DNA plasmids from each culture using the QIAprep Spin Miniprep Kit.
-
54.
Screen the DNA of several clones for the presence of the small double-stranded DNA inserts by restriction digestion and gel electrophoresis.
-
55.Confirm the presence and validate the sequence of each insert by DNA sequencing.The plasmid that contains the Term segment was named pUC19-A and the plasmid that contains the Rib segment was named pUC19-R.
-
56.
Digest the pUC19-A and pUC19-R plasmids with DraIII and KpnI restriction enzymes, sequentially, in a 37°C water bath according to the manufacturer’s protocol.
-
57.
Separate the pUC19-R DraIII/KpnI digestion by gel electrophoresis and purify the 108 bp band with QIAEX II Gel Extraction Kit.
-
58.Repeat steps 49–55 to clone the purified Rib 108 bp insert into pUC19-A.The resulting plasmid was named pUC19-B.
Realign SphI restriction site
-
59.
Linearize pUC19-B with the SphI restriction enzyme in a 37°C water bath according to the manufacturer’s protocol.
-
60.Repeat steps 33–41 to eliminate the native SphI restriction site.The resulting plasmid was named pUC19-C.
-
61.
Digest pUC19-C with DraIII and BstEII restriction enzymes, sequentially, in 37°C and 60°C water baths, respectively, according to the manufacturer’s protocol.
-
62.
Repeat steps 42–43 to anneal the forward and reverse SphI adapter primers together (Table 1).
-
63.Repeat steps 49–55 to clone the adaptor into the digested pUC19-C.The resulting plasmid was named pUC19-D.
Assemble full-length genome
-
64.
Digest pUC19-D and the mutated genomic 6.1 kb cDNA segment with SphI and BstEII, sequentially, in 37°C and 60°C water baths, respectively, according to the manufacturer’s protocol.
-
65.
Separate the mutated genomic 6.1 kb cDNA segment digestion by gel electrophoresis and purify the approximate 6 kb band using the QIAEX II Gel Extraction Kit.
-
66.
Ligate the purified mutated genomic 6.1kb cDNA segment into the digested pUC19-D using T4 DNA ligase according to manufacturer’s protocol.
-
67.
Heat inactivate the T4 DNA ligase at 65°C for 15 min in a water bath.
-
68.
Electroporate the ligated DNA into Electrocomp GeneHogs E.coli at 1.6 kV, 25 µF, and 200 Ω according to the manufacturer’s protocol using an electroporation apparatus.
-
69.
Spread transformants onto imMedia Amp Agar plates and incubate overnight in a 37°C incubator.
-
70.
Select several bacterial colonies, inoculate into 3 ml imMedia Amp Liquid cultures, and incubate overnight in a 37°C rotating incubator.
-
71.
Isolate DNA plasmids from each culture using the QIAprep Spin Miniprep Kit.
-
72.Screen the DNA of several clones for the presence of the mutated genomic 6.1kb cDNA insert by restriction digestion, gel electrophoresis, and DNA sequencing.The resulting plasmid was named pUC19-F
-
73.
Sequentially digest pUC19-F and the genomic 5.3 kb cDNA segment containing the EGFP gene with PacI followed by BstEII in 37°C and 60°C water baths, respectively, according to the manufacturer’s protocol.
-
74.
Separate the digested genomic 5.3 kb cDNA/EGFP segment by gel electrophoresis and purify the approximate 5 kb band using the QIAEX II Gel Extraction Kit.
-
75.Repeat steps 66–72 to clone the 5.3 kb cDNA/EGFP segment into the digested pUC19-F.The resulting plasmid was named pUC19-I. The addition of either the 5.3 kb or 6.1 kb genomic cDNA segment can occur in any order. However the final addition of the 4.2 kb genomic cDNA segment needs to occur last.
-
76.
Sequentially digest pUC19-I and the genomic 4.2 kb cDNA segment with DraIII and SphI, in a 37°C water bath according to the manufacturer’s protocol.
-
77.
Separate the genomic 4.2 kb cDNA segment digestion by gel electrophoresis and purify the approximate 4 kb band using the QIAEX II Gel Extraction Kit.
-
78.Repeat steps 66–72 to clone the 4.2 kb cDNA segment into the digested pUC19-I.The resulting plasmid was named pUC19-J, which represents the full-length recombinant HPIV-3 cDNA clone expressing EGFP.
-
79.
Purify transfection quality pUC19-J plasmid DNA using the EndoFree Plasmid Maxi Kit following the manufacturer’s protocol.
BASIC PROTOCOL 2
CLONE HPIV-3 SUPPORT GENES
The following protocol describes the amplification and cloning of three HPIV-3 genes that code for the nucleocapsid protein (NP), phosphoprotein (P), and large protein (L), which are necessary for viral replication and transcription. The presence of these proteins during the rescue of the recombinant virus (See basic protocol 3) is necessary to replicate and transcribe the rHPIV-3 viral RNA to stimulate a productive infection. The transcription of these genes from plasmids is initiated by a T7 promoter, which is similar to the promoter initiating the transcription of the full-length genomic cDNA but is part of the commercially available pTNT plasmid from Promega. To successfully express these proteins the orientation of the genes in relation to the T7 promoters are crucial. The start codon for each gene should be placed downstream of the T7 promoter. The T7 DNA polymerase used to transcribe these viral genes is supplied from a recombinant vaccinia virus discussed in basic protocol 3.
Materials
-
Primers (See Table 2 for sequence details)
NP-forward
NP-reverse
P-forward
P-reverse
L-forward
L-reverse
M13/pUC Sequencing Primer (−40) (NEB)
M13/pUC Reverse Sequencing Primer (−48) (NEB)
-
Enzymes
PfuTurbo Hotstart DNA polymerase (Stratagene)
T4 DNA ligase (NEB)
Thermal cycler (e.g., GENEMate)
0.1ml thin-walled PCR tubes
QIAquick PCR Purification Kit (Qiagen)
-
Plasmids
pUC19 (NEB)
pTNT (Promega)
-
Restriciton enzymes (NEB)
SmaI
KpnI
SalI
Subcloning Efficiency DH5α Chemically Competent E.coli (Invitrogen)
37°C and 65°C water baths
Electrocomp GeneHogs E.coli (Invitrogen)
Electorporation Apparatus (e.g., Gene Pulser, Bio-rad)
imMedia Amp Blue (Invitrogen)
imMedia Amp Liquid (Invitrogen)
37°C Incubators (rotating and non-rotating)
QIAprep Spin Miniprep Kit (Qiagen)
QIAEX II Gel Extraction Kit (Qiagen)
imMedia Amp Agar (Invitrogen)
EndoFree Plasmid Maxi Kit (Qiagen)
Additional reagents and equipment for performing agarose gel electrophoresis (Voytas, 2000).
Table 2.
Primers Used in the Cloning of the HPIV-3 Support Genes and PCR Condition.
| Nucleocapsid gene (NP) | Phosphoprotein gene (P)a | Large Protein gene (L) | |
|---|---|---|---|
| Forward Primer (5' to 3') |
GAAGGTCAAGAAAA GGGAACTCT |
TGATGGAAAGCG ACGCTAAA |
GCGTGCTCAGAAAA TGGACA |
| Reverse Primer (5' to 3') |
TTGATTCGATTAGTT GCTTCCA |
GGATCATTGGCAATT GTTGA |
CCTTAGGCTTAAAGATAAA GGTTAGGA |
| Primer Quantity | 20uM | 20uM | 20uM |
| Template | Genomic 5.3kb cDNA segment | Genomic 5.3kb cDNA segment | pUC19-J |
| Annealing Temp | 51°C | 51°C | 51°C |
| Extension Time | 2 min | 2 min | 7 min |
| Cycles | 30 | 30 | 30 |
| Approximate Size | 1.5kb | 1.8kb | 7.0kb |
Underlined T to C mutation to silence the expression of the virus C protein in the forward primer.
PCR amplify viral support genes
-
1.
Amplify the open reading frames of the viral NP, P, and L genes by PCR in 0.1ml thin-walled PCR tubes in a thermal cycler using the PfuTurbo Hotstart DNA polymerase enzyme following the manufacturer’s protocol. Use the experimental conditions found in Table 2.
-
2.
Check for the presence and correct length of each genomic cDNA segment by gel electrophoresis.
-
3.
Purify the PCR products with the QIAquick PCR Purification Kit.
Clone viral support genes into pUC19
-
4.
Linearize pUC19 with SmaI at room temperature according to manufacturer’s protocol.
-
5.
Purify the pUC19 digestion with the QIAquick PCR Purification Kit.
-
6.
Ligate the PCR products for each purified support gene into the digested pUC19 vector using T4 DNA ligase according to manufacturer’s protocol.
-
7.Transform the ligated DNA for the NP and P clones into Subcloning Efficiency DH5α Chemically Competent E.coli following manufactures protocol.The NP and P clones were transformed into DH5α E.coli because of the size of the inserts, which are approximately 1.5 kb and 1.8 kb, respectively.
-
8.
Heat inactivate the T4 DNA ligase used to ligate the DNA for the L clone at 65°C for 15 min in a water bath.
-
9.Electroporate the ligated DNA for the L clone into Electrocomp GeneHogs E.coli at 1.6 kV, 25 µF, and 200 Ω according to the manufacturer’s protocol using an electroporation apparatus.The L clones were electroporated into GeneHogs because of the size of the insert, approximately 7 kb.
-
10.
Spread transformants for all three support genes onto imMedia Amp Blue agar plates and incubate overnight in a 37°C incubator.
-
11.
Select several white bacterial colonies, inoculate into 3 ml imMedia Amp Liquid cultures, and incubate overnight in a 37°C rotating incubator.
-
12.
Isolate DNA plasmids from each culture using the QIAprep Spin Miniprep Kit.
-
13.
Screen the DNA of several clones for the presence of each viral support gene insert by restriction digestion and gel electrophoresis.
-
14.Sequence positive clones starting with the M13/pUC Sequencing Primer (−40) and M13/pUC Reverse Sequencing Primer (−48) and continue sequencing the complete support gene insert with gene-specific primers in both directions.The resulting positive clones were named pUC19-NP, P, or L. The insertion of each support gene into pUC19 occurred bi-directionally. Screen and select clones whose orientation resulted in the gene’s start codon downstream of the KpnI restriction site, not the SalI restriction site. When each support gene is directionally cloned into the pTNT vector in the next step, the T7 polymerase will drive the transcription of the support gene only when properly orientated.
Clone viral support genes into T7 expression plasmid
-
15.
Digest the pTNT plasmid and the plasmids containing the NP, P, and L support genes with KpnI and SalI restriction enzymes, sequentially, in a 37°C water bath according to the manufacturer’s protocol.
-
16.
Separate support gene digestions, individually, by gel electrophoresis and purify the approximate 1.5 kb band, representing the NP gene, the approximate 1.8 kb band, representing the P support gene, and the approximate 7.0 kb band, representing the L support gene, using the QIAEX II Gel Extraction Kit.
-
17.
Repeat steps 6–9 to ligate and transform the purified support genes into the digested pTNT plasmid.
-
18.
Spread transformants for all three support genes onto imMedia Amp Agar plates and incubate overnight in a 37°C incubator.
-
19.
Select several bacterial colonies, inoculate into 3 ml imMedia Amp Liquid cultures, and incubate overnight in a 37°C rotating incubator.
-
20.
Isolate DNA plasmids from each culture using the QIAprep Spin Miniprep Kit.
-
21.Screen the DNA of several clones for the presence of each viral support gene insert by restriction digestion, gel electrophoresis, and DNA sequencing.The resulting plasmids were named pTNT-NP, P, and L.
-
22.
Purify transfection quality pTNT-NP, P, and L plasmid DNA using the EndoFree Plasmid Maxi Kit following the manufacturer’s protocol.
BASIC PROTOCOL 3
RESCUE INFECTIOUS, RECOMBINANT HPIV-3 VIRUSES
The following protocol describes the process to rescue an infectious rHPIV-3 virus from a full-length genomic cDNA clone. The recombinant vaccinia virus, vTF7-3, expresses a T7 DNA polymerase, which transcribes the full-length viral genomic cDNA, basic protocol 1, and the three support plasmids, basic protocol 2. The mRNAs for the nucleocapsid protein, phosphoprotein, and large protein are further translated into proteins that replicate and transcribe the full-length viral genomic RNA, which results in the assembly of infectious rHPIV-3 virions.
To control the replication of the recombinant vaccinia virus, Ara-C is added to the medium, which inhibits DNA replication. Subsequent plaque purifications are also done to further remove residual vTF7-3 particles and prevent contamination of rHPIV-3 stocks.
Materials
HeLa cells (Cat# CCL-2, ATCC)
Cell Culture vessels (12-well plates and T-25 flasks)
Minimum Essential Medium w/ Earle’s Balanced Salts (MEM, Cat# SH30024.02, Hyclone)
Standard Fetal Bovine Serum (FBS, Cat# SH30088.03, Hyclone)
10 mM MEM Non-Essential Amino Acids Solution (NEAA, Cat# 11140050, Invitrogen)
100 mM MEM Sodium Pyruvate Solution (Cat# 11360070, Invitrogen)
Water-jacketed, humidified 37°C incubator with 5% CO2 (e.g. Isotemp, Thermo Fisher Scientific)
vTF7-3 (Dr. Bernard Moss’s lab)
Opti-MEM I Reduced-Serum Media (Cat# 11058-021, Gibco)
-
Plasmids
pUC19-J (basic protocol 1)
pTNT-NP (basic protocol 2)
pTNT-P (basic protocol 2)
pTNT-L (basic protocol 2)
Lipofectamine 2000 Transfection Reagent (Cat# 11668019, Invitrogen)
Cytosine β-D-arabinofuranoside (Ara-C, Cat# C1768, Sigma)
MA-104 cells (ATCC)
2% Agarose (See Reagents and Solutions)
2X MEM (See Reagents and Solutions)
1 ml pipet
Transfection
-
1.
Seed HeLa cells in a 12-well plate at 8 × 105 cells/well in MEM supplemented with 10% FBS, 0.1 mM NEAA, and 1 mM sodium pyruvate
-
2.
Incubate HeLa cells overnight in a water-jacketed, humidified incubator set at 37°C and 5% CO2.
-
3.
Replace growth medium with 500 µl MEM supplemented with 2% FBS, 0.1 mM NEAA, and 1 mM sodium pyruvate.
-
4.
Infect HeLa cells with vTF7-3 at a concentration of 5.4 × 105 plaque forming units/cell or 1 multiplicity of infection (MOI).
-
5.
Incubate the infected cells for 1 hour at 37°C in a CO2 (5%) incubator.
-
6.
Remove virus/media mixture and add 400 µl of Opti-MEM supplemented with 0.1 mM NEAA.
-
7.
Transfect the infected HeLa cells with 0.4 µg pUC19-J, 0.8 µg pTNT-NP, 1.6 µg pTNT-P, and 0.04 µg pTNT-L, and 5.3 µl of Lipofectamine 2000 according to the manufacturer’s protocol.
-
8.
Incubate the transfected cells for 4–5 hours at 37°C in a CO2 (5%) incubator.
-
9.
Add 500 µl of MEM supplemented with 20% FBS, 0.1 mM NEAA, 1 mM sodium pyruvate, and 250 µg/ml of Ara-C to the transfected cells.
-
10.
Incubate the transfected cells for 48 hours at 37°C in a CO2 (5%) incubator.
-
11.Scrape the transfected cells off the plate and freeze the cells suspension at −80°C.Typical HPIV-3 induced cytopathic effect (CPE) cannot be seen at the conclusion of this step. However, most cell death that is observed is due to vTF7-3 induced CPE, which is characterized by cellular rounding and sloughing, even though the Ara-C inhibitor is present in the medium.
Amplify infectious rHPIV3-EGFP
-
12.
Seed 3 × 106 MA-104 cells in a T-25 flask in MEM supplemented with 10% FBS.
-
13.
Incubate the MA-104 cells overnight at 37°C in a CO2 (5%) incubator.
-
14.
Remove the growth medium and add 800 µl MEM.
-
15.
Add 200 µl of the transfected HeLa cell lysate to the MA-104 cells.
-
16.
Incubate the MA-104 cells for 2 hours at 37°C in a CO2 (5%) incubator.
-
17.
Add 5 ml of MEM, supplemented with 2% FBS and 250 µg/ml Ara-C, to the infected MA-104 cells.
-
18.Incubate the infected MA-104 cells for 3–4 days at 37°C in a CO2 (5%) incubator.The rescued virus is now called rHPIV3-EGFP. At this point no vTF7-3 CPE should be seen. If rHPIV3-EGFP was successfully rescued then typical HPIV-3 CPE should be seen, which is characterized by syncytia formation.
-
19.
Scrape the infected MA-104 cells off the plate and freeze the cells suspension at −80°C.
Plaque purify infectious rHPIV3-EGFP
-
20.
Seed MA-104 cells in a 12-well plate at 8 × 105 cells/well in MEM supplemented with 10% FBS.
-
21.
Incubate the MA-104 cells overnight at 37°C in a CO2 (5%) incubator.
-
22.
Serially dilute the rHPIV3-EGFP virus down to 1 × 10−6 PFU/ml in 500 µl MEM.
-
23.
Remove the growth medium from the MA-104 cells and add 500 µl of MEM containing each dilution of virus into individual wells.
-
24.
Incubate the MA-104 cells for 2 hours at 37°C in a CO2 (5%) incubator.
-
25.
Remove the virus/medium mixture and replace with 500 µl of the pre-warmed (>37°C) 50:50 mixture of 2% agarose and 2X MEM.
-
26.
Incubate the infected MA-104 cells for 2–3 days at 37°C in a CO2 (5%) incubator.
-
27.
Select a well isolated virus plaque located in a well in which the 10−5 or 10−6 dilution of virus was plated. These wells should have approximately 1–20 plaques each. Remove the agarose plug directly over the isolated plaque using a 1 ml pipet and place the plug in 500 µl MEM.
-
28.
Add 25 µl of MEM to the remaining hole from where the plug was removed to extract any remaining infectious virus. Remove the 25 µl volume and add it to the 500 µl of MEM containing the agarose plug, and store at −80°C.
-
29.
Repeat steps 20–28 twice more.
-
30.
To amplify the plaque purified virus, remove the growth medium from newly plated MA-104 cells and add the 500 µl MEM containing one of the agarose plugs and virus.
-
31.
Incubate the MA-104 cells for 2 hours at 37°C in a CO2 (5%) incubator.
-
32.
Add 1.5 ml MEM supplemented with 2% FBS to the MA-104 cells.
-
33.
Incubate the infected MA-104 cells for 3–5 days at 37°C in a CO2 (5%) incubator.
-
34.Scrape the infected MA-104 cells off the plate and freeze the cells suspension at −80°C.The resulting virus rHPIV3-EGFP, which has been plaque purified a total of three times, is now free of contaminating vaccinia virus.
REAGENTS AND SOLUTIONS
2% Agarose
Bake clean glassware at 400°F for 2 hours.
Add 8 g of low-melting agarose (Thermo Fisher Scientific) to 400 ml of deionized, distilled water.
Autoclave
Store at room temperature
2X MEM
Dissolve one packet of powdered MEM (Cat# 61100-061, Invitrogen) in 400 ml of deionized, distilled water.
Add 30 ml of 7.5% Sodium Bicarbonate Solution (Invitrogen).
Adjust volume to 500 ml with deionized, distilled water.
Sterilize by passing through a 0.2-µm filter.
Store at 4°C.
COMMENTARY
Background Information
HPIV-3 is classified in the Paramyxovirinae subfamily, which are non-segmented, negative-sense, single-stranded RNA viruses. The HPIV-3 genome consists of six transcriptional gene units composed of one or more genes whose proteins are, in order, nucleocapsid protein, phosphoprotein, matrix protein, fusion protein, hemagglutinin-neuraminidase protein, and large protein. Some members of the Paramyxovirinae subfamily also express accessory proteins from the phosphoprotein gene, e.g. C, Y, W, V, D (Lamb and Kolakofsky, 2001). The transcriptional units are flanked by 3’ leader and 5’ trailer untranslated regions that are essential for viral transcription and replication regulation (Calain and Roux, 1995). Each transcriptional unit is separated by gene end, intercistronic, and gene start sequences. During viral mRNA synthesis, the viral RNA polymerase recognizes the gene end sequence and stutters, adding non-templated adenosine residues to create a poly-A tail (Lamb and Kolakofsky, 2001). The viral RNA polymerase then re-engages viral mRNA transcription at the gene start sequence for the next gene unit. The viral RNA polymerase will sometimes fail to reengage mRNA transcription for the downstream gene, which results in fewer mRNA transcripts for downstream genes compared to upstream genes transcribed from the same template. This phenomenon is called transcriptional polarity and ultimately leads to less protein expressed from the downstream gene units in a gradient fashion (Wertz et al., 1998). This phenomenon can be used advantageously for regulation purposes by cloning foreign genes into various locations on the viral genome to control the rate of expression of the foreign gene.
Several members of the Paramyxovirinae subfamily have been successfully rescued from cDNA clones, measles virus (MeV), Sendai virus (SeV), and two HPIV-3 viruses strains 47885 and JS (Durbin et al., 1997a; Garcin et al., 1995; Hoffman and Banerjee, 1997; Radecke et al., 1995). This method has been used to effectively express foreign genes from the genomes of infectious, recombinant RNA viruses for many purposes. For example, a recombinant SeV, which is non-pathogenic to humans is a prime candidate for vaccine development and gene transfer, has been used as an expression vector in gene transfer to deliver foreign eukaryotic genes to neurons and airway epithelium (Shirakura et al., 2003; Yonemitsu et al., 2000). In addition, several recombinant paramyxoviruses have been developed that express other viral surface proteins to be used as vaccines. The Ebola virus glycoprotein and the respiratory syncytial virus (RSV) fusion protein have been expressed from recombinant HPIV-3 and SeV viruses, respectively, and have resulted in protective immunity against Ebola virus and RSV, respectively (Bukreyev et al., 2006; Zhan et al., 2007).
A third area where the insertion of a foreign gene into a recombinant virus has been beneficial is for the expression of a reporter gene for the purpose of tracing the viral infection. A recombinant HPIV-3, strain JS, has been engineered to express the EGFP protein and was used to trace the infection of HPIV-3 exclusively to the apical surface of ciliated airway epithelium by attaching to α2-6-linked sialic acid receptors (Zhang et al., 2005). Another benefit of a recombinant virus that expresses a reporter gene is the ability to detect and measure virus replication in real-time, which is directly linked to the expression of the foreign reporter gene. This measurement of real-time replication has direct implications in antiviral compound screening in vitro. The traditional assays that are used for antiviral screening are labor intensive and require long incubation times that may compromise the cell controls. A detection system, such as a recombinant virus expressing a reporter gene that can be measured in real-time, may shorten the amount of time needed to complete the antiviral assay and allow direct viral replication measurement. This has been accomplished for antiviral compound screening purposes by inserting the gene for the EGFP into the genomes of recombinant Ebola virus, cytomegalovirus, and HPIV-3 (Mao et al., 2008; Marschall et al., 2000; Roth et al., 2009; Towner et al., 2005).
Critical Parameters and Troubleshooting
To ensure that a full-length viral cDNA clone can be generated, high-quality, intact viral RNA needs to be isolated. Traditional RNA isolation, e.g. Trizol extraction, can be used but assurance that reagents are nuclease-free, e.g. DEPC treated, is labor intensive and time consuming. Commercial kits are available that guarantees their components are nuclease-free and result in similar quantities of purified RNA. However, laboratory bench space and common laboratory equipment, e.g. pipets, which are used communally, should be decontaminated or, ideally, dedicated space and pipets should be set aside and reserved solely for RNA work. Gloves should also be worn at all times to prevent nuclease contamination from hands. In addition, new, clean, and nuclease-free pipet tips, preferably aerosol-resistant, and microcentrifuge tubes should also be used at all times when working with RNA. Isolated RNA should be stored at −80°C to prevent degradation.
Generating a viral cDNA clone by RT-PCR amplification is also dependent on accurate primer design. The numbers of known viral genomic sequences are increasing and can be rapidly found through GenBank. Therefore, the procedure to design primers to match the genome of the desired virus with known sequence can be easily done. For example, the genomic sequence for the virus used in this protocol, HPIV-3 strain 14702, is located in GenBank, accession no. EU424062. On the other hand, if the user of this protocol is attempting to clone a virus whose genomic sequence is unknown, then primer design can be complex and problematic. Several options may be considered. First, if the genomic sequences of other strains of the same virus the user is attempting to clone are known, then a consensus sequence of all known sequences can be assembled. This consensus will show conserved sequences in the viral genome that can be targeted by primers with a high degree of certainty of primer annealing. Second, a combination of 3’ and 5’ RACE and shotgun sequencing of the viral genome can also give insight to the actual sequence of the 3’ and 5’ genomic ends and internal genomic regions, which could be used for primer design. Once the genomic 3’ end sequence is known, the primer that will anneal to this site can be designed to contain nontemplated restriction sites and the T7 promoter sequence adjacent to the first viral nucleotide separated by two guanosine residues.
Once the primer sequence has been decided on, the production of the primers is also important. During the synthesis of primers the length of the primer ordered represents only a proportion of the primer actually in the tube. Therefore, the costly option to purify each primer, e.g. HPLC or PAGE, may be desired. This need to purify primers is especially crucial when synthesizing long primers, >30 nucleotides, and primers used for cloning purposes in PCR that contain additional, nontemplated nucleotides on the primer’s 5’ end, which may code for restriction sites. In addition, if the resulting PCR product is to be used directly in a ligation reaction, the presence of a phosphate on the 5’ end of the primer will facilitate the ligation of the product into the digested plasmid, especially if the plasmid is dephosphorylated or blunt ended. Finally, when reconstituting the primers used during the cDNA synthesis step, special consideration is needed. Since these primers will anneal to the viral genomic RNA strand, they need to be reconstituted in nuclease-free water or buffer and handled identically as an RNA sample would be handled.
A possible unforeseen and uncontrollable circumstance that may lead to the unsuccessful rescue of a recombinant virus could be the incorporation of unintentional lethal mutations in the viral genome. These mutations will most likely occur during reverse transcription of the viral genomic RNA into cDNA by the reverse transcriptase enzyme, which lacks proofreading capabilities. The size of most negative-stranded viruses, approximately 15 kb, increases the likelihood of one or more unintentional mutations; nonlethal with any luck. In addition, the relatively large size of the viral genomic RNA renders it highly unlikely to create a full-length viral cDNA in one strand because of the lack of processivity of the reverse transcriptase enzyme, which is inhibited by RNase H activity and RNA secondary structure. To counteract these inhibitors, reverse transcriptase enzymes should be obtained and tested that will lack RNase H activity and will be stable at temperatures up to 60°C in order to eliminate secondary structure. This protocol suggests the use of the ProStar First-Strand RT-PCR Kit (Stratagene) because of the lack of RNase H activity in the reverse transcriptase enzyme. An elevated incubation temperature should be used to disrupt secondary structure.
On the other hand, the advent of high-fidelity DNA polymerases with proofreading capabilities has increased the likelihood that sufficient amounts of PCR products can be obtained that are true to at least the cDNA template. This protocol suggests the use of the PfuTurbo Hotstart DNA polymerase (Stratagene) because of its high-fidelity, hotstart capabilities, and generation of blunt ends, which are needed in subsequent ligation reactions. Currently, there are additional DNA polymerases that are commercially available and possess higher fidelity rates than PfuTurbo, e.g. PfuUltra. When optimizing PCR conditions, an important aspect to consider is the primer annealing temperature. If the temperature is too low; non-specific bands appear; if too high, possibly no PCR products will be obtained. As a general rule, annealing temperatures should be 5°C below the lowest Tm of the primer pair but can be changed in either direction. The temperatures stated in this protocol are 5°C below the Tm for the primer pair and worked well with the GENEMate thermal cycler, but other annealing temperatures may give better results with different thermal cyclers. Lastly, this protocol suggests the use of the two-step approach to RT-PCR and discourages the use of the one-step approach because of the inclusion of low-fidelity DNA polymerase, compared to PfuTurbo or PfuUltra, in the super mixes.
The cloning steps in this protocol are probably the most time consuming and problematic because of the multiple steps. As long as the proper controls are run with each ligation reaction, troubleshooting should make the process less difficult. The main control that has proven to be the most useful during the cloning steps is a digested plasmid that ligates to itself in the absence of an insert. A majority of the cloning steps in this protocol involve the digestion of two restriction enzymes to allow for directional cloning. In the two enzyme system, the self ligated control will indicate whether the plasmid has been digested by both enzymes. Ideally, no transformants should be seen on the agar plate following transformation if both enzymes cut. On the other hand, if an abundance of transformants can be seen following transformation, this indicates that one of the two enzymes did not cut the plasmid of interest and that the digestions should be repeated. A majority of the time, sequential digestions cut the DNA more efficiently than simultaneous digestions, even if the manufacturer indicates the enzymes are compatible in one buffer during a double digestion. Also, extending the length of incubation time for the second digestion also increases the number of plasmids that are digested with the enzymes and increases cloning efficiencies. In addition, as the length of the genomic cDNA plasmid increases, the transformation efficiency may decrease. If no transformants are seen on the agar plates and the controls indicate that both enzymes cut the plasmid, this suggests that the ligase enzyme is functional and that the bacterial cells are competent. One should then increase the volume of the transformants plated onto the agar plates until bacterial colonies are seen.
Finally, during the rescue of the infectious, recombinant negative-stranded virus several factors are important and noteworthy. First, the use of vTF7-3 to supply the T7 RNA polymerase and drive transcription of the genomic RNA and NP, P, and L transcripts has proven very efficient. However, vTF7-3 replicates very well in HeLa and other cell lines and can cause severe virus-induced CPE that may hinder the rescue procedure. Even though the replication of vTF7-3 can be controlled with the addition of Ara-C to the medium, the high rate of replication and resulting CPE may outweigh the benefits in some rescue systems. Alternatively, the modified vaccinia virus Ankara/T7 recombinant, MVA/T7 also obtained from Dr. Bernard Moss’s lab, may be substituted for vTF7-3, which is replication-deficient in mammalian cells (Wyatt et al., 1995). However, the MVA/T7 virus is not as efficient at expressing the T7 RNA polymerase as vTF7-3, but may be overcome by increasing the MOI of the MVA/T7 during the rescue phase. Second, the amounts of the four plasmids used for transfection during the rescue can be varied. The amounts of each plasmid used in this protocol were derived from the successful rescue of the infectious, recombinant HPIV-3 strain 47885 (Hoffman and Banerjee, 1997). However, other ratios of the four plasmids may also be used to successfully rescue infectious, recombinant negative-stranded viruses, such as the ratios used to rescue HPIV-3 strain JS, SeV, and MeV (Durbin et al., 1997a; Garcin et al., 1995; Radecke et al., 1995)
Anticipated Results
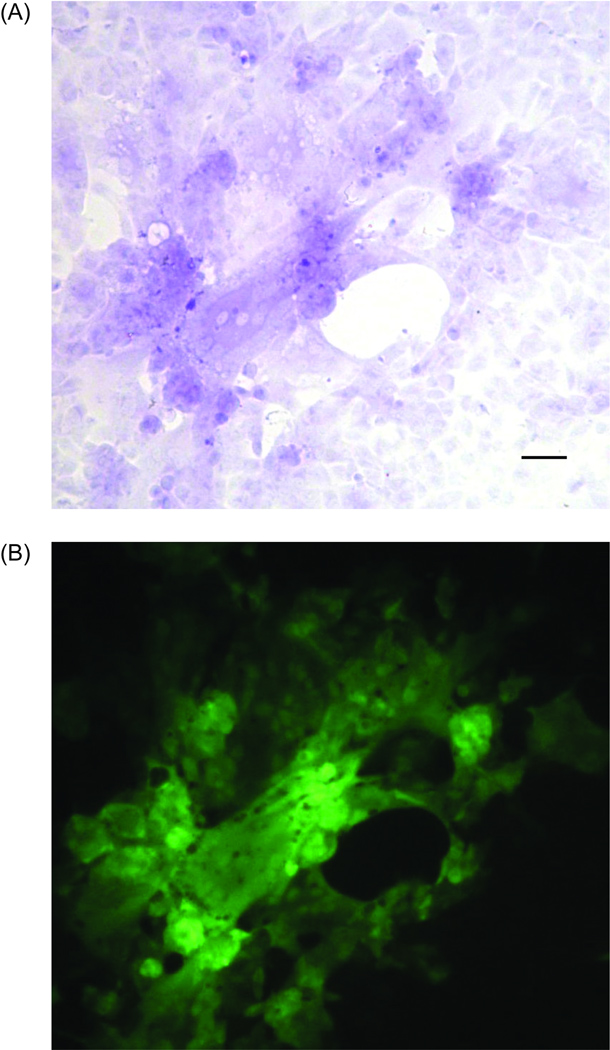
The plaques induced from the resulting rHPIV3-EGFP virus can be differentiated from wild-type HPIV-3 virus-induced plaques by visualization of green fluorescence emitted from infected cells under fluorescent microscopy (Figure 1). In addition, the replication of the rHPIV3-EGFP virus can be directly quantitated after 48 hours using a fluorometer by measuring the amount of EGFP expression in infected cells.
Figure 1.
MA-104 cells showing an rHPIV3-EGFP induced plaque. (A) Plaque stained with crystal violet and photographed with bright field microscopy. (B) Same plaque photographed using fluorescent microscopy, excitation between 450 and 490 nm and emission >515 nm. Scale bar, 50 µm.
Time Considerations
The overall procedure to rescue and purify an infectious rHPIV3-EGFP virus, strain 14702, from wild-type viral RNA can be completed in approximately 9–12 months since the viral genomic sequence is already known. However, if the reader is attempting this protocol using viral RNA isolated from the infection from a different strain of the HPIV-3 virus, more time may be needed to completely sequence the genome of the new HPIV-3 virus. Ideally, the sequence of the 5’ and 3’ ends of the virus are the most critical to know and should be sequenced first to speed up the project.
Acknowledgements
This work was supported by Contract No. N01 AI-30048 from the Virology Branch, NIAID, NIH. Joseph K.-K. Li was also supported, in part, by AES project 537/538.
Literature Cited
- Bukreyev A, Yang L, Zaki SR, Shieh WJ, Rollin PE, Murphy BR, Collins PL, Sanchez A. A single intranasal inoculation with a paramyxovirus-vectored vaccine protects guinea pigs against a lethal-dose Ebola virus challenge. J. Virol. 2006;80:2267–2279. doi: 10.1128/JVI.80.5.2267-2279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calain P, Roux L. Functional characterisation of the genomic and antigenomic promoters of Sendai virus. Virology. 1995;212:163–173. doi: 10.1006/viro.1995.1464. [DOI] [PubMed] [Google Scholar]
- Durbin AP, Hall SL, Siew JW, Whitehead SS, Collins PL, Murphy BR. Recovery of infectious human parainfluenza virus type 3 from cDNA. Virology. 1997a;235:323–332. doi: 10.1006/viro.1997.8697. [DOI] [PubMed] [Google Scholar]
- Durbin AP, Siew JW, Murphy BR, Collins PL. Minimum protein requirements for transcription and RNA replication of a minigenome of human parainfluenza virus type 3 and evaluation of the rule of six. Virology. 1997b;234:74–83. doi: 10.1006/viro.1997.8633. [DOI] [PubMed] [Google Scholar]
- Friedman H, Macarak E, MacGregor R, Wolfe J, Kefalides N. Virus infection of endothelial cells. J. Infect. Dis. 1981;143:266–273. doi: 10.1093/infdis/143.2.266. [DOI] [PubMed] [Google Scholar]
- Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinski MS. Annotated nucleotide and protein sequences for selected paramyxoviridae. In: Kingsbury DW, editor. The Paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 537–568. [Google Scholar]
- Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MA, Banerjee AK. An infectious clone of human parainfluenza virus type 3. J. Virol. 1997;71:4272–4277. doi: 10.1128/jvi.71.6.4272-4277.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Sakai Y, Shioda T, Kondo T, Nakanishi M, Nagai Y. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells. 1996;1:569–579. doi: 10.1046/j.1365-2443.1996.d01-261.x. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Kolakofsky D. Paramyxoviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fundamental Virology. Philadelphia, PA: Lippincott Williams & Wilkins; 2001. pp. 689–724. [Google Scholar]
- Mao H, Thakur CS, Chattopadhyay S, Silverman RH, Gudkov A, Banerjee AK. Inhibition of human parainfluenza virus type 3 infection by novel small molecules. Antiviral Res. 2008;77:83–94. doi: 10.1016/j.antiviral.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall M, Freitag M, Weiler S, Sorg G, Stamminger T. Recombinant green fluorescent protein-expressing human cytomegalovirus as a tool for screening antiviral agents. Antimicrob. Agents Chemother. 2000;44:1588–1597. doi: 10.1128/aac.44.6.1588-1597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa K, Yamada A, Takeuchi K, Watanabe Y, Miyata H, Sato H. Genetic characterization of parainfluenza virus 3 derived from guinea pigs. J. Vet. Med. Sci. 1998;60:919–922. doi: 10.1292/jvms.60.919. [DOI] [PubMed] [Google Scholar]
- Perrotta AT, Been MD. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter MA. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JP, Li JK, Smee DF, Morrey JD, Barnard DL. A recombinant, infectious human parainfluenza virus type 3 expressing the enhanced green fluorescent protein for use in high-throughput antiviral assays. Antiviral Res. 2009;82:12–21. doi: 10.1016/j.antiviral.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakura M, Fukumura M, Inoue M, Fujikawa S, Maeda M, Watabe K, Kyuwa S, Yoshikawa Y, Hasegawa M. Sendai virus vector-mediated gene transfer of glial cell line-derived neurotrophic factor prevents delayed neuronal death after transient global ischemia in gerbils. Exp. Anim. 2003;52:119–127. doi: 10.1538/expanim.52.119. [DOI] [PubMed] [Google Scholar]
- Stokes A, Tierney EL, Murphy BR, Hall SL. The complete nucleotide sequence of the JS strain of human parainfluenza virus type 3: comparison with the Wash/47885/57 prototype strain. Virus Res. 1992;25:91–103. doi: 10.1016/0168-1702(92)90102-f. [DOI] [PubMed] [Google Scholar]
- Storey D, Dimock K, Kang C. Structural characterization of virion proteins and genomic RNA of human parainfluenza virus 3. J. Virol. 1984;52:761–766. doi: 10.1128/jvi.52.3.761-766.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapparel C, Maurice D, Roux L. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J. Virol. 1998;72:3117–3128. doi: 10.1128/jvi.72.4.3117-3128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- Voytas D. Agarose gel electrophoresis. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. Hoboken, N.J.: John Wiley & Sons; 2000. pp. 2.5A.1–2.5A.9. [Google Scholar]
- Wertz GW, Perepelitsa VP, Ball LA. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3501–3506. doi: 10.1073/pnas.95.7.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt LS, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- Yonemitsu Y, Kitson C, Ferrari S, Farley R, Griesenbach U, Judd D, Steel R, Scheid P, Zhu J, Jeffery PK, Kato A, Hasan MK, Nagai Y, Masaki I, Fukumura M, Hasegawa M, Geddes DM, Alton EW. Efficient gene transfer to airway epithelium using recombinant Sendai virus. Nat. Biotechnol. 2000;18:970–973. doi: 10.1038/79463. [DOI] [PubMed] [Google Scholar]
- Zhan X, Hurwitz JL, Krishnamurthy S, Takimoto T, Boyd K, Scroggs RA, Surman S, Portner A, Slobod KS. Respiratory syncytial virus (RSV) fusion protein expressed by recombinant Sendai virus elicits B-cell and T-cell responses in cotton rats and confers protection against RSV subtypes A and B. Vaccine. 2007;25:8782–8793. doi: 10.1016/j.vaccine.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Bukreyev A, Thompson CI, Watson B, Peeples ME, Collins PL, Pickles RJ. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 2005;79:1113–1124. doi: 10.1128/JVI.79.2.1113-1124.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]