Abstract
Intricate interactions between kinetochores and microtubules are essential for the proper distribution of chromosomes during mitosis. A crucial long-standing question is how vertebrate kinetochores generate chromosome motion while maintaining attachments to the dynamic plus ends of the multiple kinetochore MTs (kMTs) in a kinetochore fibre. Here, we demonstrate that individual kMTs in PtK1 cells are attached to the kinetochore outer plate by several fibres that either embed the microtubule plus-end tips in a radial mesh, or extend out from the outer plate to bind microtubule walls. The extended fibres also interact with the walls of nearby microtubules that are not part of the kinetochore fibre. These structural data, in combination with other recent reports, support a network model of kMT attachment wherein the fibrous network in the unbound outer plate, including the Hec1–Ndc80 complex, dissociates and rearranges to form kMT attachments.
Kinetochores are proteinaceous complexes that perform at least four functions vital for accurate chromosome segregation during mitosis: attachment of chromosomes to the mitotic spindle; control of kMT dynamics; generation of force for chromosome movement; and generation of a cell-cycle checkpoint that delays anaphase onset until all chromosomes are attached to the mitotic spindle and aligned at the spindle equator1-5. In Saccharomyces cerevisae, the kinetochore is a molecularly well-defined structure that binds a single kMT1,6-8. However, in most other eukaryotic organisms, including vertebrates, individual kinetochores bind multiple kMTs9-11. The prevailing repeat-subunit model postulates that vertebrate kinetochores are constructed from 30 or more kMT ‘docking sites’, each of which is functionally equivalent to the S. cerevisae kMT binding site6,12,13. Thus, each of the putative kMT docking sites is envisioned as having a complete set of the kinetochore functions. Thus far, however, there has been no ultrastructural visualization of a kMT binding site in the kinetochore.
We tested the repeat-subunit model by using electron tomography to construct a high-resolution structural map of the kinetochore outer plate in PtK1 cells (Figs 1 and 2, and see Supplementary Information, Movies 1 and 2). In vertebrates, the kinetochore outer plate is a distinctive, disk-shaped structure that forms just outside the surface of the centromere during mitosis (Fig. 1a–d)4,9. As ~90% of the kMT plus ends terminate in the outer plate, it is the logical location for putative kMT docking sites14. Electron tomography reveals that before microtubule binding the outer plate is a distinct network of crosslinked fibres about 10 nm in diameter. Some of the fibres are at least 80–90 nm in length (Figs 1e–h and 2a–d, and see Supplementary Information, Movies 1 and 2). These long fibres are roughly parallel to one another and are aligned in the plane of outer plate. There are nodes on the long fibres where cross-links occur. Analysis of neighbouring regions of the tomographic reconstruction demonstrates that the network of long, crosslinked fibres is unique to the outer plate (see Supplementary Information, Fig. S1 and Movie 1).
Figure 1.
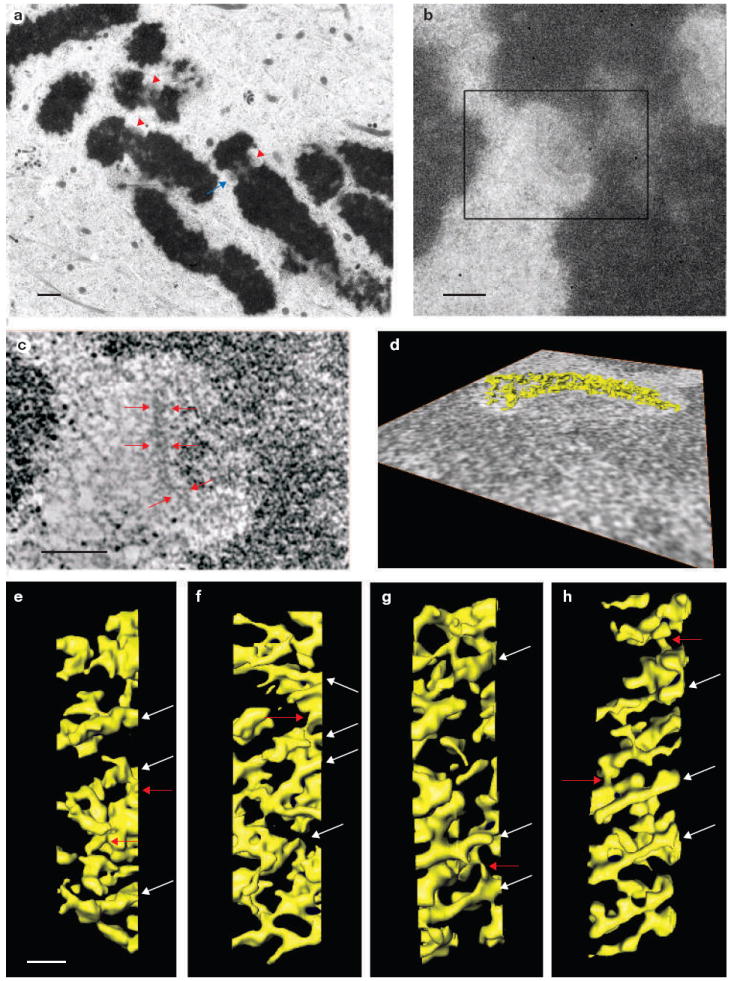
Electron tomography reveals that the unbound kinetochore outer plate is a fibrous network. (a) Lower magnification image of a nocodazole-treated mitotic PtK1 cell prepared by high-pressure freezing and freeze-substitution (see Methods). Red arrowheads indicate kinetochores visible in the 200-nm thick plastic section. The blue arrow indicates the kinetochore shown in b–d. (b) A higher magnification image reveals the fine structure of the kinetochore indicated by the blue arrow in a. The kinetochore outer plate appears as a fibrous mat with a thickness of 50–75 nm. (c) A 1.6-nm thick edge-view slice from the three-dimensional tomographic reconstruction of the boxed area in b. Red arrows indicate the fibrous outer plate. (d) Surface rendering of the outer plate. The outer plate fibres were segmented from the rest of the three-dimensional reconstruction and displayed as a surface-rendered volume using the Amira software. The viewing direction of the surface rendering is indicated by an overlay with the 1.6-nm slice shown in c (see Supplementary Information, Movies 1 and 2). (e–h) Face views of surface renderings of the outer plates from four of the 12 unbound kinetochores examined. The outer plate consistently appears as a network formed from crosslinked fibres that are aligned in the outer plate disk, and often parallel to one another. White arrows indicate long fibres and red arrows indicate crosslinks. The scale bars present 1 μm in a, 400 nm in b, 200 nm in c and 50 nm in e–h.
Figure 2.
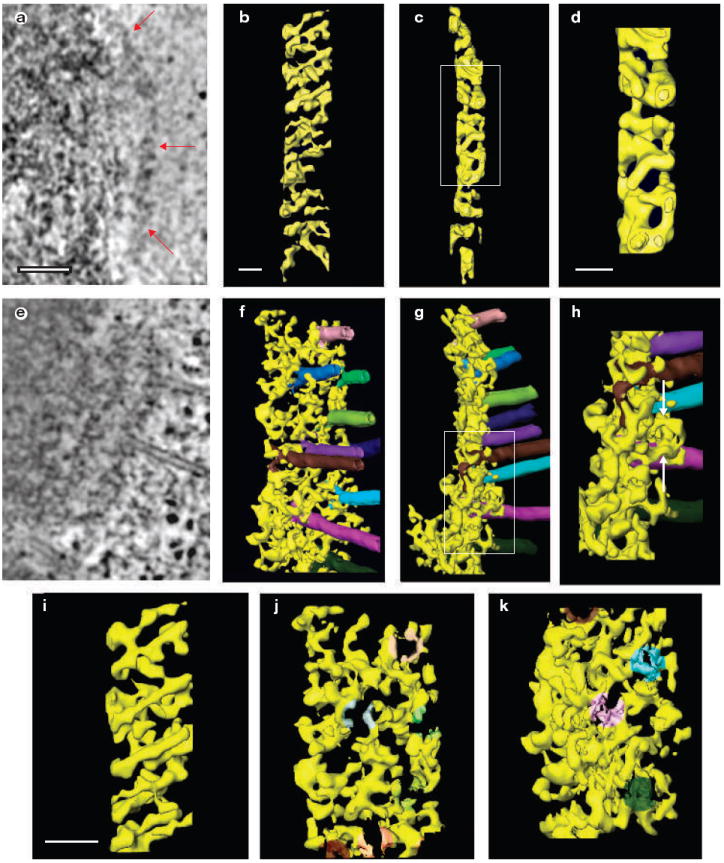
The fibrous network of the outer plate rearranges when microtubules bind. (a–h) Edge-view slices (1.6 nm thick) from the three dimensional reconstructions of kinetochore outer plates in cells treated (a), or untreated (e) with nocodazole to remove all microtubules. Red arrows indicate the outer plate in a. Surface rendered views of kinetochore outer plates in cells treated (b, c) or untreated (f, g) with nocodazole. Face views, as observed from the cytoplasmic side, are shown in b and f. The view in f is 15° from an exact face view. Edge views are shown in c and g. The fibrous network of unbound kinetochore becomes more disorganized and less well aligned in the outer plate when microtubules bind (see Supplementary Information, Movies 2 and 4). Higher-magnification views of boxed areas in c and g are shown in d and h, respectively. White arrows in h indicate fibres that orient along the microtubule axis and bind along the microtubule wall. (i–k) Higher magnification face views of unbound (i) and bound (j, k) kinetochores. The image in i is taken from the upper portion of b, and j and k are taken from the upper and lower portions of f. The kMTs have been digitally removed from j and k to visualize the rearrangement of outer plate fibres on microtubule attachment. The shading in j and k indicates the location of the kMT of the same colour in f. In general, the long fibres seem to disassemble into shorter units and rearrange their orientation on kMT attachment. The scale bars represent 100 nm in a, and 50 nm in b, d and i.
The fibrous nature of the outer plate is consistent with the rod shape observed for the Ndc80 complex (a known molecular component of the outer plate) and the elongated shapes predicted for other possible outer plate proteins8,15-20. The general arrangement of the outer-plate fibres that was observed is also largely consistent with the results of our previous electron tomography study of the unbound kinetochore outer plate, except that due to improved methods for specimen preparation (see Methods) a relatively open network with large channels and gaps between the fibres was observed (Figs 1e–h and 2b, and see Supplementary Information, Movie 2)21,22.
We do not find any evidence for a series of unit motifs that could serve as the microtubule docking sites proposed by the repeat-subunit model6,13. Even when the outer plate seems to be constructed from distinct subunits in some two-dimensional slices of the three-dimensional reconstructions, this observation only extends for around 10–12 nm in the volume. Surface-rendered views demonstrated that the subunit appearance arises from sections of fibres that border wide channels or rings in the network (see Supplementary Information, Fig. S2a–c and Movie 3). Similarly, in contrast with previous reports, structural differences between the inner and outer surfaces of the outer plate were not detected, and the three-dimensional structure of outer plate did not exhibit a double-layer organization (see Supplementary Information, Fig. S2c–e and Movie 3)9, 21. We also found no evidence of the 50-nm rings that are formed around the microtubules in vitro by the Dam1–DASH from S. cerevisae, either in the kinetochore outer plate or on bound kMTs23,24. Small rings (13 ± 2 nm in diameter) were consistently observed, generally embedded at nodes in the outer plate network (see Supplementary Information, Fig. S3), but these rings were smaller in diameter than microtubules, and microtubule binding did not decrease the number of rings observed.
When microtubules bound, the outer-plate meshwork becomes less organized (Fig. 2a, b, e and f) and seemed to be thicker when viewed edge-on (Fig. 2c, d, g and h). The thicker appearance arose from fibres extending from the outer plate and binding to the sides of microtubules (Fig. 2h and see Supplementary Information, Movie 4). Other fibres changed their orientation within the plane of outer plate to form a radial pattern of attachments to the tips of microtubule plus ends (Fig. 3a, b and d, and see Supplementary Information, Movies 4 and 5). Figure 3c shows a striking example of the two types of binding. In agreement with previous serial-section electron microscopy, the average spacing between bound kMTs was 71 ± 15 nm, with occasional kMTs spaced as close as 40 nm (range = 40–92 nm; n = 26 kMTs in three kinetochores)25.
Figure 3.
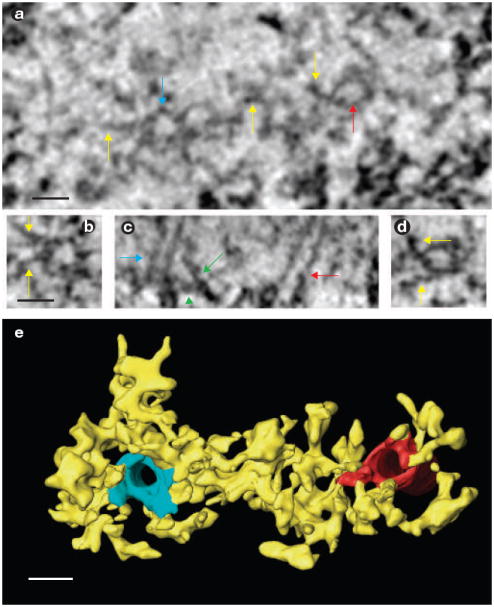
Two distinct mechanisms for kMT attachment in a kinetochore. (a) A face-view slice from the tomographic reconstruction of an en face section (see Methods) of a bound kinetochore. The viewing direction is looking towards the spindle pole. The plus-end tips of two microtubules are indicated by blue and red arrows, and outer plate fibres are indicated by yellow arrows. (b) High-magnification window of the microtubule plus end indicated by the blue arrow in a. Yellow arrows indicate the fibres on the outer plate that bind to the kMT tips in a radial array. (c) An edge-view slice through the three-dimensional reconstruction. Blue and red arrows indicate the same microtubules as in a. The green arrow indicates a fibre extending out of the outer plate to bind the microtubule wall. The green arrowhead indicates the fibres within the outer plate attaching to a kMT plus-end. (d) High-magnification window of the microtubule plus end indicated by the red arrow in a. Yellow arrows indicate the fibres on the outer plate that bind to the kMT tips in a radial array. (e) Surface rendering of the volume viewed from approximately the same orientation as in a. Fibres are attached to the kMT plus ends and link adjacent kMTs. The scale bars represent 25 nm in a, b and e.
The semi-regular spacing of kMTs was similar to the semi-ordered arrangement of long fibrous elements found in the unbound outer plate (Figs 1e–h and 2b). Except for the occasional large gaps, the spacing between outer plate fibres was 47 ± 6 nm with a range of 39–57 nm (n = 11 in four kinetochores). These results suggest that minimum spacing between kMTs is limited by the minimum spacing between outer plate fibres. Figure 2i–k and Movie 6 show that a fibrous pattern is still present in the outer plate after kMT attachment, but that many of the fibres are shorter and radially arranged around kMT tips. Thus, the minimum spacing between kMTs seems to be determined by the radial arrangement of outer plate fibres that bind kMT tips, which in turn is determined by the spacing of fibres in the unbound outer plate.
In addition to two types of binding for end-on kMT attachments, the outer plate formed attachments to the walls of microtubules that pass close enough to surface of outer plate for such interactions to form (Fig. 4a–d). In contrast with end-on kMTs attachments, these laterally associated microtubules passed tangential to one surface or edge of the outer plate without embedding in, or penetrating through, the outer plate. In general, the laterally associated microtubules terminate outside of the reconstruction area, and can even be part of another kinetochore fibre26. The ratio of end-on bound to laterally associated microtubules was 4.5–5.5 in both control and taxol-treated cells (Fig. 4). As most vertebrate kinetochores bind 15–30 microtubules, we estimate that on average 3–8 microtubules are laterally associated with every vertebrate kinetochore9-11. Laterally associated microtubules were attached to the kinetochore by groups of short outer plate fibres that were similar to the fibres that attach to the walls of the end-on bound kMTs (Fig. 4c–f). These observations suggest that a set of kinetochore fibres extends from the outer plate and binds to microtubule walls without distinguishing between end-on attached kMTs and other microtubules in the vicinity. In contrast with end-on bound kMTs, laterally associated microtubules were not stabilized against brief nocodazole treatment (Fig. 4). Thus, stabilization of kMTs arises from the radial array of outer plate fibres attached to the kMT tips, and these fibres have a distinct and independent function from those attached to kMT walls.
Figure 4.
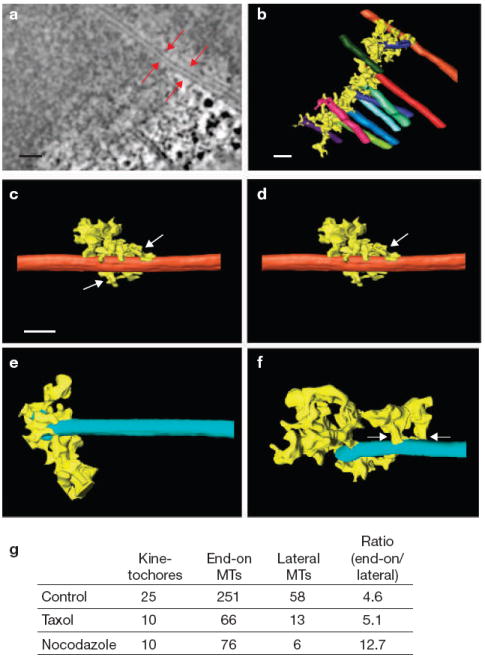
Comparison of the lateral and end-on outer plate attachments to microtubules. (a) A 1.6-nm edge-view slice from the three-dimensional reconstruction of a bound kinetochore, with red arrows indicating a laterally bound microtubule located near the edge of kinetochore. This microtubule is distinct from other kMTs because it extends well beyond the kinetochore without ever going directly through the outer plate. (b) A surface-rendered view of the kinetochore shown in a. The laterally bound microtubule is shown in orange at the top of the figure. (c) Higher-magnification view of the laterally bound microtubule shown in a and b. Outer plate attachments to the microtubule are indicated by white arrows. (d) A laterally bound microtubule from a different kinetochore, with outer plate fibres forming short attachments on the walls (white arrow). (e, f) Microtubules bound end-on, with outer plate fibres attached to the tip of plus ends (e) and on walls (f, white arrows). Note that the outer plate forms similar attachments on the walls of end-on and laterally bound microtubules. (g) The numbers of end-on bound microtubules and laterally bound microtubules found in electron tomographic reconstructions of kinetochores. The scale bars represent 100 nm in a, and 50 nm in b and c.
Although our study focuses on the structure of the outer plate, the kinetochore–kMT interactions must communicate with elements of the inner kinetochore1,2,27. In agreement with our earlier electron tomographic studies, current three-dimensional reconstructions showed that the outer plate is a distinct and freestanding entity that is connected to the inner centromere by fibrous links (see Supplementary Information, Fig. S4)21. We speculate that these linkers are composed of members of the CENP H–I complex that interact with both inner-centromere proteins and putative outer-plate proteins, such as Spc24/25, KNL-1 and the Mis12 complex28.
Our structural analysis found no evidence to support the repeat-subunit model, despite the use of optimal methods for specimen preparation and electron tomography. The repeat-subunit model is primarily derived from immunofluorescence microscopy data showing that kinetochores fragment into patches when under mechanical stretching, or when they enter into mitosis with an unreplicated genome12,13. However, electron microscopy has shown that these fragments are irregular in size and typically capable of binding several microtubules12,29. Therefore, the multiple patches observed by immunofluorescence microscopy seem to arise from the kinetochore outer plate breaking apart under tension at points where the structure is weaker because of large channels (for example, see Figs 1e–h and 2b). We think it is unlikely that the high concentration of motor and checkpoint proteins recruited to the kinetochore during prolonged nocodazole treatment masks unit microtubule-binding sites in our reconstructions of the unbound outer plate, as motor and checkpoint proteins rapidly turnover and are primarily located in the corona2. Furthermore, the long fibrous elements and their putative dissociated components are visible in the bound kinetochore where concentrations of checkpoint and motor proteins are much lower (Fig 2i–k and see Supplementary Information, Movie 6). Thus, although dynamic components (such as motor and checkpoint proteins) probably contribute to the irregularities observed in the unbound outer plate, it is unlikely that they make significant structural contributions that mask a repeating structural motif.
As an alternative to the repeat-subunit model, we propose that the kinetochore outer plate is a flexible network resembling a spider’s web, which captures or interacts with the microtubules it encounters. Our flexible-network model differs radically from current models that generally envision the kinetochore outer plate as a series of discrete microtubule-binding sites containing sleeves and/or rings6,13,24. However, in addition to being consistent with the structural data, our flexible-network model postulates a more efficient system for microtubule capture because the microtubules do not have to orient precisely into a docking site. Microtubules can contact the sticky network at a variety of angles, as is frequently observed (see Supplementary Information, Fig. S4d and Movie 6)25. The network model also solves the problem of how the kinetochore converts from lateral to end-on binding without falling off the spindle, because lateral attachments can simply remain intact while end-on attachments are added. Finally, the network model more readily accommodates data suggesting that various kinetochore functions are independent and can be uncoupled.
Technical barriers currently preclude directly placing molecular markers on the structural features of our tomographic reconstructions, but it is possible to postulate a credible, albeit speculative, model for the molecular architecture of the outer plate in vertebrate kinetochores (Fig. 5). Numerous studies have shown that the Hec1–Ndc80 complex is required for kMT attachment and chromosome alignment, is a 40–55 nm long rod, and is located in the outer plate2,15-17,20. KNL-1 and the Mis12 complex (predicted to be extended and globular structures, respectively) form a super complex with the Hec1–Ndc80 complex in vitro and are required for recruitment of the Hec1–Ndc80 complex to the kinetochore in vivo17-19. Taken together, these observations suggest that KNL-1 and the Hec1–Ndc80 complex are part of the long fibrous elements we detect in the unbound kinetochore outer plate, and that the Mis12 complex is a component of the nodes and/or other non-fibrous elements (Figs 1e –h, 2b and 5). We place Mis12 in the outer plate at the junction of KNL-1 and the Spc24/25 end of the Hec1–Ndc80 complex because Mis12 interacts with these components and helps recruit them to the outer plate17,19. In addition, our structural map contains contributions from CENP-F, a large fibrous molecule that is located in the outer plate30. It is likely that other kinetochore proteins also located in the outer plate contribute to the structural map.
Figure 5.
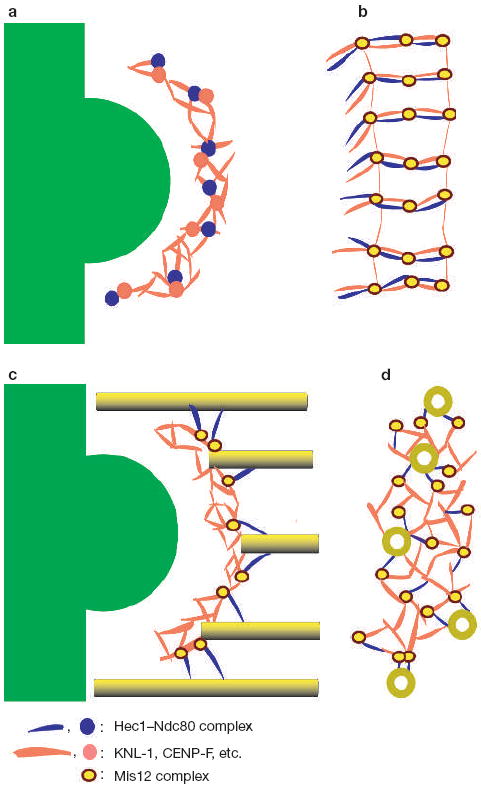
Schematic representation of a network model of the vertebrate kinetochore outer plate. (a, b) Unbound outer plate in the edge (a) and face (b) views. In b, the outer plate appears as a series of roughly parallel bundles of fibres that include the Hec1–Ndc80 complex (blue), KNL-1 (red) and other extended proteins, such as CENP-F (also red). The globular Mis12 complex (yellow circle with a brown border) is shown at the junction between Hec1–Ndc80 and KNL-1 because both the Mis12 complex and KNL-1 are required for recruiting the Hec1–Ndc80 complex to the kinetochore, and to the KNL-1–Mis12–Ndc80 super complex in vitro17,18. The outer plate undoubtedly contains other globular components corresponding to some of the features in our tomographic reconstructions. From the edge view (a), most of the bundles appear as dots. (c, d) Bound outer plate in the edge (c) and face (d) views. When microtubules attach, the long bundles of fibres dissociate and the Hec1–Ndc80 complex reorients from being in the plane of the outer plate to binding to the microtubule wall at a shallow angle to the microtubule axis (c). Other components in the long bundles (including KNL-1) reorient within the plane of the outer plate to form a radial array of attachments to the plus end tips of kMTs (d). Presumably, the Hec1–Ndc80 complex is also able to form attachments to the walls of laterally associated microtubules. The chromosome, heterochromatin and inner centromere are shown in green in a and c.
Hec1–Ndc80, KNL-1 and CENP-F all exhibit weak microtubule binding, and there is evidence of synergy when more than one putative outer plate component is present18,30. It has been postulated that each kMT is bound by an array of low-affinity components, rather than a few high-affinity components18. It has been suggested that this arrangement provides the flexibility required to maintain stable attachment to dynamic kMT plus ends. Our finding that the outer plate forms multiple attachments to both the tip and wall of each kMT supports this view (Figs 3b–d and 5). The low-affinity array model is also supported by the finding that metaphase kinetochores in S. cerevisae contain three copies of CENP-F, five copies of KNL-1 and eight copies of Ndc80 in a single kMT binding site6. Interestingly, vertebrate kinetochores have approximately 800–1200 copies of Ndc80 and Mis12, which, according to the S. cerevisae results, is 3–10-fold excess over what is required to bind the15–30 kMTs typically found on vertebrate kinetochores19. This supports our contention that kMT binding activity is dispersed over a network, rather than being efficiently packaged into unit binding sites.
We hypothesize that the Hec1–Ndc80 complex is one of the fibres extending out to attach kMT walls (Fig. 5) because of the similarity of our images with images of the Hec1–Ndc80 complex binding to microtubules in vitro (Fig. 3c and ref. 18). This assignment is also in agreement with high-resolution immunofluorescence microscopy data indicating that, on bi-oriented chromosomes, the rod-shaped Hec1–Ndc80 complex is oriented approximately perpendicular to the plane of the outer plate, with the Spc24/24 end towards the inner centromere and the microtubule binding Ndc80–Nuf2 end towards the cytoplasm31. However, our tomographic reconstructions make the additional prediction that the Hec1–Ndc80 complex only acquires this orientation on binding to kMTs because before kMT attachment all fibrous elements are oriented in the plane of the outer plate (Fig. 2). There is unpublished immunofluorescence microscopy data that supports this hypothesis (DeLuca, J. and Salmon, E.D., unpublished observations).
Further efforts to identify molecular components corresponding to the structural features in the kinetochore outer plate will require combining electron tomographic reconstruction with immunolocalization, molecular depletion, point mutations resulting in loss of protein function and high-resolution light microscopy methods. The structural data presented here provide a framework for designing and interpreting these future studies, which will ultimately lead to a molecular understanding of the structure and function of the kinetochore outer plate.
METHODS
Specimen preparation
PtK1 cells were cultured on sapphire disks (3.0 × 0.05 mm; Rudolf Brugger, Minusio, Switzerland) coated with carbon and fibronectin and prepared for electron microscopy as described previously14. Some cells were treated with 10 μM nocodazole for 3 h to provide kinetochores without microtubules. At 60–80% confluency, disks with mitotic cells were briefly immersed in media containing 10% Ficoll (relative molecular molecular mass, 70,000; Sigma, St Louis, MO), placed in a specimen holder and immediately high-pressure frozen in a Bal-Tec HP10 high-pressure freezer. Freeze-substitution was carried out in a Bal-Tec FSU 010 at −90 °C for approximately 24 h in 0.5% glutaraldehyde with 0.1% tannic acid, followed by 24 h in 1% OsO4 with 0.1% uranyl acetate. At the end of freeze-substitution, the sapphire disks were brought to room temperature over 5–6 h, placed overnight in 50% Epon, embedded in 100% Epon and polymerized at 60 °C for 48 h. Flat-embedded specimens were remounted onto plastic blocks, cut into 150–250 nm thick serial sections with a diamond knife, and collected on formvar-coated copper grids.
For cells sectioned en face, a phase-contrast microscope was used to examine flat-embedded specimens and to locate mitotic cells. The embedded monolayer was then trimmed to a small square block so that one edge was close in proximity and parallel to the chromosomes in the mitotic spindle. The trimmed block was then tipped on its side so that the cell of interest was on one side, near the top, and the block was then remounted onto an Epon peg. The block was trimmed down to include the cell of interest and sectioned at 200–250 nm, with the mitotic spindle oriented perpendicular to the cutting direction.
Electron tomography
Serial sections of mitotic cells were scanned at low magnification (5,000×) in a Zeiss 910 transmission electron microscope. Kinetochores with microtubules bound were identified by the location of microtubule plus-end attachments, whereas the unbound kinetochores were identified by the ribosome-exclusion zones and accompanying plate-like structures located at the primary constriction of the chromosome. A Tecnai F20 transmission electron microscope (FEI, Eindhoven, The Netherlands) with a Gatan 2Kx 2K CCD camera was used to record the tomographic tilt series at 200 kV with a pixel size of 1.6 nm. Double-tilt series were collected over an angular range of −70° to +70°, and the angular interval was varied according to the cosine of the tilt angle, with a 2.0° increment at the untilted image32. Tomographic reconstructions were computed as previously described14,32.
Surface rendering and data analysis
Segmentation of the kinetochore outer plate was achieved using a three-dimensional interactive tool in the AMIRA software package (TGS Inc., San Diego, CA). Accurate boundaries of the outer plate components were set on each slice of the volume and verified from an orthogonal direction. Surface rendering was carried out on the stack of segmented slices.
Supplementary Material
Acknowledgments
We thank R. Glaser, M. Koonce and T. Wagenknecht and for helpful comments on the manuscript. This work was supported by National Institutes of Health R01 GM06627 to B.F.M and GM59363 to A.K. We also acknowledge National Institutes of Health P41 RR01219 and National Science Foundation DBI9871347 to J. Frank to support the Wadsworth Center’s Resource for Visualization of Biological Complexity and the FEI Tecnai F20 electron microscope. Finally, we are grateful for technical support from the Wadsworth Center’s Core Facility for Electron Microscopy.
Footnotes
AUTHOR CONTRIBUTIONS B.F.M. directed the work. Y.D. and B.F.M. were responsible for experimental planning. K.J.V., Y.D. and B.F.M. preprared the specimens. K.J.V. and X.M. were responsible for data collection. Y.D. and K.J.V. computed tomographic reconstructions and Y.D. segemented the volumes. Y.D., B.F.M. and A.K. were responsible for data analysis and writing.
COMPETING FINANCIAL INTERESTS The authors declare that they have no competing financial interests.
Note: Supplementary Information is available on the Nature Cell Biology website.
References
- 1.Tanaka TU, Stark MJ, Tanaka K. Kinetochore capture and bi-orientation on the mitotic spindle. Nature Rev Mol Cell Biol. 2005;6:929–942. doi: 10.1038/nrm1764. [DOI] [PubMed] [Google Scholar]
- 2.Maiato H, Deluca J, Salmon ED, Earnshaw WC. The dynamic kinetochore-microtubule interface. J Cell Sci. 2004;117:5461–5477. doi: 10.1242/jcs.01536. [DOI] [PubMed] [Google Scholar]
- 3.McIntosh JR, Grishchuk EL, West RR. Chromosome-microtubule interactions during mitosis. Annu Rev Cell Dev Biol. 2002;18:193–219. doi: 10.1146/annurev.cellbio.18.032002.132412. [DOI] [PubMed] [Google Scholar]
- 4.Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 6.Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nature Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheeseman IM, Drubin DG, Barnes G. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J Cell Biol. 2002;157:199–203. doi: 10.1083/jcb.200201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinkley BR, Stubblefield E. The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma. 1966;19:28–43. doi: 10.1007/BF00332792. [DOI] [PubMed] [Google Scholar]
- 10.McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol. 1997;137:1567–1580. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McEwen BF, et al. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol Biol Cell. 2001;12:2776–2789. doi: 10.1091/mbc.12.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.VandenBeldt KJ, et al. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr Biol. 2006;16:1217–1223. doi: 10.1016/j.cub.2006.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciferri C, et al. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–29095. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 17.Cheeseman IM, et al. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 19.Emanuele MJ, McCleland ML, Satinover DL, Stukenberg PT. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol Biol Cell. 2005;16:4882–4892. doi: 10.1091/mbc.E05-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLuca JG, et al. Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell. 2005;16:519–531. doi: 10.1091/mbc.E04-09-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McEwen BF, Arena JT, Frank J, Rieder CL. Structure of the colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography. J Cell Biol. 1993;120:301–312. doi: 10.1083/jcb.120.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL. A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma. 1998;107:366–375. doi: 10.1007/s004120050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nature Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 24.Westermann S, et al. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 25.Rieder CL. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 1981;84:145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- 26.Kapoor TM, et al. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311:388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 28.Okada M, et al. The CENP–H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nature Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 29.Khodjakov A, Cole RW, McEwen BF, Buttle KF, Rieder CL. Chromosome fragments possessing only one kinetochore can congress to the spindle equator. J Cell Biol. 1997;136:229–240. doi: 10.1083/jcb.136.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng J, Huang H, Yen TJ. CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma. 2006;115:320–329. doi: 10.1007/s00412-006-0049-5. [DOI] [PubMed] [Google Scholar]
- 31.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 32.McEwen BF, Marko M, Hsieh CE, Mannella C. Use of frozen-hydrated axonemes to assess imaging parameters and resolution limits in cryoelectron tomography. J Struct Biol. 2002;138:47–57. doi: 10.1016/s1047-8477(02)00020-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


