Abstract
Background
Diagnosis of eosinophilic esophagitis (EoE) requires quantification of esophageal eosinophilia.
Aims
The aims of this study were to assess inter- and intraobserver reliability for measuring esophageal eosinophil counts and to validate a novel method of determining tissue eosinophil density using digitized histopathology slides.
Methods
Patients were selected from the University of North Carolina EoE clinicopathologic database. Glass slides were de-identified and scanned to create digitized slides. Using a set protocol, 40 slides were read by each of three pathologists for interobserver measures, and were also reread by one pathologist as traditional glass slides. Different sets of 20 unique slides were read twice by each pathologist for intraobserver measures. Correlation and agreement were calculated with Pearson’s rho and the κ statistic.
Results
There was excellent correction between digitized images and glass slides (r = 0.91–0.95, P < 0.001). For maximum eosinophil densities, interobserver correlations were 0.91, 0.76, and 0.79. For mean densities, interobserver correlations were 0.90, 0.89, and 0.85. Intraobserver correlations for maximum densities were 0.99, 0.94, and 0.96, and for mean densities were 0.97, 0.87, and 0.89 (P < 0.001 for all correlations). Agreement was in the “substantial” to “near-perfect” range for pathologists using several diagnostic cut-points for EoE.
Conclusions
Both inter- and intraobserver correlations were excellent for determining eosinophil densities and counts. A method of using digitized slides was valid when compared with traditional glass slides. This protocol could be adopted for research and clinical purposes to further standardize the diagnostic process for EoE.
Keywords: Eosinophilic esophagitis, Interobserver reliability, Intraobserver reliability, Histopathology
Introduction
Eosinophilic esophagitis (EoE) is a clinicopathologic condition ultimately diagnosed by demonstrating abnormal eosinophilia within esophageal mucosal biopsies in a suggestive clinical setting [1]. Typical symptoms include dysphagia, food impaction, refractory heartburn, chest pain, and in children, feeding intolerance [2–5]. Endoscopic hallmarks include esophageal rings, linear furrows, white plaques or exudates, and “crêpe-paper” mucosa which tears easily, but a proportion of cases may have an endoscopically normal esophagus [5–8].
While the incidence and prevalence of EoE have increased over the past decade [9–13], formal diagnostic criteria have only recently been proposed [1]. The histopathologic portion of these criteria requires the presence of ≥15 eosinophils in at least one high-power field (eos/hpf). To date, however, there has been significant variability in diagnostic criteria used throughout the EoE literature [14]. Moreover, the size of a high-power field (hpf) used to determine eosinophil counts is not standardized between microscopes [14], implying that the threshold level of 15 eos/hpf on one microscope may not be replicated on another. Finally, the reliability of count determination has not been well described [14, 15], and it is unknown whether obtaining counts (in eos/hpf) or densities (in eos/mm2) would be preferable.
This study had two objectives: (1) to assess inter- and intraobserver reliability for measuring maximum and mean esophageal eosinophil counts, and (2) to describe and validate a novel method of determining tissue eosinophil density by digitizing histopathologic slides and counting eosinophils on a computer monitor. We hypothesized that digitized counts would have a high correlation with traditional glass slide counts, that reliability would be good to excellent, and that calculating eosinophil densities would permit back-calculation to eos/hpf and allow comparison with previously published studies that specified a particular hpf size.
Methods
Patients and Pathology Samples
The University of North Carolina (UNC) EoE clinico-pathologic database was used for this study. This database contains clinical, endoscopic, and pathologic characteristics on over 1,200 patients with esophageal eosinophilia from any cause from 2000 to 2007. It also contains information on patients in whom esophageal eosinophilia was excluded with a normal esophageal biopsy. Because this study was designed primarily to test reliability of determining eosinophil counts, subjects were selected based on their initial eosinophil counts (eos/hpf) performed for clinical care, and specifically chosen to represent a wide range of esophageal eosinophil counts, from 0 to >300 eos/hpf. The esophageal eosinophilia could have been from any cause, including EoE or gastroesophageal reflux disease, and clinical characteristics were not relevant for the purposes of this study. After identification of appropriate patients, archived pathology slides were pulled for review. This study was approved by the University of North Carolina Institutional Review Board.
Slide Scanning and Reading Protocol
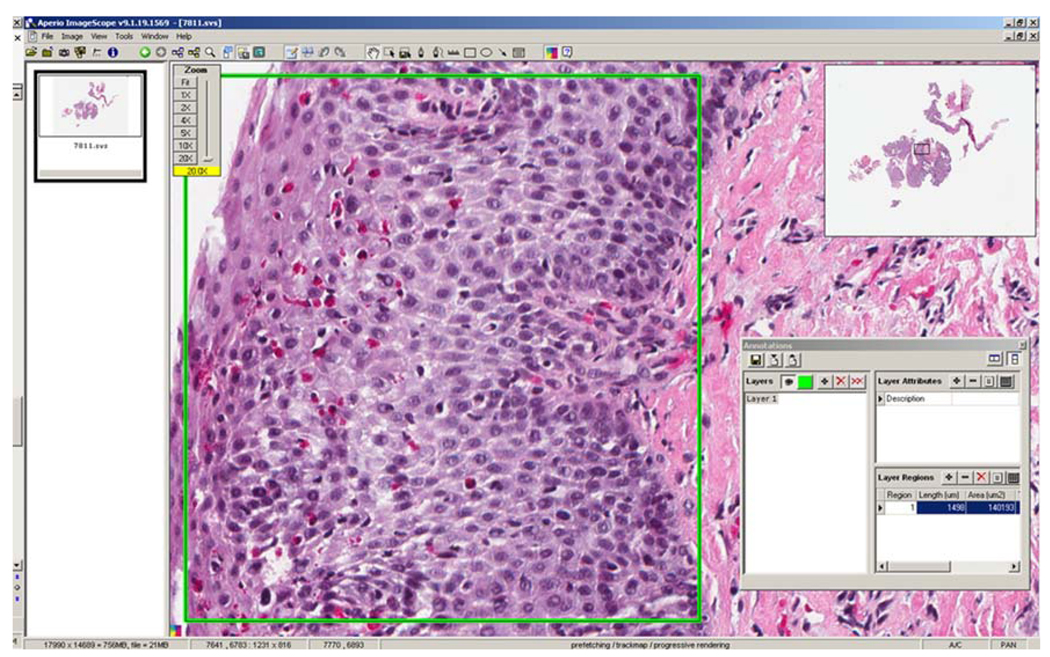
All glass slides were scanned using the Aperio ScanScope® CS slide scanner (Aperio Technologies, Vista, CA) to create digitized slides for use with Aperio ImageScope viewing software (version 9.1.19.1569; Aperio Technologies, Vista, CA; http://www.aperio.com/download.asp). This software allows high-resolution digitized pathology slides to be examined on a computer monitor, with the ability to rapidly scan and zoom to any area of interest (Fig. 1).
Fig. 1.
View of a virtual esophageal biopsy slide using the Aperio ImageScope software. Note that the “Zoom” dialogue box (upper left portion of the screen) indicates a 5 × zoom, while the “Thumbnail” box (upper right portion of the screen) shows the location of the zoomed-in area in relation to the remainder of the biopsy specimen. (Permission for screenshot granted by Aperio Technologies.)
Each scanned slide was blinded and read by three pathologists (T.C.R., K.J.F., and J.T.W.) with a special interest in gastrointestinal pathology. The counting protocol is outlined in Table 1, illustrated in Fig. 1 and Fig. 2, and yielded the outcomes of maximum and mean eosinophil density.
Table 1.
Slide reading and eosinophil counting protocol
| Review the entire digitized slide at low to medium power to identify the areas, if any, of prominent eosinophil infiltration (Fig. 1). |
| Select the area with the highest eosinophil density, zoom to full image resolution (20×), and outline with the “rectangle tool” to yield a measurement of the selected area in square microns (µm2; Fig. 2)a. |
| Count the number of eosinophils within the sampled area to obtain the maximum eosinophil density (primary outcome). |
| Actively degranulating eosinophils are counted; eosinophilic granules in isolation are not. |
| In areas of dense inflammation with overlapping cell borders, attempt to count each eosinophil nucleus. |
| Do not count eosinophils at the edge of the biopsy specimen, as eosinophils in this area can degranulate due to biopsy tissue trauma [15]. |
| Record the presence of degranulated eosinophils (yes/no) in the selected area. |
| Record the presence of eosinophilic microabscesses (clusters of ≥4 eosinophils) [15] in the selected area. |
| Repeat this protocol for four more areas: one area judged to be the next most densely infiltrated with eosinophils, and three that are representative of the biopsy specimen overall. |
| Calculate the mean eosinophil density from all five areas counted. |
| Make a global determination of whether the biopsy findings are consistent with eosinophilic esophagitis (EoE) “in the correct clinical context”b. |
The goal was to select an area approximately 150,000–300,000 µm2 in size, which corresponds to a mid-sized microscopy field [14]
A “global determination” was chosen because pathologists may use a variety of indicators, including number and distribution of cells, presence of activated cells, type of inflammation, and epithelial and stromal changes when assessing a biopsy specimen for EoE. Because clinical characteristics of the individual slides were not provided to the pathologists for this study, their response was not correlated with clinical parameters
Fig. 2.
View of Aperio software with an area of high eosinophil density selected with the “rectangle” tool. In this example, the “Annotations” box, seen in the lower right corner of the figure, displays the selected area as 140,193 µm2. In this example, there are approximately 51 eosinophils within the selected area, for a eosinophil density of 364 eosinophils/mm2, or an eosinophil count of 87 eos/hpf assuming a high-power field area of 0.24 mm2, a typical size reported in the literature. (Permission for screenshot granted by Aperio Technologies.)
Validation of Digitized Reading Protocol
In order to validate the eosinophil counts obtained from the digitized slides, the original 40 glass slides that all three pathologists read were reread by one pathologist on a microscope (Olympus BX-41; hpf diameter = 0.55 mm; hpf area = 0.24 mm2) using the same protocol. The number of eos/hpf was converted to density (eos/mm2) by dividing the count by the hpf area size. This read was correlated with the same pathologist’s read from the digitized slides, as well as with a “consensus” read, calculated by averaging all three pathologists’ maximum count for each digitized slide.
Statistical Analysis and Power Calculation
Summary statistics were used to characterize the counts. For counts obtained using digitized images, eosinophil density was converted from eos/µm2 to eos/mm2 by multiplying the eosinophil count by 1,000,000 µm2/mm2 and dividing by the measured area in µm2. Density (eos/mm2) was converted to eos/hpf by multiplying by the hpf size of interest (in mm2). Pearson’s correlation (r) was performed for continuous measures and the Kappa statistic (κ) was used to assess agreement for dichotomous measures. A κ of 0.0 or less is considered to represent poor agreement, 0.01– 0.20 slight agreement, 0.21–0.40 fair, 0.41–0.60 moderate, 0.61–0.80 substantial, and 0.81–1.00 near-perfect agreement [16]. This measure was also used to assess agreement on typical eosinophil cut-points used for EoE diagnosis in the literature (i.e., 15, 20, 24, and 30 eos/hpf).
The study was powered for the primary outcome of maximum eosinophil density. For interobserver correlation, assuming an α of 0.05 and a power of 0.80, correlations as low as r = 0.43 could be detected with 40 samples. Therefore, a set of 40 slides was generated to assess interobserver agreement. For intraobserver correlation, assuming an α of 0.05 and a power of 0.80, correlations as low as r = 0.59 could be detected with 20 samples. Therefore, three sets of 20 additional slides were generated (60 unique patients in total), and one set was provided to each of the three pathologists to independently read twice to assess intraobserver agreement. These slides were randomly mixed with other esophageal slides that were being read as part of another study, so that slides were reread by the same pathologist at least 2 weeks after the original reads.
Results
Correlation Between Digitized and Glass Slides
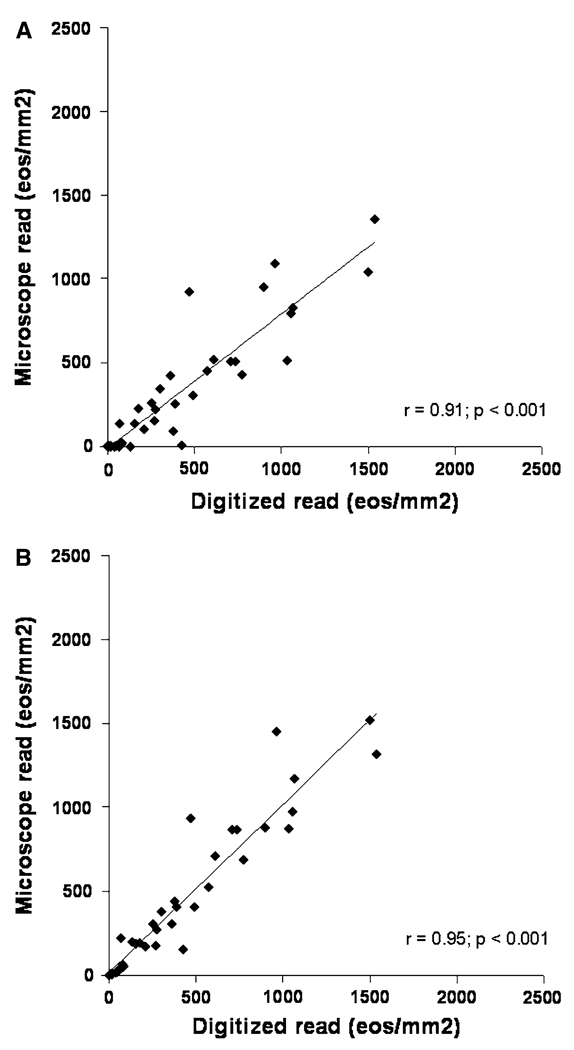
The correlation between maximum eosinophil densities as read on the digitized slides as compared with the glass slides was excellent, with r = 0.91 (P < 0.001) for the single pathologist who reread the glass slides (Fig. 3a). The correlation for the mean count was r = 0.92 (P < 0.0011). It took a mean of 3.4 ± 1.1 min per glass slide to perform the counts. A similar result (r = 0.95; P < 0.001) was seen when the glass slide read was correlated with the average maximum count from all of the pathologists (Fig. 3b), and when mean counts were examined (r = 0.92; P < 0.001).
Fig. 3.
Correlation between the digitized slides and traditional glass slides for determination of maximum eosinophil density (eos/mm2). a Digitized and microscope reads on the same slide set by pathologist 1. b Comparison of the microscope read with the average of all the maximum counts from all three pathologists
Agreement between the glass and digitized slides was κ = 0.73 (P < 0.001) for assessing degranulation, κ = 0.54 (P < 0.001) for microabscesses, and κ = 0.89 (P < 0.001) for determining whether the biopsy was consistent with EoE.
Interobserver Correlation
The maximum and mean eosinophil densities (eos/mm2) as determined by each of the three pathologists are presented in Table 2. For maximum densities, interobserver correlation was r = 0.91 (P < 0.001) for pathologists 1 and 2, r = 0.79 (P < 0.001) for pathologists 1 and 3, and r = 0.76 (P < 0.001) for pathologists 2 and 3 (Fig. 4a–c). For mean densities, interobserver correlation was r = 0.90 (P < 0.001) for pathologists 1 and 2, r = 0.89 (P < 0.001) for pathologists 1 and 3, and r = 0.85 (P < 0.001) for pathologists 2 and 3. To complete the reading protocol the mean time spent was 6.4 ± 4.4 min for pathologist 1, 8.2 ± 3.4 min for pathologist 2, and 10.0 ± 5.7 min for pathologist 3.
Table 2.
Maximum and mean esophageal eosinophil densities for determination of interobserver correlation
| Subject number |
Pathologist 1 | Pathologist 2 | Pathologist 3 | |||
|---|---|---|---|---|---|---|
| Maximum density (eos/mm2) | Mean density (eos/mm2)a | Maximum density (eos/mm2) | Mean density (eos/mm2)a | Maximum density (eos/mm2) | Mean density (eos/mm2)a | |
| 1 | 388 | 112 | 524 | 154 | 299 | 88 |
| 2 | 78 | 30 | 74 | 28 | 30 | 26 |
| 3 | 174 | 112 | 211 | 129 | 196 | 128 |
| 4 | 961 | 748 | 1,103 | 825 | 2,291 | 1,443 |
| 5 | 133 | 51 | 354 | 157 | 105 | 53 |
| 6 | 50 | 10 | 10 | 3 | 0 | 0 |
| 7 | 373 | 118 | 485 | 168 | 458 | 131 |
| 8 | 468 | 234 | 1,430 | 828 | 899 | 471 |
| 9 | 570 | 288 | 560 | 353 | 448 | 457 |
| 10 | 772 | 371 | 671 | 339 | 619 | 394 |
| 11 | 1,034 | 426 | 1,076 | 478 | 500 | 404 |
| 12 | 64 | 27 | 148 | 36 | 452 | 256 |
| 13 | 609 | 302 | 963 | 523 | 562 | 454 |
| 14 | 1,538 | 676 | 1,669 | 682 | 746 | 536 |
| 15 | 7 | 3 | 10 | 4 | 6 | 4 |
| 16 | 361 | 204 | 368 | 201 | 186 | 187 |
| 17 | 297 | 175 | 413 | 227 | 423 | 400 |
| 18 | 37 | 9 | 15 | 4 | 4 | 3 |
| 19 | 1,052 | 647 | 1,265 | 716 | 608 | 531 |
| 20 | 251 | 133 | 367 | 191 | 299 | 334 |
| 21 | 733 | 374 | 971 | 524 | 894 | 726 |
| 22 | 896 | 432 | 944 | 469 | 802 | 880 |
| 23 | 63 | 28 | 68 | 36 | 3 | 3 |
| 24 | 152 | 79 | 240 | 143 | 169 | 122 |
| 25 | 426 | 149 | 19 | 5 | 15 | 15 |
| 26 | 74 | 28 | 77 | 31 | 22 | 17 |
| 27 | 79 | 17 | 81 | 17 | 0 | 0 |
| 28 | 268 | 68 | 156 | 51 | 94 | 59 |
| 29 | 705 | 258 | 1,052 | 441 | 853 | 441 |
| 30 | 1,499 | 768 | 1,393 | 667 | 1,673 | 1,001 |
| 31 | 490 | 235 | 403 | 190 | 324 | 154 |
| 32 | 206 | 104 | 162 | 99 | 189 | 149 |
| 33 | 0 | 0 | 5 | 2 | 0 | 0 |
| 34 | 274 | 125 | 266 | 194 | 268 | 186 |
| 35 | 0 | 0 | 5 | 1 | 0 | 0 |
| 36 | 14 | 4 | 19 | 7 | 2 | 1 |
| 37 | 0 | 0 | 5 | 2 | 0 | 0 |
| 38 | 14 | 3 | 9 | 3 | 0 | 0 |
| 39 | 7 | 1 | 4 | 1 | 0 | 0 |
| 40 | 1,064 | 581 | 749 | 482 | 1,704 | 1,175 |
Mean densities are the average of the five high-power fields counted for each subject
Fig. 4.
Graphs of interobserver correlation for determination of maximum eosinophil density (eos/mm2) as calculated with Pearson’s rho. a Pathologist 1 versus pathologist 2. b Pathologist 1 versus pathologist 3. c Pathologist 2 versus pathologist 3
Agreement between pathologists 1 and 2, 1 and 3, and 2 and 3 for assessing degranulation was κ = 0.73, 0.78, and 0.94, respectively (P < 0.001 for each). Agreement between the same pairs of pathologists for eosinophil microabscesses was κ = 0.47, 0.63, and 0.61, respectively (P < 0.001 for each).
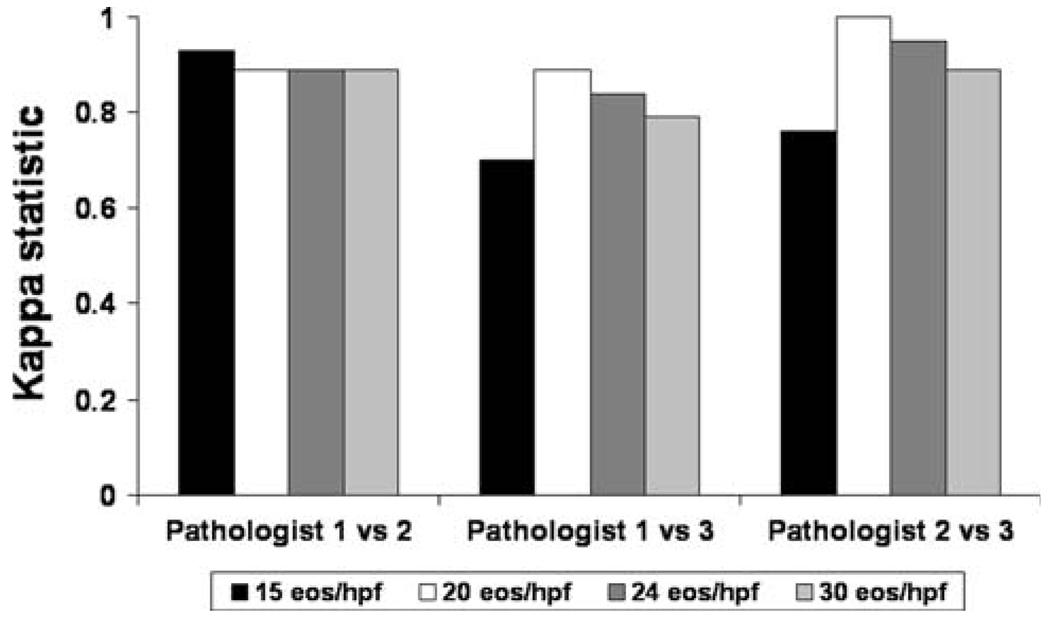
For determining whether the biopsy was consistent with EoE “in the correct clinical context,” agreement between pathologists 1 and 2, 1 and 3, and 2 and 3 was κ = 0.76, 0.78, and 0.71, respectively (P < 0.001 for each). These values further improved when the densities were converted to tissue counts (eos/hpf; Fig. 5).
Fig. 5.
Interobserver agreement measured with the κ statistic for selected diagnostic cut-points for EoE. The tissue count (eos/hpf) was back-calculated from the measured eosinophil density (eos/mm2) for an assumed microscope hpf of 0.24 mm2
Intraobserver Correlation
The maximum eosinophil densities for the first and second read of 20 individual slides for each pathologist are presented in Table 3. Intraobserver correlation was r = 0.99 (P < 0.001) for pathologist 1, r = 0.94 (P < 0.001) for pathologist 2, and r = 0.96 (P < 0.001) for pathologist 3 (Fig. 6a–c). For mean densities, intraobserver correlation was r = 0.97 (P < 0.001) for pathologist 1, r = 0.87 (P < 0.001) for pathologist 2, and r = 0.89 (P < 0.001) for pathologist 3. Slide reading times were 5.4 ± 3.6 and 5.0 ± 1.7 min for each of the reads for pathologist 1, and 6.5 ± 3.1 and 6.0 ± 2.0 min for pathologist 2. Reading times were not reported for pathologist 3.
Table 3.
Maximum esophageal eosinophil densities for determination of intraobserver correlation
| Pathologist 1 | Pathologist 2 | Pathologist 3 | ||||||
|---|---|---|---|---|---|---|---|---|
| Subject | First read | Second read | Subject | First read | Second read | Subject | First read | Second read |
| Maximum eosinophil counts (eos/mm2) | ||||||||
| 1 | 760 | 787 | 21 | 552 | 401 | 41 | 74 | 70 |
| 2 | 0 | 0 | 22 | 383 | 689 | 42 | 10 | 0 |
| 3 | 72 | 86 | 23 | 1,513 | 1,041 | 43 | 9 | 2 |
| 4 | 164 | 175 | 24 | 1,513 | 1,255 | 44 | 0 | 0 |
| 5 | 181 | 192 | 25 | 24 | 19 | 45 | 55 | 13 |
| 6 | 216 | 181 | 26 | 1,179 | 1,428 | 46 | 337 | 218 |
| 7 | 289 | 245 | 27 | 133 | 161 | 47 | 309 | 118 |
| 8 | 1,995 | 1,775 | 28 | 101 | 89 | 48 | 5 | 0 |
| 9 | 117 | 103 | 29 | 61 | 119 | 49 | 0 | 0 |
| 10 | 393 | 283 | 30 | 110 | 32 | 50 | 21 | 12 |
| 11 | 170 | 36 | 31 | 319 | 492 | 51 | 451 | 278 |
| 12 | 71 | 44 | 32 | 5 | 10 | 52 | 39 | 30 |
| 13 | 34 | 21 | 33 | 29 | 75 | 53 | 241 | 183 |
| 14 | 7 | 0 | 34 | 297 | 360 | 54 | 0 | 0 |
| 15 | 372 | 247 | 35 | 694 | 660 | 55 | 0 | 0 |
| 16 | 278 | 216 | 36 | 432 | 492 | 56 | 36 | 18 |
| 17 | 55 | 49 | 37 | 731 | 723 | 57 | 1,154 | 623 |
| 18 | 237 | 174 | 38 | 10 | 19 | 58 | 297 | 270 |
| 19 | 29 | 173 | 39 | 19 | 18 | 59 | 0 | 0 |
| 20 | 0 | 0 | 40 | 5 | 5 | 60 | 349 | 353 |
Note that each pathologist read 20 unique cases twice, for a total of 60 unique cases represented in this table
Fig. 6.
Intraobserver correlation for determination of maximum eosinophil density (eos/mm2) as calculated with Pearson’s rho. a Pathologist 1. b Pathologist 2. c Pathologist 3
Agreement between reads for pathologists 1, 2, and 3 for degranulation was κ = 0.04, 0.89, and 0.60, respectively (P = 0.42,<0.001, and 0.63, respectively). Agreement for eosinophil microabscesses was κ = 0.46, 0.60, and 0.76, respectively (P = 0.007,<0.001, and <0.001, respectively).
For determining whether the biopsy was consistent with EoE, agreement between reads for pathologists 1, 2, and 3 was κ = 0.80, 0.86, and 0.63, respectively (P < 0.001 for each).
Discussion
Because the clinical symptoms and endoscopic findings in EoE are nonspecific, diagnosis requires demonstration of brisk esophageal eosinophilia on histopathologic examination. While the recent consensus guidelines have suggested that a maximum count ≥15 eos/hpf in one hpf is sufficient to make the diagnosis in the correct clinical context [1], these recommendations are largely based on expert opinion because so much diagnostic variability exists in the literature [14]. Moreover, these recommendations include the assumption that counting eosinophils in a tissue specimen is reliable. While it seems that it would be a straightforward task to count eosinophils, in practice and for rigorous research methodology, the issue is actually quite complex.
First, where on the biopsy specimen should a pathologist look? Should only the field with the most eosinophils be reported, or should every field be counted and a mean generated? Should a set number of fields be counted, and the maximum and mean determined? In the literature, many such permutations has been reported [14], and the counting process likely changes the reported result. Second, the size of a hpf on one microscope model differs from that on another. For any given tissue eosinophil density, an eosinophil count on one microscope might be above the diagnostic threshold but below the threshold on a different microscope [14]. Third, no study has previously examined as a primary outcome whether eosinophil counts can be made reliably. If this were not to be the case, much of the existing literature would be uninterpretable.
The present study set out to determine inter- and intraobserver reliability for quantifying tissue esophageal eosinophilia using a clearly defined protocol. It also aimed to validate a new methodology of using digitized slides for determining eosinophil density. Overall, we found excellent inter- and intraobserver correlation for determining both maximum and mean densities. To put the correlation coefficients we observed (in the 0.8 to >0.9 range) into context, it is helpful to examine values observed for other clinical tests. The correlation for determining blood pressure manually compared with automatic cuffs ranges from 0.28 to 0.39 [17]. Correlation between senior cardiologists and critical-care nurses for measuring pulmonary artery wedge pressure is 0.67 [18]. Interobserver correlation for measuring the ankle-arm blood pressure index using an ultrasound probe ranges from 0.88 to 0.92 [19].
In addition, we found that the counts and densities determined on the digitized slides correlated extremely well with the counts made using traditional glass slides and a microscope. This result validates the use of the digitized slides for counting eosinophils in a research setting using this particular counting protocol.
Our analysis for dichotomous variables showed “substantial” to “near-perfect” agreement between the pathologists for eosinophil degranulation, “moderate” to “substantial” agreement for microabscesses, and “substantial” agreement for classifying the slide as a case of EoE “in the correct clinical context.” When we back-calculated eosinophil counts from densities and assessed several potential EoE diagnostic cut-points, we found agreement almost exclusively in the “near-perfect” range. To place these κ values into context, it is again helpful to compare with other tests. Typical κ values for mammographic findings such as mass shape, margin, and calcification are 0.48, 0.48, and 0.32, respectively [20]. Values for determining Gleason scores for prostate cancer range from 0.47 to 0.64 [21]. For Barrett’s esophagus, the κ for distinguishing high-grade dysplasia from intramucosal adenocarcinoma ranges from 0.30 to 0.42 [22, 23], and the κ values for low-grade dysplasia and indeterminate changes are 0.32 and 0.15 [24].
Our results are comparable to the limited published data available on this topic. In 1995, Kelly and colleagues reported the results of ten pediatric patients receiving elemental nutrition for presumed EoE [25]. As a subanalysis, two pathologists performed a joint assessment of eosinophil counts (in eos/hpf) on ten blinded specimens, showing a correlation of 0.96. In 1996, as part of an autopsy study, Lowichik and Weinberg determined eosinophil counts throughout the gastrointestinal tract with the exception of the esophagus [26]. The interobserver correlation was 0.96; intraobserver correlation was not reported. Despite the recently increasing interest in EoE, we were unable to identify other studies examining measurement reliability in this population.
There are several limitations to our methodology. First, there is an extra step required to scan each glass slide, though this only took approximately 2 min per slide. While this may be cumbersome for routine clinical use, it provides no difficulty in the research setting. Second, while the digitized slides have outstanding image and color fidelity as compared with the glass slides, the focal plane of the digitized slide cannot be altered. Therefore, subtle focal adjustments through the plane of the tissue cannot be made, making it difficult to distinguish an eosinophil from a neutrophil in areas of dense inflammation. Additionally, we observed one outlying κ value for intraobserver agreement for eosinophil degranulation, so degranulation may not be a consistently reproducible finding. Other potential limitations of this study include the tertiary care center setting, utilizing pathologists with an interest in gastrointestinal (GI) diseases, and the fact that knowledge of their participation in a research study may have influenced behavior. Whether our results can be replicated in a community setting is unclear.
Despite these limitations, there are several strengths to this study which support using the protocol presented here for future studies. First, our methodology demonstrates excellent inter- and intraobserver reliability for determining eosinophil densities in a large number of clinical specimens spanning a wide range of esophageal eosinophilia. It is also valid when compared with counts obtained from traditional glass slides. Second, once the biopsy slides are scanned, they are a permanent and easily accessible archived resource. Not only can they be de-identified and shared between centers, but if methodological questions arise, repeat analysis is trivial. These images could even be posted on journal websites as supplemental material. There is also the possibility of developing algorithms for computer- assisted eosinophil recognition which, if successful, would remove much of the tedium of determining exact counts manually.
Finally, this study shows that it is both feasible and practical to characterize esophageal eosinophils in density rather than as discrete counts. Because of the issue of variability of hpf between microscopes, counts may not be comparable from center to center [14], or even within centers. Reporting eosinophil density readily allows comparison between all investigators. It will, however, require an extra step to convert the count to density, as well as an awareness of how eosinophil densities relate to previously obtained counts (Table 4). It will take ongoing research to determine a standard eosinophil density that might be used for future diagnostic guidelines. In addition, because eosinophil count alone is only one of several findings of EoE on histopathologic examination, we note that the count must be taken in the context of other findings such as eosinophil degranulation, microabscesses, basal zone hyperplasia, and eosinophil distribution through both the mucosa and the entire biopsy specimen [15].
Table 4.
Conversions between eosinophil density and eosinophil count for typically encountered high-power field sizes
| Eosinophil count (eos/hpf) |
Eosinophil density (eos/mm2) by size of high-power field |
||||
|---|---|---|---|---|---|
| 0.12 mm2 | 0.20 mm2 | 0.24 mm2 | 0.30 mm2 | 0.44 mm2 | |
| 1 | 8 | 5 | 4 | 3 | 2 |
| 5 | 42 | 25 | 21 | 17 | 11 |
| 15 a | 125 | 75 | 63 | 50 | 34 |
| 20 | 167 | 100 | 83 | 67 | 45 |
| 24 | 200 | 120 | 100 | 80 | 55 |
| 30 | 250 | 150 | 125 | 100 | 68 |
| 50 | 417 | 250 | 208 | 167 | 114 |
| 100 | 833 | 500 | 417 | 333 | 227 |
Current diagnostic cut-point as recommended by the consensus guidelines [1]
Bold values represent the current standards for diagnosis
In conclusion, this study found excellent inter- and intraobserver correlation for determining eosinophil densities and counts using digitized slides. Additionally, this method is valid when compared with traditional glass slides. We recommend that this protocol be adopted for research purposes, and be considered for clinical purposes, to help to further standardize the diagnostic process for EoE.
Acknowledgments
This work is funded, in part, by support from the National Institutes of Health training grant T32 DK007634, and award number KL2RR025746 from the National Center for Research Resources.
Abbreviations
- eos/hpf
Eosinophils per high-power field
- hpf
High-power field
- µm2
Square millimeters
- µm2
Square microns
Footnotes
Conflict of Interest No conflicts of interest pertaining to this study exist for any of the authors.
Contributor Information
Evan S. Dellon, Email: edellon@med.unc.edu, Center for Esophageal Diseases and Swallowing, Center for Gastrointestinal Biology and Disease, Division of Gastroenterology and Hepatology, Department of Medicine, University of North Carolina School of Medicine, UNC-CH, CB#7080, Bioinformatics Building, 130 Mason Farm Rd., Chapel Hill, NC 27599-7080, USA.
Karen J. Fritchie, Department of Pathology and Laboratory Medicine, University of North Carolina School of Medicine, Chapel Hill, NC, USA
Tara C. Rubinas, Department of Pathology and Laboratory Medicine, University of North Carolina School of Medicine, Chapel Hill, NC, USA
John T. Woosley, Department of Pathology and Laboratory Medicine, University of North Carolina School of Medicine, Chapel Hill, NC, USA
Nicholas J. Shaheen, Center for Esophageal Diseases and Swallowing, Center for Gastrointestinal Biology and Disease, Division of Gastroenterology and Hepatology, Department of Medicine, University of North Carolina School of Medicine, UNC-CH, CB#7080, Bioinformatics Building, 130 Mason Farm Rd., Chapel Hill, NC 27599-7080, USA
References
- 1.Furuta GT, Liacouras CA, Collins MH, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–1363. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 2.Desai TK, Stecevic V, Chang CH, et al. Association of eosinophilic inflammation with esophageal food impaction in adults. Gastrointest Endosc. 2005;61:795–801. doi: 10.1016/s0016-5107(05)00313-5. [DOI] [PubMed] [Google Scholar]
- 3.Putnam PE. Eosinophilic esophagitis in children: clinical manifestations. Gastrointest Endosc Clin North Am. 2008;18:11–23. doi: 10.1016/j.giec.2007.09.007. vii. [DOI] [PubMed] [Google Scholar]
- 4.Katzka DA. Demographic data and symptoms of eosinophilic esophagitis in adults. Gastrointest Endosc Clin North Am. 2008;18:25–32. doi: 10.1016/j.giec.2007.09.005. viii. [DOI] [PubMed] [Google Scholar]
- 5.Sgouros SN, Bergele C, Mantides A. Eosinophilic esophagitis in adults: a systematic review. Eur J Gastroenterol Hepatol. 2006;18:211–217. doi: 10.1097/00042737-200602000-00015. [DOI] [PubMed] [Google Scholar]
- 6.Straumann A, Rossi L, Simon HU, et al. Fragility of the esophageal mucosa: a pathognomonic endoscopic sign of primary eosinophilic esophagitis? Gastrointest Endosc. 2003;57:407–412. doi: 10.1067/mge.2003.123. [DOI] [PubMed] [Google Scholar]
- 7.Straumann A, Spichtin HP, Bucher KA, Heer P, Simon HU. Eosinophilic esophagitis: red on microscopy, white on endoscopy. Digestion. 2004;70:109–116. doi: 10.1159/000080934. [DOI] [PubMed] [Google Scholar]
- 8.Fox VL. Eosinophilic esophagitis: endoscopic findings. Gastrointest Endosc Clin North Am. 2008;18:45–57. doi: 10.1016/j.giec.2007.09.015. viii. [DOI] [PubMed] [Google Scholar]
- 9.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004;351:940–941. doi: 10.1056/NEJM200408263510924. [DOI] [PubMed] [Google Scholar]
- 10.Straumann A, Simon HU. Eosinophilic esophagitis: escalating epidemiology? J Allergy Clin Immunol. 2005;115:418–419. doi: 10.1016/j.jaci.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Ronkainen J, Talley NJ, Aro P, et al. Prevalence of oesophageal eosinophils and eosinophilic oesophagitis in adults: The population-based Kalixanda study. Gut. 2006 doi: 10.1136/gut.2006.107714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonsalves N. Eosinophilic esophagitis: history, nomenclature, and diagnostic guidelines. Gastrointest Endosc Clin North Am. 2008;18:1–9. doi: 10.1016/j.giec.2007.09.010. vii. [DOI] [PubMed] [Google Scholar]
- 13.Chehade M, Sampson HA. Epidemiology and etiology of eosinophilic esophagitis. Gastrointest Endosc Clin North Am. 2008;18:33–44. doi: 10.1016/j.giec.2007.09.002. viii. [DOI] [PubMed] [Google Scholar]
- 14.Dellon ES, Aderoju A, Woosley JT, Sandler RS, Shaheen NJ. Variability in diagnostic criteria for eosinophilic esophagitis: A systematic review. Am J Gastro. 2007;102:2300–2313. doi: 10.1111/j.1572-0241.2007.01396.x. [DOI] [PubMed] [Google Scholar]
- 15.Collins MH. Histopathologic features of eosinophilic esophagitis. Gastrointest Endosc Clin North Am. 2008;18:59–71. doi: 10.1016/j.giec.2007.09.014. viii–ix. [DOI] [PubMed] [Google Scholar]
- 16.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 17.Pannarale G, Bebb G, Clark S, et al. Bias and variability in blood pressure measurement with ambulatory recorders. Hypertension. 1993;22:591–598. doi: 10.1161/01.hyp.22.4.591. [DOI] [PubMed] [Google Scholar]
- 18.Al-Kharrat T, Zarich S, Amoateng-Adjepong Y, Manthous CA. Analysis of observer variability in measurement of pulmonary artery occlusion pressures. Am J Respir Crit Care Med. 1999;160:415–420. doi: 10.1164/ajrccm.160.2.9808082. [DOI] [PubMed] [Google Scholar]
- 19.Aboyans V, Lacroix P, Lebourdon A, et al. The intra- and interobserver variability of ankle-arm blood pressure index according to its mode of calculation. J Clin Epidemiol. 2003;56:215–220. doi: 10.1016/s0895-4356(02)00584-x. [DOI] [PubMed] [Google Scholar]
- 20.Lazarus E, Mainiero MB, Schepps B, Koelliker SL, Livingston LS. BI-RADS lexicon for US and mammography: interobserver variability and positive predictive value. Radiology. 2006;239:385–391. doi: 10.1148/radiol.2392042127. [DOI] [PubMed] [Google Scholar]
- 21.Allsbrook WC, Jr, Mangold KA, Johnson MH, et al. Interobserver reproducibility of Gleason grading of prostatic carcinoma: urologic pathologists. Hum Pathol. 2001;32:74–80. doi: 10.1053/hupa.2001.21134. [DOI] [PubMed] [Google Scholar]
- 22.Ormsby AH, Petras RE, Henricks WH, et al. Observer variation in the diagnosis of superficial oesophageal adenocarcinoma. Gut. 2002;51:671–676. doi: 10.1136/gut.51.5.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Downs-Kelly E, Mendelin JE, Bennett AE, et al. Poor Interobserver Agreement in the Distinction of High-Grade Dysplasia and Adenocarcinoma in Pretreatment Barrett’s Esophagus Biopsies. Am J Gastroenterol. 2008 doi: 10.1111/j.1572-0241.2008.02020.x. [DOI] [PubMed] [Google Scholar]
- 24.Montgomery E, Bronner MP, Goldblum JR, et al. Reproducibility of the diagnosis of dysplasia in Barrett esophagus: a reaffirmation. Hum Pathol. 2001;32:368–378. doi: 10.1053/hupa.2001.23510. [DOI] [PubMed] [Google Scholar]
- 25.Kelly KJ, Lazenby AJ, Rowe PC, et al. Eosinophilic esophagitis attributed to gastroesophageal reflux: improvement with an amino acid-based formula. Gastroenterology. 1995;109:1503–1512. doi: 10.1016/0016-5085(95)90637-1. [DOI] [PubMed] [Google Scholar]
- 26.Lowichik A, Weinberg AG. A quantitative evaluation of mucosal eosinophils in the pediatric gastrointestinal tract. Mod Pathol. 1996;9:110–114. [PubMed] [Google Scholar]