CONSPECTUS
Protein aggregation can be defined as the sacrifice of stabilizing intrachain contacts of the functional state that are replaced with interchain contacts to form non-functional states. The resulting aggregate morphologies range from amorphous structures without long-range order typical of nondisease proteins involved in inclusion bodies to highly structured fibril assemblies typical of amyloid disease proteins. In this Account, we describe the development and application of computational models for the investigation of nondisease and disease protein aggregation as illustrated for the proteins L and G and the Alzheimer’s Aβ systems.
In each case, we validate the models against relevant experimental observables and then expand on the experimental window to better elucidate the link between molecular properties and aggregation outcomes. Our studies show that each class of protein exhibits distinct aggregation mechanisms that are dependent on protein sequence, protein concentration, and solution conditions. Nondisease proteins can have native structural elements in the denatured state ensemble or rapidly form early folding intermediates, which offers avenues of protection against aggregation even at relatively high concentrations. The possibility that early folding intermediates may be evolutionarily selected for their protective role against unwanted aggregation could be a useful strategy for reengineering sequences to slow aggregation and increase folding yield in industrial protein production. The observed oligomeric aggregates that we see for nondisease proteins L and G may represent the nuclei for larger aggregates, not just for large amorphous inclusion bodies, but potentially as the seeds of ordered fibrillar aggregates, since most nondisease proteins can form amyloid fibrils under conditions that destabilize the native state.
By contrast, amyloidogenic protein sequences such as Aβ1–40,42 and the familial Alzheimer’s disease (FAD) mutants favor aggregation into ordered fibrils once the free-energy barrier for forming a critical nucleus is crossed. However, the structural characteristic and oligomer size of the soluble nucleation species have yet to be determined experimentally for any disease peptide sequence, and the molecular mechanism of polymerization that eventually delineates a mature fibril is unknown. This is in part due to the limited experimental access to very low peptide concentrations that are required to characterize these early aggregation events, providing an opportunity for theoretical studies to bridge the gap between the monomer and fibril end points and to develop testable hypotheses. Our model shows that Aβ1–40 requires as few as 6–10 monomer chains (depending on sequence) to begin manifesting the cross-β order that is a signature of formation of amyloid filaments or fibrils assessed in dye-binding kinetic assays. The richness of the oligomeric structures and viable filament and fibril polymorphs that we observe may offer structural clues to disease virulence variations that are seen for the WT and hereditary mutants.
Introduction
Evolution has guided the design of amino acid sequences such that globular proteins reliably assume a specific functional native state, precisely bringing together residues to form, for example, catalytic sites in enzymes or specific binding site architectures for protein complexation and signaling. The ability of the protein to find and maintain the native state is therefore dependent on an amino acid sequence that gives rise to a structural ensemble that is thermodynamically stable at the physiological pressures and temperatures and solution conditions in the normal cellular or extracellular environment. Destabilizing sequence mutations,1 chemical modification,2 or changes in protein concentration and solution environment of the protein3 can shift the equilibrium from the native state in favor of aggregates, that is, misfolded states with interchain contacts made with other proteins. These aggregates range from structurally amorphous collections of misfolded proteins often found in inclusion bodies when proteins are overexpresssed in bacterial hosts4 to fibrils with regular and repeating structure associated with a number of human diseases.1 In order to change deleterious aggregation outcomes, it is of critical importance to develop an understanding of the molecular driving forces for early and late aggregation events, which in turn might be reversed to prevent disease proteins from nucleating thermodynamically stable aggregate assemblies or to break up inclusion bodies to recover functional protein.
Though the gross morphology of large fibril aggregates can be investigated with current biochemical or protein structural experimental techniques,1,5 these are more limited in application to early aggregation events involving small and likely disordered oligomers at very dilute concentration. Molecular simulations currently offer great promise of directly observing the entire aggregation process in molecular detail. In this Account, we show how judicious use of coarse-grained models, validated against appropriate experimental observables, can characterize the aggregation thermodynamics and kinetic pathways at a level of detail and insight not possible with experiment alone. We use these models to quantify molecular mechanistic differences in aggregation outcomes for nondisease proteins L and G and the Aβ peptide indicted in Alzheimer’s disease.
Folding and Aggregation for Nondisease Proteins
Experimental evidence suggests that there is an increased propensity to aggregate for proteins that fold through kinetic intermediates.6 Since these states do not adopt the full complement of intrachain contacts made in the folded state, interchain attraction can develop between partially formed proteins. However, most proteins typically fold through intermediates due to the, on average, large size (>200 amino acids) and corresponding greater folding complexity. Furthermore, there is competition between the folding of protein monomers and the formation of oligomeric protein aggregates that derive from association of protein denatured states.7,8 Since folding and aggregation are thought to occur in parallel, it is assumed that at low protein concentration the possibly faster monomer folding pathway dominates,9 while at sufficiently high concentration, the folding protein is trapped into an oligomeric phase irreversibly or much more slowly converts aggregates to native monomer.8,9
However, if cellular thermodynamic conditions in the crowded cell were similar to the folding temperature midpoint used to study folding in vitro, in which ~50% of the protein population is unfolded or occupying stable intermediates, aggregation would be the far more common and detrimental outcome without protective mechanisms in place. While the unfolded protein response such as rescue by chaperonins and ubiquitin targeting for proteasomal degradation does exist to protect the cell against the build up of misfolded protein, a sustained and costly cellular level response in order for a given protein to reach a functional native state would seem to be a rather serious evolutionary flaw. That is, it would appear more likely that proteins would reliably fold despite intermediates and slow-folding kinetic phases.
The nondisease immunoglobulin (IgG)-binding proteins L and G make excellent targets for understanding the role of intermediates and unfolded ensembles on protein aggregation, since they have little sequence homology but high structural homology and fold through distinctly different mechanisms. Experimental evidence shows that protein L is a two-state folder, with formation of a transition state involving only native β-hairpin 1.10 Protein G on the other hand, folds through an early intermediate, followed by a rate-limiting step that involves formation of β-hairpin 2.11 The question that we set out to address was whether structural characteristics of the denatured and intermediate ensembles and the time scales of folding of these two different proteins might explain aggregation outcomes.12
We have developed a coarse-grained (CG) protein model that uses only the α-carbon centers to represent the protein, in which structural details of the amino acid side chains and aqueous solvent are replaced with effective bead–bead interactions.13–16 Figure 1 compares the native structure of the protein L and G models and that determined from the solution NMR structures (2PTL17 and 2GB1,18 respectively). This is one of the simplest models capable of representing a real protein to medium resolution and tractable enough to fully characterize the thermodynamics and kinetics of folding and aggregation.
FIGURE 1.
Comparison of the structural fidelity of the protein L and G models compared with experiment:15 (a) protein L model (right) vs experiment17 (left); (b) protein G model (right) vs experiment18 (left). Reproduced by permission from ref 15. Copyright 2008 Wiley.
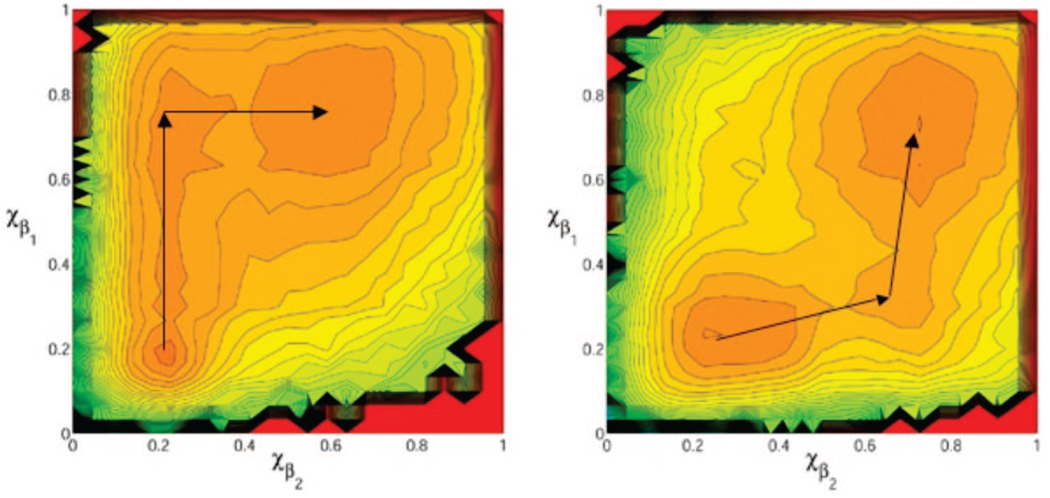
We begin by showing that our CG model can differentiate the experimental folding mechanisms of proteins L and G.19 The L and G sequences were mapped onto the CG reduced letter code, and secondary structure dihedral angle assignments were based on their PDB structures.17,18 At this level of sequence resolution, it is revealed that L and G share far higher sequence similarity (~60–70%) than the full chemical sequences suggest. However, analysis shows that protein L has more stabilizing interactions in β-hairpin 1 and a net loss of stabilizing interactions in β-hairpin 2, while the protein G sequence introduces net stabilization into β-hairpin 2.19 This difference is reflected in the free-energy projections along order parameters for native hairpin structure, χβ1 and χβ2 (Figure 2), in which there is a minimum free-energy path through formation of β-hairpin 1 and then β-hairpin 2 for protein L or β-hairpin 2 and then β-hairpin 1 for protein G.
FIGURE 2.
Free-energy contour plot as a function of native-state similarity of χ β1 and χ β2 19 for protein L (left) and protein G (right). Contour lines are spaced 1k B T apart. Arrows show the lowest free-energy path to folding along the reaction coordinates.
While thermodynamics are suggestive of the folding mechanism, we need to characterize the folding trajectories of proteins L and G to confirm the true kinetic mechanisms from the model. We found that the mean first passage time to the folded state of protein L conforms to two-state kinetics, with the presence of a transition state ensemble with a well-formed β-hairpin 1, consistent with experiment.19 Similar analysis of protein G showed that it folds through two pathways. One pathway exhibits two-state kinetics and folds through a transition-state ensemble with a well-formed β-hairpin 2 as per experiment.19
The second pathway for protein G gives rise to three-state kinetics, and involves an intermediate that precedes the rate-limiting step in folding. Figure 3a shows the intrachain contacts made in the native state (black contour) and the intrachain contacts made in the folding intermediate (maroon contours) for protein G. The intermediate shows hydrophobic contacts between β-strands 1, 2, and 3; this would be representative of most early folding intermediates that are typically formed by hydrophobic collapse. To confirm that we correctly identified the intermediate ensemble, the simulation trajectories were successfully fit to a reversible two-step U ⇔ I ⇔ N kinetic model to summarize the folding for protein G (Figure 3b).19
FIGURE 3.
(a) Contact map comparing the structure of the native (black) and intermediate (maroon) for the slow folding pathway of protein G.19 The contours outline which amino acids and their associated secondary structure elements are in spatial proximity to each other. (b) Kinetic data (symbols) and kinetic fits (lines) for U ⇔ I ⇔ N folding mechanism for protein G’s slow folding pathway. Reproduced with permission from ref 19. Copyright 2004 The Protein Society.
Next we simulated three chains of proteins L and G to relate differences in aggregation kinetics to differences in folding mechanism.12 When considering the time course for disappearance of the unaggregated population, we found that protein G aggregates more slowly than protein L.12 For protein L and the fast folding pathway for protein G, the time scales for folding are comparable to the aggregation time scale, whereas the protein G folding intermediate forms on time scales that are an order of magnitude faster than that for aggregation.12 We found that the structural signatures of the denatured state ensemble (DSE) for protein L and the intermediate state ensemble (ISE) for protein G and their time scales for folding provide complete insight into their aggregation pathways and kinetics.
In Figure 4, we display contact maps of the DSE for protein L, as well as the ISE for the slow folding pathway of protein G (both in red contours). These figures show that nativelike contacts made in the DSE of protein L are more localized (they do not show up in all or as extensively in the native structural elements given by the black contour) relative to that exhibited in the ISE of protein G. We also display in the contact maps the self-chain contacts (green contours) made in the aggregated ensemble for proteins L and G. For each protein, it is evident that the intrachain contacts of the aggregated ensemble resemble contacts formed in the DSE or ISE of the related protein monomer. Because stable intrachain structural elements are localized for protein L, the corresponding aggregate is much richer in interchain β-strand association. By contrast, protein G, with its more extensive native structural elements in the ISE, shows a reduced propensity for domain swapping and largely exhibits only interchain association of β-strands 3 and 3′. Because the third β-strand is the most hydrophobic segment of protein G, its rapid protection in the folding mechanism as an early intermediate (Figure 3a) minimizes the destructive tendency of protein G to aggregate. By determining the structural signatures of the DSE or ISE of a protein, then one can propose mutations that introduce additional native contacts across the entire protein fold to ameliorate aggregation.12
FIGURE 4.
Comparisons of contacts made in the folded monomer and aggregated ensembles for protein L and G.12 Native (black) and denatured state (red) of the monomer and intrachain contacts in the aggregated ensemble (green) for protein L (top left) and protein G (bottom left) and representative structures for aggregation of protein L (top, right) and protein G (bottom, right) are shown. Reproduced from ref 12. Copyright 2005 The Protein Society.
Aggregation and Alzheimer’s Disease
The aggregation of peptides or proteins into amyloid fibrils is associated with Alzheimer’s, Parkinson’s, type II diabetes, and other human diseases.1 Although the proteins that comprise the disease-related aggregates are dissimilar with respect to amino acid sequence, the aggregates take on consistent morphologies of unbranched fibrils 7–10 nm in diameter rich in β-strands orthogonal to the fibril axis, organizing into intermolecular β-sheets that can extend to micrometers in length.1 Alzheimer’s disease is characterized by the appearance in the brain of these fibril deposits, which are comprised primarily of amyloid-β (Aβ) peptide, created by proteolytic cleavage of the amyloid precursor protein (APP) as Aβ1–40, or Aβ1–42.2 Although early attention focused on the amyloid fibrils as the cause of Alzheimer’s disease, it is now hypothesized that Aβ oligomers formed during early aggregation may be the primary cytotoxic species.20
A physical separation of the oligomer and fibril regimes may be gleaned from the fibrillization kinetics that follow a nucleation-dependent polymerization mechanism21,22 in which the observed lag phase is due to the formation of a critical nucleus, the assembly into an oligomer corresponding to the largest free-energy barrier, beyond which a gradient of favorable free-energy results in a “down-hill” polymerization into a mature fibril. However, the structural characteristics and oligomer size of the soluble nucleating species have yet to be determined experimentally for any disease peptide sequence, and the molecular mechanism of polymerization that eventually delineates a mature fibril is unknown.
Solid-state NMR (SS-NMR) work by Tycko and co-workers23,24 has provided detailed experimental models as to the “folded state” of the Aβ1–40 monomer in the context of the mature “agitated” prepared fibril (Figure 5). It is composed of “U-shaped” monomers that form intermolecular N-terminal and C-terminal in-register parallel β-sheets orthogonal to the fibril axis, which we refer to as “filaments”. The SS-NMR restraints indicate that the N- and C- terminal β-strands interdigitate to form side-chain contacts between the C-termini of monomer i and the N-termini of the i − 2 monomer, introducing a geometric “stagger” in the individual filament structure (STAG(−2)).23 The early SS-NMR proposed two quaternary structures involving the relative orientation of two filaments24 based on approximate C2 symmetry around the fibril axis (C2z) and orthogonal to the fibril axis (C2x), and later it was determined that the agitated fibril was the C2z form.23 By contrast, Luhrs and co-workers25 found only filament order for Aβ1–42 with STAG(−1), but the mutation to methionine sulfoxide in position 35 would likely explain the lack of fibril order, since the mutation would likely destabilize the filament pair interface. While both experimental models may be relevant for insight into the disease state–both Aβ1–40 and Aβ1–42 are present as are oxidative stresses in the cell–we explore the implications of the SS-NMR model of Tycko and co-workers here.
FIGURE 5.
Summary of the solid-state NMR models for the Aβ1–40 monomer in the context of the mature “agitated” filaments and fibrils.23,24,26 Reproduced in part with permission from ref 26. Copyright 2007 Elsevier. Reproduced in part with permission from ref 23. Copyright 2006 American Chemical Society.
Using a more recent CG model that incorporates backbone hydrogen bonding,15 we built a 40-chain fibril fully consistent with the static NMR model of the two symmetry forms proposed by the early SS-NMR data, albeit with a preference for STAG(−1).26–28 With this validation, we characterize the stability of different lengths of the fibril for the C2x and C2z forms of WT Aβ1–40 to determine the critical nucleus.26 To accomplish this, we systematically shorten the fibril by retaining the innermost chains for sizes ranging between 20 to 4 monomer chains. For each size, we run 50–100 independent simulations and measure the final structural integrity of the fibril seeds by evaluating a quantity χ f that measures fibril order over the entire cross-section ends.26
Based on the ensemble of final structures for a given size, n, we can calculate the equilibrium populations of structurally stable and unstable fibrils based on a χ f cutoff value, χ c. The fraction of trajectories that correspond to χ f > χ c measures a population, Pn, of an ordered fibril with intact end monomers. This population is in equilibrium with the remaining P n–1 population corresponding to a loss of structural order of one end cross-section. We can calculate the change in free energy, ΔG, per unit cross-section as
| (1) |
Integrating eq (1) over n leads to free-energy changes as a function of n-chain fibril ordering, and we determine a critical nucleus size of ~10 chains for both C2x and C2z within the CG model26 (Figure 6). For aggregate sizes >8 chains, we observe that there are reversible changes in χ f, but for <8 chains, the structures consistent with a fibril are so disfavored that we see fewer instances of reversibility. This makes the free-energy curve along the fibril reaction coordinate below 8 chains ill-defined, and thus the barrier height difference between C2x and C2z is not meaningful since the free-energy curves are not on an absolute scale.
FIGURE 6.
Free energy for free monomer and fibril equilibrium for C2x and C2z(left) and representative structures for the different ordered regimes (right):26 (a) below the critical nucleus, (b) at the critical nucleus, and (c) the stable fibril. Reproduced with permission from ref 26. Copyright 2007 Elsevier.
Below the critical nucleus, we find that while there is some β-strand structure in the Aβ1–40 oligomers, they do not organize even at the level of filaments. At concentrations near the critical nucleus where the free energy reaches a maximum, we find that there are well-formed filaments, but the two filaments lack structural definition at their C-terminal interface, so the two filaments do not align to define a fibril axis. Past the free-energy barrier, the nucleation of a well-defined fibril axis arises when the entropy advantage for disorder at the interface of filaments is finally compensated by favorable enthalpic interactions. The primary enthalpic driver is the burial of the exposed hydrophobic plane of the C-terminal interface of the two filaments. At the critical nucleus, most hydrophobic contacts are satisfied regardless of the orientation of the two filament interfaces; however, as the fibril continues to lengthen and accumulate hydrophobic density along the direction of the filament axis, rotations of the two filaments to nonfibril orientations are now highly unfavorable due to the loss of the enthalpic stabilization. Eventually the hydrophobic density saturates at some fibril length so that successive cross-section addition results in a ΔΔG that is a constant, which occurs in our model at ~16 chains, and the protofibril exhibits the structural integrity of a mature fibril.
Given a mature fibril size, we use it for characterizing fibril growth mechanisms between the C2x and C2z forms under two assumptions: (1) that the addition unit for growth is a single monomer chain and (2) that the Aβ1–40 monomer exists in a largely random coil configuration. These assumptions are minimal in the sense that there is no definitive experimental measurement of preferred structure for the monomer, and while fibrils in vitro and in vivo may incorporate disordered oligomers that only later take on cross-β structure, the relative ability of the mature fibril to order these peptides is probed by this experiment. Given those assumptions, we seed the ends of the fibril, for each symmetry case, with monomers at distances that are close enough to not be diffusion-limited but far from van der Waals contact. Again, we run large numbers of independent simulations to collect an ensemble of fibril growth probabilities.
The probability for successful monomer addition, defined as the ratio of in-register parallel β-strand addition to growth-halting antiparallel addition, is found to be highest for one end of the C2z fibril, while the other end of the C2z fibril and both ends of the C2x fibril show significantly lower probabilities for successful addition. The primary reason for this difference arises from the structural symmetry (C2x) vs asymmetry (C2z) at the ends of the fibrils (Figure 7), which arises from the interplay of the stagger within the protofilaments, and the symmetry axis of the C2x and C2z fibril.26,28
FIGURE 7.
Effect of axis symmetry and stagger on terminating fibril ends of Aβ1–40.26 A schematic of 16 chain fibrils is shown with N-terminal region colored in teal and C-terminal region colored in orange: STAG(−1) C2x and STAG(−1) C2z(top); a mixed C2x and C2z STAG(−1/+1) (bottom). Reproduced with permission from ref 26. Copyright 2007 Elsevier.
For C2z, the N-terminal region spatially projects an amino acid patterning that better specifies in-register parallel addition and more importantly fewer growth-halting antiparallel additions, resulting in unidirectional growth of the C2z fibril but bidirectional growth for C2x. However, the NMR data restraints for Aβ1–40 do not rule out the possibility of a mixed stagger, that is, +N stagger for one filament and –N stagger for the other filament. Using our model, we can build a mixed stagger structure (Figure 7),26 showing that it is possible to reverse the structural end symmetries of the two quaternary forms and potentially their elongation mechanism.
We see that polymorphs of the mature fibers arise from different organizations of at least two filaments that, combined with stagger in the β-sheets, can affect fibril growth patterns.26,29,30 This is a supercategory for the eight classes of steric zippers describing interaction permutations between covalent structures noted by Eisenberg and co-workers in their work on microcrystals of short peptides.31 We note that the finite length of our simulations makes the absolute percentages of any type of correct monomer addition rather low (~3%). This suggests that incorrect additions might eventually anneal out and reconfigure to create a new viable end structure on longer time scales, as suggested by AFM observations of fibril maturation.32 It also opens up the question as to whether the Aβ monomer is the dominant unit for fibril elongation or whether in fact small oligomers are more viable addition units for fibril lengthening.33
Familial Alzheimer’s Disease Mutants
Clues to spontaneous forms of Alzheimer’s disease can be gleaned by contrasting its behavior to familial Alzheimer’s disease (FAD) mutants, including the Flemish (A21G),34 Arctic (E22G),35 and Dutch mutants (E22Q),36 all of which have been characterized for both Aβ1–40 and Aβ1–42. Differences among the WT and FAD mutants are evident for in vitro studies of fibrillization kinetics; the Dutch mutant nucleates and fibrillizes more readily than WT, while the Arctic mutation has a higher propensity to nucleate protofibrils, although subsequent fibrillization rates are comparable to WT.35 The nucleation and rate of fibril formation is greatly reduced for the Flemish mutant relative to WT.35
We emphasize that experiments are highly unspecific in regards to what structural order is accumulating in the kinetic profiles. The kinetics of the Arctic Aβ peptides have been quantified by chromatographic methods that measure rates of disappearance of monomer and appearance of oligomer assemblies based on their mass and not their structures.35 Although Congo Red or Thioflavin T dye-binding fluorescence are thought to measure the disappearance of monomer into fibril assemblies, no definitive experimental evidence exists to confirm that they can differentiate order accumulation at the level of filaments or fibrils, since both have cross β-strand order.
We have used our CG model study to address the clear differences in the kinetics of the formation of fibril assemblies of the Dutch, Flemish, and Arctic FAD mutants, using the WT C2z morphology as the reference fibril structure and reevaluating the free-energy trends along the fibril reaction coordinate as a function of fibril size.37 We take as our measure for greater ease of nucleation a shift in the critical nucleus to lower number of peptides and hence more accessible at lower concentration. We take as our measure of faster fibrillization kinetics a change in the free-energy slope for large ordered assemblies, that is, that |ΔAG mutant| > |ΔG WT|. Again we evaluate the populations that achieve χ f order over the whole fibril cross-section using the WT reference fibril. We also use an additional order parameter, P f, that measures the “nativeness” of individual filament cross-sections relative to the WT filament.
Despite the locality of the mutation, substantial free-energy differences and structural ensembles exist among the four different Aβ sequences measured as filaments (using P f) or fibrils (using χ f) (Figure 8). We find that both the Arctic and Flemish sequences promote greater disorder of the β-turn region, which results in lower order as measured by P f for both mutants relative to WT. However, the difference in sequence position of the glycine mutation for the Arctic and Flemish cases radically alters fibril order stability as measured by χ f.
FIGURE 8.
Free energy profile for free monomer vs fibrils (left) and filaments (right) for WT (black), Arctic (green), Dutch (WT fibril reference in aqua and new polymorph in blue), and Flemish (red) mutants. Reproduced with permission from ref 37. Copyright 2008 Biophysical Society.
The A21G mutant disrupts the N-terminal β-strands, and regardless of the detection method (Pf or χ f) for cross β-sheet structure, the dynamic equilibrium strongly favors the monomeric peptide (Figure 9a). The greater resistance of the Flemish mutant to order into fibril assemblies of any size suggests that it is capable of both fragmentation into smaller oligomers and promoting amorphous aggregation to yield large plaques, given its lack of any definitive filament or fibril morphology state.37 By contrast, the E22G mutation is enough removed from the β-strands so that the Arctic mutant retains β-strand order (Figure 9b), and the more flexible turn can now form new contacts that allow little rotation between the filaments beyond six chains.37 While new stabilizing contacts favor smaller fibrils than those found for WT (Figure 8), they could slow or even block the addition reaction to create larger fibril assemblies. Our observation of distinctly different fibril properties of the Arctic mutant may be an example where disordered hydrophobic collapse is now relatively more favorable than ordered hydrogen bond formation.38 Furthermore, the constant negative slope indicative of reaching a stable fibril regime is the same for the Arctic mutant and WT, consistent with chromatography methods that measure more rapid disappearance of monomer into protofibrils for E22G relative to WT37 but finding little difference in rates of forming fibrils.35
FIGURE 9.
Representative fibril structure of the Arctic (green) and Flemish (red) mutants. Reproduced with permission from ref 37. Copyright 2008 Biophysical Society.
The Dutch mutant shows the smallest critical nucleus size based on measures of filament order but not fibril order (Figure 8). Perhaps the Dutch mutant with its more negative slope beyond the critical nucleus relative to all other sequences favors a filament form such as that found for Aβ1–42.25 This may explain its significantly enhanced fibrillization kinetics using dye-binding assays of cross β-sheet structure but measuring accumulation of filaments only. Another possible reason is that the Dutch mutation eliminates charge repulsion between peptides on the same filament, resulting in a more exaggerated twist down the filament axis compared with WT. This in turn requires a reorganization of the two-filament interface to define a new polymorph of fibril order that is distinct from the WT agitated fibril morphology (Figure 10).37 When the alternative fibril polymorph for the Dutch mutant is added as a reference, there is a qualitative shift for preference for fibril order (Figure 8a).
FIGURE 10.
Comparison of the Dutch fibril polymorph (blue) with respect to WT sequence (black).37 The yellow spheres represent amino acid 33 on each monomer chain. Reproduced with permission from ref 37. Copyright 2008 Biophysical Society.
Conclusion
We have used a coarse-grained model of proteins15 to examine the molecular factors that differentiate nondisease and disease aggregation. By characterizing in silico the aggregation of proteins at high concentration, akin to the environment of overexpressed proteins that aggregate into inclusion bodies,4 our investigations on proteins L and G suggest that protective structure in the DSE or ISE and time scales of functional folding can set up protective mechanisms that help avoid deleterious aggregation.12 Whether any protein uses early intermediates in folding for protection against unwanted aggregation in vivo may involve evolutionary selection that depends on a given protein’s cellular conditions. In vitro, protein sequences could be reengineered to manifest an early folding intermediate as a strategy to increase folding yield in industrial protein production. The observed nondisease aggregates may represent the soluble nuclei for larger aggregates, not just for inclusion bodies, but potentially as the seeds of ordered fibrillar assemblies, since most nondisease proteins3,39,40 can be induced to form amyloid fibrils.
Do protective folding mechanisms break down altogether for disease-related sequences such as Aβ1–40 or Aβ1–42? While diminished structure in the DSE may promote interchain aggregation, the enhancement of a specific type of collapsed structure involving exposed β-strands has been suggested to be the aggregate seed for Aβ.41 Recently we have shown using all-atom molecular dynamics simulations that reproduce high-field solution ROESY spectra42 that the WT Aβ21–30 monomer fragment shows no evidence of a dominant population of stable β-strands. Recent theoretical studies,43 validated against experimentally determined three bond scalar coupling constants, showed that the longer Aβ1–42 disease peptide sequence is highly flexible but with some β-hairpin formation in the C-terminal region. However, scalar coupling constants are insensitive to subpopulations of ordered structure that are better picked up by NOESY/ROESY experiments, combined with molecular dynamics to interpret the NMR populations.42 We are currently conducting new NOESY experiments and molecular simulations on the Aβ1–40,42 sequences to address these issues.
While most studies favor the origin of cytotoxicity as arising from soluble oligomers,44 the evidence for insoluble fibrils as also being a cytotoxic agent are still compelling. Experiments have shown that different polymorphs of the mature Aβ1–40 fibril can contribute to variation in cell viability,29 and synaptic activity is greatly impaired in the presence of the insoluble plaque.45 Cognitive deficits arising from the Arctic mutant were traced to a nonfibrillar form, whereas the severity of memory loss symptoms for carriers of the Dutch mutation were consistent with interference from the mature fibrillar species.20 In our studies, we find that the morphologies of the fibril state are highly varied within the WT Aβ1–40 sequence itself, in which two symmetry forms of the “agitated” fibril are equally viable.26 The FAD mutants investigated here show very different concentration regimes needed to nucleate ordered filament and/or fibril assemblies and even new polymorphs.37 Thus the fibril regimes for the WT and FAD mutants remain an important line of investigation for understanding the Alzheimer’s disease process.
Finally, in vitro studies are only part of the larger in vivo complexity of degenerative aggregation disease processes that indicate an overall system failure. For example, alternative FAD mutations of APP outside the Aβ sequence affect ratios of Aβ1–42/Aβ1–40 due to processing errors by β- and γ-secretases2 and therefore disease severity depending on the abundance of the more virulent Aβ1–42. The location of the amyloid plaque deposits in the brain defines an important aspect of the neuropathology of the disease state.46 Carriers of the Arctic mutation exhibit deposits primarily of Aβ1–42 in brain tissue and typical AD dementia symptoms,35 whereas the Dutch mutation carriers show deposition of Aβ1–42 in blood vessels that contribute to cerebral amyloid angiopathy (CAA) with vascular dementia symptoms.36,46 Carriers of the Flemish mutation are distinct by having the largest plaque cores centered on blood vessels and dominated by Aβ1–40, resulting in both AD dementia and CAA features.46 Recent work has shown that differences in ganglioside binding of the FAD mutants, an important constituent of cell membrane in the central nervous system, might explain the region-specific Aβ deposition in the brain.47 These provide examples of the need for theory to push toward more complex problems that confront the disease process, with the goal of demonstrable success in the development of theoretical models that have predictive power.
Acknowledgments
We thank the DOE Computational Science (K.L.K.) and Whitaker Foundation (N.L.F.) for graduate fellowships and the Guidant Foundation (Y.O.)for a summer research fellowship. This work was supported by NIH Grant GM070919 and NERSC (Grant DE-AC03-76SF00098).
Biographies
Nicolas Lux Fawzi (B.S. 2002, U. Pennsylvania; Ph.D. 2007, UC Berkeley) completed his doctorate at UC Berkeley on theoretical studies of protein aggregation and is now an NIH postdoctoral researcher.
Enghui Yap (B.S., M.S., U. Illinois, Urbana–Champaign, 2002) is pursuing a Ph.D. at UC Berkeley developing multiscale methods for biomolecular assembly.
Yuka Okabe (B.S. 2006, UC Berkeley) did undergraduate research in the Head-Gordon laboratory and is now a Bioengineering graduate student at UC Irvine.
Kevin L. Kohlstedt (B.S. 2003, U. Kansas) is pursuing a Ph.D. at Northwestern and did a practicum in the Head-Gordon laboratory in 2006.
Scott Brown (B.S. Chemistry 1995, U. Utah; Ph.D. 2000, Colorado State University) is an Associate Research Computational Scientist at Abbott Laboratories working on computer-aided drug design.
Teresa Head-Gordon (Ph.D. 1989, Carnegie Mellon, 1990–1992, Postdoctoral Member of Technical Staff, AT&T Bell Laboratories) leads a group at UC Berkeley that develops theoretical/experimental methods to study biomaterials assembly and bulk and hydration water properties.
REFERENCES
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Goedert M, Spillantini MG. A century of Alzheimer’s disease. Science. 2006;314:777–781. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- 3.Cellmer T, Douma R, Huebner A, Prausnitz J, Blanch H. Kinetic studies of protein L aggregation and disaggregation. Biophys. Chem. 2007;125:350–359. doi: 10.1016/j.bpc.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Clark ED. Protein refolding for industrial processes. Curr. Opin. Biotechnol. 2001;12:202–207. doi: 10.1016/s0958-1669(00)00200-7. [DOI] [PubMed] [Google Scholar]
- 5.Tycko R, Petkova A, Oyler N, Chan CC, Balbach J. Probing the molecular structure of amyloid fibrils with solid-state NMR. Biophys. J. 2002;82:187A. [Google Scholar]
- 6.King J, Haasepettingell C, Robinson AS, Speed M, Mitraki A. Thermolabile folding intermediates-inclusion body precursors and chaperonin substrates. FASEB J. 1996;10:57–66. doi: 10.1096/fasebj.10.1.8566549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uversky VN, Li J, Fink AL. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J. Biol. Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 8.Silow M, Tan YJ, Fersht AR, Oliveberg M. Formation of short-lived protein aggregates directly from the coil in two-state folding. Biochemistry. 1999;38:13006–13012. doi: 10.1021/bi9909997. [DOI] [PubMed] [Google Scholar]
- 9.Chiti F, Taddei N, Baroni F, Capanni C, Stefani M, Ramponi G, Dobson CM. Kinetic partitioning of protein folding and aggregation. Nat. Struct. Biol. 2002;9:137–143. doi: 10.1038/nsb752. [DOI] [PubMed] [Google Scholar]
- 10.Kim DE, Fisher C, Baker D. A breakdown of symmetry in the folding transition state of protein L. J. Mol. Biol. 2000;298:971–984. doi: 10.1006/jmbi.2000.3701. [DOI] [PubMed] [Google Scholar]
- 11.Park SH, Shastry MCR, Roder H. Folding dynamics of the B1-domain of protein G explored by ultrarapid mixing. Nat. Struct. Biol. 1999;6:943–947. doi: 10.1038/13311. [DOI] [PubMed] [Google Scholar]
- 12.Fawzi NL, Chubukov V, Clark LA, Brown S, Head-Gordon T. Influence of denatured and intermediate states of folding on protein aggregation. Protein Sci. 2005;14:993–1003. doi: 10.1110/ps.041177505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Head-Gordon T, Brown S. Minimalist models for protein folding and design. Curr. Opin. Struct. Biol. 2003;13:160–167. doi: 10.1016/s0959-440x(03)00030-7. [DOI] [PubMed] [Google Scholar]
- 14.Brown S, Fawzi N, Head-Gordon T. Coarse-grained sequences for protein folding and design. Proc. Natl. Acad. Sci. U.S.A. 2003;100:10712–10717. doi: 10.1073/pnas.1931882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yap EH, Fawzi NL, Head-Gordon T. A coarse-grained alpha-carbon protein model with anisotropic hydrogen-bonding. Proteins. 2008;70:626–638. doi: 10.1002/prot.21515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thirumalai D, Klimov DK. Deciphering the timescales and mechanisms of protein folding using minimal off-lattice models. Curr. Opin. Struct. Biol. 1999;9:197–207. doi: 10.1016/S0959-440X(99)80028-1. [DOI] [PubMed] [Google Scholar]
- 17.Wikstrom M, Drakenberg T, Forsen S, Sjobring U, Bjorck L. Three-dimensional solution structure of an immunoglobulin light chain-binding domain of protein-L. Comparison with the IgG-binding domains of protein G. Biochemistry. 1994;33:14011–14017. doi: 10.1021/bi00251a008. [DOI] [PubMed] [Google Scholar]
- 18.Gronenborn AM, Filpula DR, Essig NZ, Achari A, Whitlow M, Wingfield PT, Clore GM. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein-G. Science. 1991;253:657–661. doi: 10.1126/science.1871600. [DOI] [PubMed] [Google Scholar]
- 19.Brown S, Head-Gordon T. Intermediates and the folding of proteins L and G. Protein Sci. 2004;13:958–970. doi: 10.1110/ps.03316004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klyubin I, Walsh DM, Cullen WK, Fadeeva JV, Anwyl R, Selkoe DJ, Rowan MJ. Soluble Arctic amyloid beta protein inhibits hippocampal long-term potentiation in vivo. Eur. J. Neurosci. 2004;19:2839–2846. doi: 10.1111/j.1460-9568.2004.03389.x. [DOI] [PubMed] [Google Scholar]
- 21.Dolphin GT, Dumy P, Garcia J. Control of amyloid beta-peptide protofibril formation by a designed template assembly. Angew. Chem., Int. Ed. 2006;45:2699–2702. doi: 10.1002/anie.200600395. [DOI] [PubMed] [Google Scholar]
- 22.Ferrone F. Analysis of protein aggregation kinetics. Methods Enzymol. 1999;309:256–274. doi: 10.1016/s0076-6879(99)09019-9. [DOI] [PubMed] [Google Scholar]
- 23.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, et al. A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doöbeli H, Schubert D, Riek R. 3D structure of Alzheimer’s amyloid-β(1−42) fibrils. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fawzi NL, Okabe Y, Yap EH, Head-Gordon T. Determining the critical nucleus and mechanism of fibril elongation of the Alzheimer’s Aβ(1−40) peptide. J. Mol. Biol. 2007;365:535–550. doi: 10.1016/j.jmb.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buchete NV, Tycko R, Hummer G. Molecular dynamics simulations of Alzheimer’s beta-amyloid protofilaments. J. Mol. Biol. 2005;353:804–821. doi: 10.1016/j.jmb.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 28.Buchete NV, Hummer G. Exploring the structural stability and the mechanism of dissociation of Alzheimer’s amyloid fibrils. Biophys. J. 2007:194A. [Google Scholar]
- 29.Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 30.Paravastu AK, Petkova AT, Tycko R. Polymorphic fibril formation by residues 10−40 of the Alzheimer’s beta-amyloid peptide. Biophys. J. 2006;90:4618–4629. doi: 10.1529/biophysj.105.076927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen AÃ, Riekel C, Eisenberg D. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 32.Ban T, Yamaguchi K, Goto Y. Direct observation of amyloid fibril growth, propagation, and adaptation. Acc. Chem. Res. 2006;39:663–670. doi: 10.1021/ar050074l. [DOI] [PubMed] [Google Scholar]
- 33.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 34.Huet A, Derreumaux P. Impact of the mutation A21G (Flemish variant) on Alzheimer’s beta-amyloid dimers by molecular dynamics simulations. Biophys. J. 2006;91:3829–3840. doi: 10.1529/biophysj.106.090993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nilsberth C, Westlind-Danielsson A, Eckman CB, Condron MM, Axelman K, Forsell C, Stenh C, Luthman J, Teplow DB, Younkin SG, Naslund J, Lannfelt L. The ‘Arctic’ APP mutation (E693G) causes Alzheimer’s disease by enhanced Aβ protofibril formation. Nat. Neurosci. 2001;4:887–893. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- 36.Levy E, Carman MD, Fernandez-Madrid IJ, Power MD, Lieberburg I, van Duinen SG, Bots GT, Luyendijk W, Frangione B. Mutation of the Alzheimer’s disease amyloid gene in hereditary cerebral hemorrhage, Dutch type. Science. 1990;248:1124–1126. doi: 10.1126/science.2111584. [DOI] [PubMed] [Google Scholar]
- 37.Fawzi NL, Kohlstedt KL, Okabe Y, Head-Gordon T. Protofibril assemblies of the Arctic, Dutch and Flemish mutants of the Alzheimer’s A{beta}1−40 peptide. Biophys. J. 2008;94:2007–2016. doi: 10.1529/biophysj.107.121467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheon M, Chang I, Mohanty S, Luheshi LM, Dobson CM, Vendruscolo M, Favrin G. Structural reorganization and potential toxicity of oligomeric species formed during the assembly of amyloid fibrils. PLoS Comp. Biol. 2007;3:1727–1738. doi: 10.1371/journal.pcbi.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byeon IJL, Louis JM, Gronenborn AM. A protein contortionist: Core mutations of GBI that induce dimerization and domain swapping. J. Mol. Biol. 2003;334:605–605. doi: 10.1016/s0022-2836(03)00928-8. [DOI] [PubMed] [Google Scholar]
- 40.Ramirez-Alvarado M, Cocco MJ, Regan L. Mutations in the B1-domain of protein G that delay the onset of amyloid fibril formation in vitro. Protein Sci. 2003;12:567–576. doi: 10.1110/ps.0227403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grant MA, Lazo ND, Lomakin A, Condron MM, Arai H, Yamin G, Rigby AC, Teplow DB. Familial Alzhemier’s disease mutations alter the stability of the amyloid-beta-protein monomer folding nucleus. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16522–16527. doi: 10.1073/pnas.0705197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fawzi NL, Phillips AHP, Ruscio JZ, Doucleff M, Wemmer DE, Head-Gordon T. Structure and dynamics of the Aβ21−30 peptide from the interplay of NMR experiments and molecular simulations. J. Am. Chem. Soc. 2008;130:6145–6158. doi: 10.1021/ja710366c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sgourakis NG, Yan YL, McCallum SA, Wang CY, Garcia AE. The Alzheimer’s peptides Abeta40,42 adopt distinct conformations in water: A combined MD/NMR study. J. Mol. Biol. 2007;368:1448–1457. doi: 10.1016/j.jmb.2007.02.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng IH, Scearce-Levie K, Legleiter J, Palop JJ, Gerstein H, Bien-Ly N, Puolivali J, Lesné S, Ashe KH, Muchowski PJ, Mucke L. Accelerating amyloid-β fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J. Biol. Chem. 2007;282:23818–23828. doi: 10.1074/jbc.M701078200. [DOI] [PubMed] [Google Scholar]
- 45.Stern EA, Bacskai BJ, Hickey GA, Attenello FJ, Lombardo JA, Hyman BT. Cortical synaptic integration in vivo is disrupted by amyloid-beta plaques. J. Neurosci. 2004;24:4535–4540. doi: 10.1523/JNEUROSCI.0462-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang-Nunes SX, Maat-Schieman MLC, van Duinen SG, Roos RAC, Frosch MP, Greenberg SM. The cerebral beta-amyloid angiopathies: hereditary and sporadic. Brain Pathol. 2006;16:30–39. doi: 10.1111/j.1750-3639.2006.tb00559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kakio A, Nishimoto S, Yanagisawa K, Kozutsumi Y, Matsuzaki K. Interactions of amyloid-β protein with various gangliosides in raft-like membranes: Importance of GM1 ganglioside-bound form as an endogenous seed for Alzheimer-amyloid. Biochemistry. 2002;41:7385–7390. doi: 10.1021/bi0255874. [DOI] [PubMed] [Google Scholar]