Abstract
The nucleotide receptor P2X7 is an immunomodulatory cation channel and a potential therapeutic target. P2X7 is expressed in immune cells such as monocytes/macrophages and is activated by extracellular ATP following tissue injury or infection. Ligand binding to P2X7 can stimulate ERK1/2, the transcription factor CREB, enzymes linked to the production of reactive oxygen species and interleukin-1 isoforms, and the formation of a non-specific pore. However, little is known about the biochemistry of P2X7, including whether the receptor is N-linked glycosylated and if this modification affects receptor function. Here we provide evidence that P2X7 is sensitive to the glycosidases EndoH and PNGase F, and that the human receptor appears glycosylated on N187, N202, N213, N241 and N284. Mutation of N187 results in diminished P2X7 agonist-induced phosphorylation of ERK1/2, CREB, and p90 ribosomal S6 kinase, as well as decreased pore formation. In further support of a role for glycosylation in receptor function, treatment of RAW 264.7 macrophages with the N-linked glycosylation synthesis inhibitor tunicamycin attenuates P2X7 agonist-induced, but not phorbol ester-induced, ERK1/2 phosphorylation. Interestingly, residue N187 belongs to an N-linked glycosylation consensus sequence found in six of the seven P2X family members, suggesting this site is fundamentally important to P2X receptor function. To address the mechanism whereby N187 mutation attenuates receptor activity, we developed a live cell proteinase K digestion assay that demonstrated altered cell surface expression of P2X7 N187A. This is the first report to map human P2X7 glycosylation sites and reveal residue N187 is critical for receptor trafficking and function.
Keywords: P2X7, N-linked glycosylation, cell surface expression, ERK1/2, macrophages
The nucleotide ATP can act as an important extracellular signaling molecule that regulates multiple processes, including neurotransmission and immune response mediator production (1,2). ATP is released in millimolar concentrations from cells following infection or tissue injury and can stimulate cells in the microenvironment by binding to the P2 nucleotide receptors (3). The P2 nucleotide receptors have been divided into two major subfamilies: the G protein-coupled P2Y receptors and the ionotropic P2X cation channels (4).
The cation channel P2X7 is considered an important component of the inflammatory response (5). Activation of P2X7 by extracellular nucleotides leads to the processing of interleukin-1β and the production of reactive oxygen species through the NADPH oxidase complex (6,7). The P2X7 receptor stimulates a number of downstream targets including mitogen activated protein kinases (MAPK) and several transcription factors, including cyclic-AMP response element-binding protein (CREB) and activating transcription factor 1 (ATF1) (7–11). Upon ligand binding, P2X7 mediates Ca2+ and Na+ influx and K+ efflux. Prolonged stimulation of P2X7 can also promote the formation of a non-specific pore, allowing for molecules up to 900 Da to enter the cell (4).
It has been proposed that P2X7 is an attractive therapeutic target for inflammatory diseases (5). Animal studies have shown that pharmacological targeting of P2X7 may be used to treat certain types of arthritis (12,13), and a recent clinical study has correlated P2X7 activity with virus-induced loss of asthma control (14).
Although numerous P2X7 studies have focused on ATP-induced cell signaling, relatively few reports have examined the biochemical properties of the receptor. One aspect of P2X7 biology that is poorly understood involves the glycosylation status of the receptor and how glycosylation contributes to receptor function. Such an analysis is important given that glycosylation of plasma membrane-bound proteins is often critical for numerous processes, including protein folding, cell adhesion, and pathogen recognition of host cells (15,16).
In the present study, we report that P2X7 is N-linked glycosylated on five residues and this post-translational modification is important for P2X7 agonist-stimulated signaling and pore formation. We present the first experimental evidence that human P2X7 is glycosylated on residues 187, 202, 213, 241 and 284, and that mutation at the conserved amino acid 187 results in decreased nucleotide-induced signaling events. In addition, we propose P2X7 N187A exhibits attenuated activity because its expression on the plasma membrane is altered. A recent report has shown that two P2X7 naturally-occurring single nucleotide polymorphisms (SNPs) located near residue N187, E186K and L191P, exhibit decreased channel and pore activities (17). Those findings and this current report demonstrate that the extracellular region of P2X7 surrounding the N187 glycosylation site is critical for receptor function.
MATERIALS AND METHODS
Reagents
The potent P2X7 agonist, 2′ (3′)-O-(4-benzoylbenzoyl)-ATP (BzATP), and tunicamycin were obtained from Sigma (St. Louis, MO). EndoH and PNGase F were purchased from New England Biolabs (Ipswich, MA), and proteinase K was purchased from Promega (Madison, WI). The anti-c-Myc antibody (cat. no. sc-40), the anti-P2X7 antibody (cat. no. sc-25698) used for immunofluorescence and the immunoblot in Fig. 7, as well as the anti-EGFR antibody (cat. no. sc-03), were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-PDI antibody (cat. no. SPA-891) was obtained from Stressgen (Ann Arbor, MI). The anti-ERK1/2 antibody (cat. no. 06-182) was purchased from Millipore (Billerica, MA), the anti-pERK1/2 antibody (cat. no. 44680G) was purchased from Invitrogen (Carlsbad, CA), and the anti-P2X7 antibody (cat. no. 550694) used for the immunoblots in Figs. 3, 6 and 10 was purchased from BD Biosciences (San Jose, CA). The anti-CREB (cat. no. 9197), anti-pCREB (cat. no. 9191), anti-pp90RSK (cat. no. 9344) and anti-β-tubulin (cat. no. 2146) antibodies were purchased from Cell Signaling Technology (Danvers, MA).
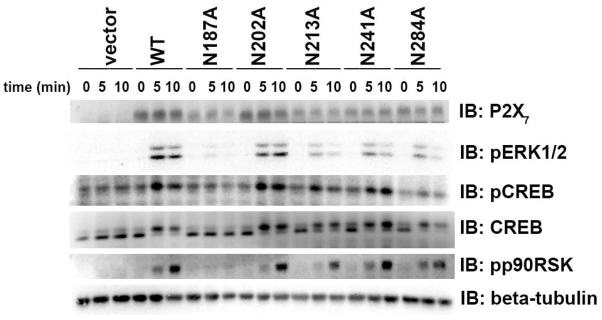
Fig. 7. Mutation of P2X7 N-linked glycosylation sites results in attenuated BzATP-induced signaling.
Wild-type P2X7 and the Asn to Ala mutants were stably-expressed in HEK293 cells by selecting G418 sulfate-resistant populations. The cells were treated with 250 μM BzATP for 5 or 10 min, and cell lysates were prepared and immunoblotted with anti-P2X7, anti-pERK1/2, anti-pCREB, anti-pp90RSK, and anti-β-tubulin antibodies. The β-tubulin immunoblot is the loading control for the anti-pERK1/2 immunoblot. These data are representative of three experiments.
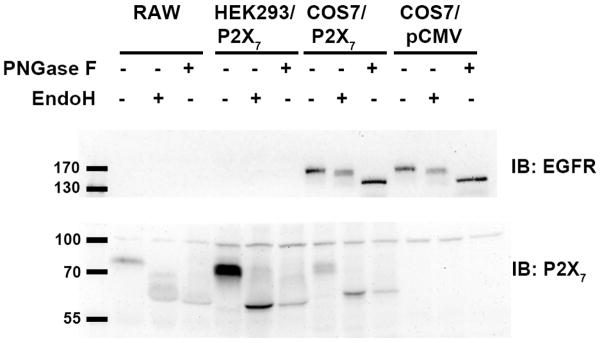
Fig. 3. Endogenous and exogenous P2X7 is sensitive to glycosidase treatment.
Proteins from murine RAW 264.7 macrophages naturally expressing wild-type P2X7 as well as human HEK293 and monkey COS7 cells transfected with human wild-type P2X7 were denatured and treated with either EndoH or PNGase F for 1 h at 37°C as described under Materials and Methods. The proteins were then resolved on polyacrylamide gels containing SDS and immunoblotted for P2X7. Cleavage of EGFR was used as a positive control to show EndoH and PNGase F were active. These data are representative of three or more experiments.
Fig. 6. Treatment of RAW 264.7 macrophages with tunicamycin results in attenuated BzATP-induced ERK1/2 activation.
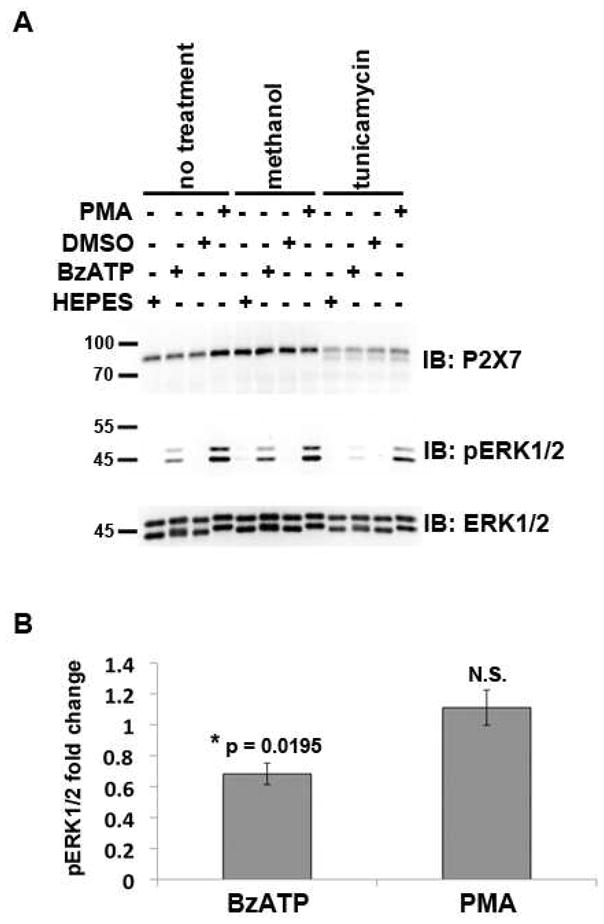
A) RAW 264.7 macrophages were treated with the N-linked glycosylation synthesis inhibitor tunicamycin (5 μg/mL) for 42 h, and the cells were stimulated with either 250 μM BzATP or 1 μg/mL PMA for 5 min. Cell lysates were then immunoblotted with anti-P2X7, anti-pERK1/2 and anti-ERK1/2 antibodies. Methanol is the vehicle control for tunicamycin, HEPES is the buffer control for BzATP, and DMSO is the vehicle control for PMA. These data are representative of four experiments. B) The fold changes between tunicamycin-treated cells stimulated with BzATP or PMA versus tunicamycin-treated cells stimulated with buffer control were quantified and graphed. The band intensities of the phospho-ERK1/2 bands were normalized to total ERK1/2, and the percent activation of ERK1/2 with either BzATP or PMA in comparison to the appropriate buffer control was calculated. Student’s t-tests were performed to determine the p values. N.S. = not significant.
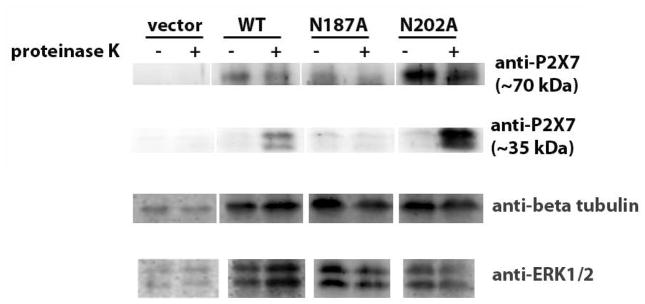
Fig. 10. P2X7 N187A expression on the cell surface is altered.
HEK293 cells stably expressing pcDNA3 vector control, P2X7 WT, P2X7 N187A and P2X7 N202A were treated with 333 μg/mL proteinase K for 90 min as described under Materials and Methods. The cells were then boiled for 10 min to inactivate the enzyme, diluted in 2X sample buffer, sonicated, and immunoblotted with anti-P2X7, anti-ERK1/2 and anti-β-tubulin antibodies. These data are representative of three or more experiments.
Cell Culture, Harvesting and Protein Assays
Human HEK293 and monkey COS7 cells (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% cosmic calf serum (Mediatech, Herndon, VA), 1% L-glutamine and 100 U/ml penicillin/streptomycin, and the murine macrophage RAW 264.7 and RAW 264.7 SF cells were cultured in RPMI-1640 media supplemented with 5% cosmic calf serum, 2 mM sodium pyruvate, and 1% L-glutamine. The cells were incubated at 37 °C under 5% CO2. RAW 264.7 SF is a mutant cell line that contains a Ser to Phe mutation at amino acid 342 in the extracellular domain of P2X7, rendering the receptor non-functional and the cell line as a useful negative control (8,18,19). Cell lysates were generally prepared by harvesting the cells in 2X sample buffer (20 mM Tris, 2 mM EDTA, 2 mM DTT, 1 mM Na3VO4, 2% SDS, 20% glycerol) followed by boiling and sonicating the samples. Total protein concentrations of lysates were quantified using a BCA protein assay kit obtained from Thermo Scientific (Waltham, MA).
P2X7 Constructs
Full-length human P2X7 (accession number AAH11913) was subcloned into pcDNA3 and pCMV-Myc. Subcloning P2X7 into pCMV-Myc yielded an N-terminally tagged receptor. The P2X7 point mutants used in these studies were generated by site-directed mutagenesis using the following primers and their reverse complements:
P2X7 N187A 5′-CAGTGCCGAAGCCTTCACTGTGCTC-3′,
P2X7 N187H 5′-CAGTGCCGAACACTTCACTGTGCTC-3′,
P2X7 N187Q 5′-CAGTGCCGAACAGTTCACTGTGCTC-3′,
P2X7 N202A 5′-CCGGCCACGCCTACACCACG-3′,
P2X7 N213A 5′-CCAGGTTTAGCCATCACTTGTACC-3′,
P2X7 N241A 5′-GAAACAGGCGATGCTTTTTCAG-3′,
P2X7 N284A 5′-CAAGACCACCGCCGTGTCCTTGTAC-3′.
Glycosidase Assays
Cell lysates were treated with EndoH or PNGase F according to the manufacturer’s instructions. Briefly, ~30 μg of cell extract protein was denatured and then treated with 250 units of glycosidase at 37°C for 1 h.
Immunofluorescent Staining
RAW 264.7 macrophages and COS7 cells were cultured on glass cover slips, fixed with 4% paraformaldehyde, permeabilized with 1% Triton X-100, and stained with the indicated antibodies. The cells were then stained with the following secondary antibodies from Molecular Probes (Eugene, OR): Alexa Fluor® 488 donkey anti-rabbit (used with anti-P2X7 in Figs. 4a and 9), Alexa Fluor® 488 donkey anti-mouse (used with anti-PDI), and Alexa Fluor® 594 donkey anti-rabbit (used with anti-P2X7 in Fig. 4c). The samples were imaged with either a Zeiss Axioplan 2 microscope (Figs. 4a and 9) or a Bio-Rad Radiance 2100 MP Rainbow confocal microscope (Fig. 4c). The ImageJ software co-localization plug-in was used to generate the co-localized image in Fig. 4c.
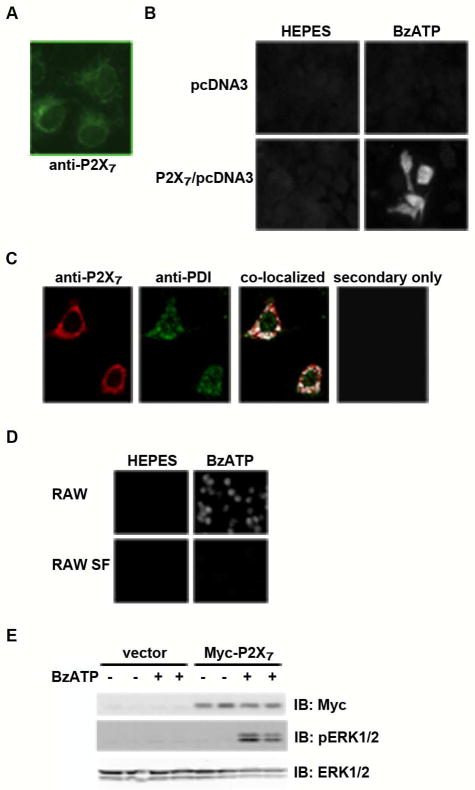
Fig. 4. P2X7 is highly localized in intracellular compartments but is activated in response to extracellular BzATP administration.
A) The human nucleotide receptor P2X7 was expressed in COS7 cells, and the cells were stained with an anti-P2X7 antibody and visualized with a Zeiss Axioplan 2 microscope. This image is representative of >6 experiments. B) COS7 cells were transfected with either P2X7/pcDNA3 or pcDNA3 and were stimulated with either 250 μM BzATP or HEPES buffer control in the presence of 1 μM YO-PRO-1. The cells were fixed and relative dye uptake was imaged with a Zeiss Axioplan 2 microscope. These data are representative of >6 experiments. C) RAW 264.7 macrophages were fixed, stained with anti-P2X7 and anti-PDI antibodies and visualized with a Bio-Rad Radiance 2100 MP Rainbow confocal microscope. As a negative control, the cells were stained with secondary antibody only. The represented images are of one z-stack. The anti-P2X7 and anti-PDI images were merged using the ImageJ co-localization plug-in. These data are representative of two experiments. D) RAW 264.7 and RAW 264.7 SF macrophages were stimulated with either BzATP or HEPES buffer control in the presence of 1 μM YO-PRO-1 and visualized as live cells using an Olympus IX Fluorescence Inverted Microscope. These YO-PRO-1 dye uptake assays are representative of five experiments. E) HEK293 cells were transfected with either P2X7/pCMV-Myc or pCMV-Myc and were stimulated with either 250 μM BzATP or HEPES buffer control for 5 min. Cell lysates were prepared and immunoblotted using anti-Myc, anti-pERK1/2 and anti-ERK antibodies. These data are representative of >8 experiments.
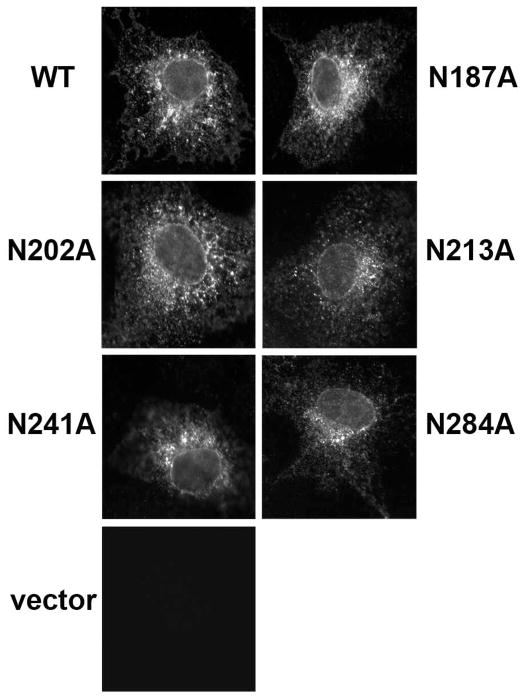
Fig. 9. Wild-type and mutant P2X7 display similar localization patterns.
Wild-type P2X7, the Asn to Ala mutants, and vector control were expressed in COS7 cells. Approximately 24 h after the cells were transfected, they were fixed with paraformaldehyde, stained with an anti-P2X7 antibody, and imaged with a Zeiss Axioplan 2 microscope. Similar data were obtained in two separate experiments.
YO-PRO-1 Dye Uptake Assay
COS7 cells were cultured on glass cover slips and were transfected with the indicated constructs using FuGENE™ 6 transfection reagent according to the manufacturer’s instructions (Roche). The COS7 and RAW 264.7 cells were stimulated at room temperature in potassium-glutamate buffer (130 mM K-glutamate, 20 mM HEPES-KOH (pH 7.4), 5 mM KCl, 0.1% BSA, 10 mM glucose) with 250 μM BzATP or HEPES buffer control for 10 or 20 min in the presence of 1 μM YO-PRO-1 (Molecular Probes). Potassium-glutamate buffer is known to facilitate robust YO-PRO-1 uptake in response to BzATP, as previously reported (20,21). The cells were then treated with 10 mM MgCl2 to close the pore, fixed with 4% paraformaldehyde, and imaged with either a Zeiss Axioplan 2 microscope or an Olympus IX Fluorescence Inverted Microscope (20,21).
ERK1/2, CREB and p90RSK Phosphorylation/Activation Assays
HEK293 cells were transfected with pcDNA3, P2X7/pcDNA3 or the P2X7 Asn to Ala mutants using FuGENE™ 6 transfection reagent, and G418 sulfate-resistant populations were selected. For the transient transfection experiments, HEK293 cells were transfected with pCMV-Myc, P2X7/pCMV-Myc, P2X7 N187H/pCMV-Myc, P2X7 N187Q/pCMV-Myc, or the Myc-tagged P2X7 Asn to Ala mutants. The cells were serum starved for 1–4 h, treated with 250 μM BzATP for 5 or 10 min, harvested, and cell extracts were immunoblotted as detailed below.
Immunoblotting
Proteins from cell lysates (~10–30 μg) were separated by electrophoresis at 15–20 mA on 10% SDS-polyacrylamide gels, transferred to polyvinylidene fluoride (PVDF) membranes, and blocked in 5% nonfat dry milk. The membranes were probed with the indicated primary antibodies, incubated with secondary antibodies conjugated to horseradish peroxidase (Santa Cruz Biotechnology), and visualized by enhanced chemiluminescence.
Proteinase K Digestion Assays
HEK293 cells stably expressing wild-type or mutant P2X7 were washed off the plate with media and pelleted. The cells were suspended in serum-free DMEM and treated with 333 μg/mL proteinase K or buffer control for 90 min at 37°C. The samples were then boiled for 10 min, diluted in 2X sample buffer, sonicated and immunoblotted.
RESULTS
P2X7 is sensitive to glycosidase treatment
The NetNGlyc program (Technical University of Denmark) was used to identify putative N-linked glycosylation sites in the seven human P2X family members (Figs. 1 and 2). N-linked glycosylation sites contain the consensus sequence Asn-X-Ser/Thr, where X is any amino acid except proline (22). Cell lysates from RAW 264.7 macrophages, which express endogenous P2X7 (8), and HEK293 and COS7 cells expressing exogenous human receptor were treated with the glycosidases EndoH or PNGase F and immunoblotted for P2X7. We chose to include COS7 and HEK293 cells in our studies because they do not express detectable levels of endogenous P2X7 (23). The enzyme EndoH cleaves high mannose and some hybrid N-linked glycosylation modifications while PNGase F is less specific and can also cleave complex oligosaccharides with di-, tri-, and tetra-antennary arms (24). Both endogenous and exogenous P2X7 are sensitive to EndoH and PNGase F as indicated by faster electrophoretic migration (Fig. 3). The epidermal growth factor receptor (EGFR) was immunoblotted as a positive control to demonstrate the EndoH and PNGase F enzymes were functional (25).
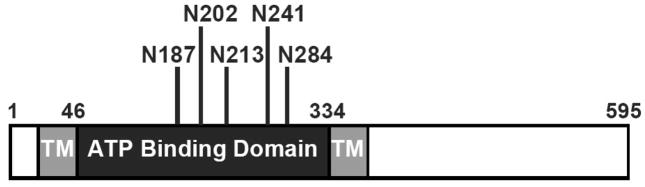
Fig. 1. Graphical representation of the major P2X7 domains and its putative N-linked glycosylation sites.
The nucleotide receptor P2X7 has two predicted transmembrane domains (TM) and an extracellular ATP binding domain. The extracellular portion of P2X7 contains putative N-linked glycosylation sites at amino acids 187, 202, 213, 241 and 284. The PredictProtein program was used to determine where the putative transmembrane domains and N-linked glycosylation sites are located (31).
Fig. 2. All human P2X family members contain putative N-linked glycosylation sites.
The protein sequences of the seven human P2X family members were aligned using ClustalW (32), and the NetNGlyc 1.0 program was used to identify potential N-linked glycosylation sites (marked in red boxes). Identical residues are marked as black and chemically similar residues are marked as gray. The following protein accession numbers were used in the alignment: P2X1 AAC24494, P2X2 Q9UBL9, P2X3 NP_002550, P2X4 NP_002551, P2X5 AAH39015, P2X6 AAF13303, and P2X7 AAH11913.
Endogenous P2X7 in RAW 264.7 and transfected P2X7 in HEK293 and COS7 cells are functional
EndoH cleaves N-linked glycosylation modifications that are processed in the Golgi, and proteins that are insensitive to this enzyme are thought to have trafficked out of the Golgi and possibly to the plasma membrane (26). Because we did not observe a large EndoH-resistant population of P2X7 in the HEK293, COS7 and RAW 264.7 cells, we tested the ideas that P2X7 is primarily localized at intracellular sites and a small population of the receptor is expressed on the cell surface that is capable of eliciting P2X7 agonist-induced events. As assessed by immunofluorescence, both exogenous P2X7 in COS7 cells and endogenous P2X7 in RAW 264.7 macrophages appear to be highly represented at intracellular sites (Fig. 4a,c). In addition, P2X7 in RAW 264.7 and COS7 cells co-localizes with the endoplasmic reticulum (ER) marker protein disulfide isomerase (PDI) (Fig. 4c and data not shown). Although P2X7 is not readily detected on the plasma membrane via immunofluorescence even in naturally P2X7-expressing RAW 264.7 macrophages, we observed that both the P2X7-transfected COS7 and RAW 264.7 cells, but not the P2X7-defective cell line RAW 264.7 SF, are capable of taking up the fluorescent dye YO-PRO-1 in the presence of BzATP (Fig. 4b,d). BzATP is a synthetic ATP derivative that is used to activate P2X7, and the YO-PRO-1 dye uptake assay is a method of assessing the ability of P2X7 to stimulate the formation of a pore (20). To demonstrate that exogenously-expressed P2X7 in HEK293 cells is functional, BzATP-induced ERK1/2 phosphorylation tests were employed (Fig. 4e). The kinases ERK1 and ERK2 are phosphorylated/activated by BzATP when wild-type P2X7 is expressed but not when the cells are transfected with the empty vector control.
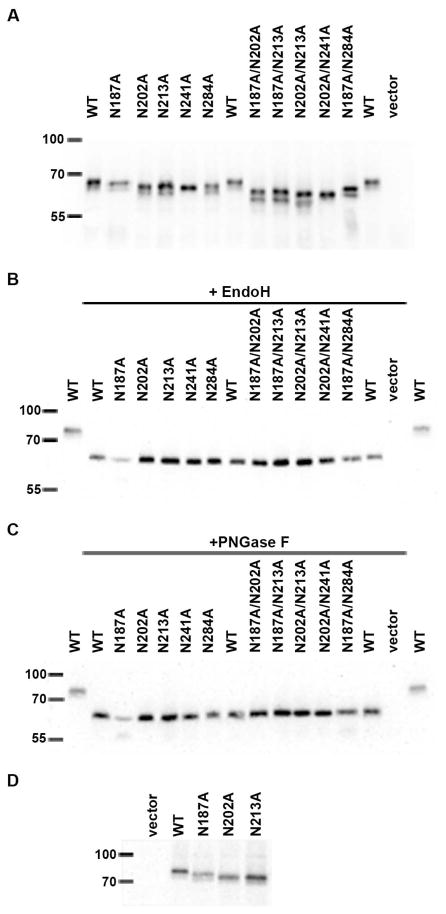
Mutation of human P2X7 at N187, N202, N213, N241 and N284 results in increased electrophoretic mobility
To identify the residues that likely contain a glycosylation modification, the five predicted N-linked glycosylation sites were mutated to alanine either individually or doubly, and their electrophoretic mobility was examined (Fig. 5). All of the Asn to Ala single and double mutants migrated faster than wild-type P2X7. To illustrate that these migratory differences were likely due to an altered glycosylation profile and not protein degradation, lysates were treated with EndoH or PNGase F and immunoblotted for P2X7. After cleavage with either glycosidase, the mutants had comparable apparent molecular weights as enzyme-treated wild-type receptor (Fig. 5b,c). In addition, we observed that P2X7 N187A appears to have a slightly higher molecular weight than receptors containing the N202A, N213A, N241A or N284A mutations (Fig. 5a,d) as assessed by SDS-polyacrylamide gel electrophoresis. It is possible that this site (N187) is differentially glycosylated compared to the other sites and/or that the protein is altered in its folding and interaction with SDS.
Fig. 5. P2X7 is glycosylated on N187, N202, N213, N241 and N284.
A) Wild-type P2X7 (WT) and the Asn to Ala mutants were expressed in HEK293 cells and cell lysates were immunoblotted with anti-Myc antibody. The five single Asn to Ala mutants exhibited faster electrophoretic mobility than WT receptor, and the double mutants migrated further than the single mutants. The proteins were denatured and treated with B) EndoH or C) PNGase F for ~1 h at 37°C and immunoblottred with anti-Myc antibody. The data shown in panels A-C are representative of two or more experiments. D) P2X7 WT and Asn to Ala mutants were expressed in HEK293 cells and lysates were immunoblotted with anti-Myc. The P2X7 N187A mutant does not migrate as fast as the other Asn to Ala mutants but does migrate faster than WT. These data are representative of >10 experiments.
Treatment of RAW 264.7 macrophages with tunicamycin results in attenuated BzATP-induced ERK1/2 activation
To determine whether N-linked glycosylation is required for P2X7 agonist-induced ERK1/2 phosphorylation/activation, RAW 264.7 macrophages were treated with the N-linked glycosylation synthesis inhibitor tunicamycin for 42 h, and the cells were stimulated with either BzATP or phorbol 12-myristate 13-acetate (PMA) for 5 min to activate ERK1/2. As shown in Fig. 6, treatment of cells with 5 μg/mL tunicamycin decreases BzATP but not PMA-stimulated ERK1/2 phosphorylation. Treatment with tunicamycin results in the formation of faster migratory P2X7 proteins, which are presumably newly synthesized P2X7 receptors that do not contain glycosylation modifications. In addition, less full-length receptor is observed in the cells treated with the inhibitor, providing further evidence that a smaller pool of P2X7 is N-linked glycosylated in these samples (Fig. 6a). It should be noted that a cell viability assay was conducted to determine whether treatment with 5 μg/mL tunicamycin for 42 h results in significant cell death, and we found 85% of the tunicamycin-treated cells were metabolically active in comparison to the methanol control-treated cells (data not shown) indicating that decreases observed after tunicamycin treatment were not due to cytotoxicity. The fold change in ERK1/2 phosphorylation between tunicamycin-treated and buffer control-treated cells is depicted in Fig. 6b. Treatment with tunicamycin results in approximately a 30% reduction in BzATP-induced ERK1/2 phosphorylation but no significant change in PMA-stimulated ERK1/2 phosphorylation.
Mutation of P2X7 N-linked glycosylation sites results in attenuated BzATP-induced signaling and pore formation
To determine which N-linked glycosylation sites of P2X7 are required for its activity, we examined BzATP-induced ERK1/2, CREB and p90RSK phosphorylation/activation in cells stably-expressing the wild-type receptor or the Asn to Ala mutants. The phosphorylation of CREB was assessed by immunoblotting cell lysates with an antibody that detects phosphorylated/activated CREB and by analyzing mobility shifts on an anti-CREB antibody immunoblot. Cells expressing P2X7 N187A do not detectably exhibit ERK1/2, CREB or p90RSK phosphorylation in the presence of the P2X7 agonist BzATP (Fig. 7). Mutation of P2X7 N213, N241 or N284 also resulted in an apparent reduction in BzATP-induced ERK1/2 phosphorylation, but not to the same extent as the P2X7 N187A mutation. Conversely, mutation of P2X7 N202 to Ala did not result in attenuated ligand-stimulated ERK1/2, CREB or p90RSK phosphorylation in either the stably-expressing cells (Fig. 7) or in transient transfection experiments (data not shown). The data from three separate experiments revealed that the fold change in ERK1/2 phosphorylation following 10 minutes of BzATP treatment in wild-type P2X7-expressing cells ranged from 8.2 to 29 fold. In contrast, the change in BzATP-induced ERK1/2 activation ranged from 1.5 to 4.8 fold for P2X7 N213A, 3.4 to 11 fold for P2X7 N241A, and 3.8 to 13 fold for P2X7 N284A. In addition, attenuated BzATP-induced ERK1/2 activation was also observed in HEK293 cells transiently expressing N213A, N241A and N284A (data not shown). Similar trends in CREB activation were also obtained, but smaller fold changes were achieved in comparison to ERK1/2 (data not shown). For example, in three separate experiments, the average increase in CREB activation in BzATP-treated cells expressing wild-type P2X7 was 2.0-fold. These decreases in BzATP-stimulated responses observed in cells expressing a mutant P2X7 wherein the Asn at position 284 has been altered to alanine complements a prior report demonstrating that the introduction of an Asn residue at the equivalent residue in mouse P2X7 (D284), confers increased sensitivity to BzATP and ATP (27). These data support the idea that P2X7 N284 may also be important for human P2X7 function.
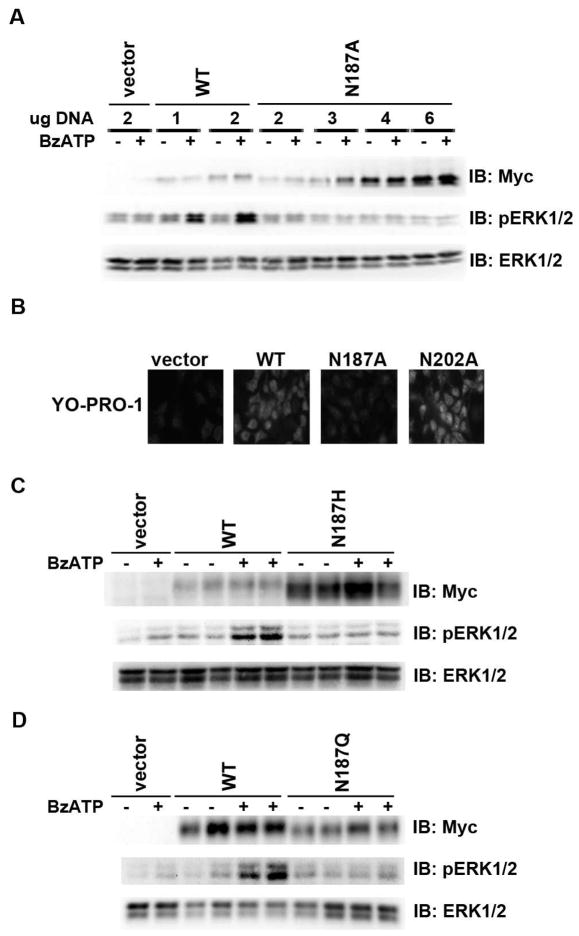
Because P2X7 N187A generally exhibited lower expression levels than other Asn to Ala mutants, we increased the amount of P2X7 N187A/pCMV-Myc DNA used in transient transfections in an effort to drive its expression to a level that is comparable to wild-type receptor. Even when P2X7 N187A is expressed at a similar or higher level as wild-type receptor, the mutant receptor does not exhibit BzATP-induced ERK1/2 activation (Fig. 8a). In addition, COS7 cells transiently expressing P2X7 N187A exhibit reduced BzATP-induced YO-PRO-1 uptake in comparison to cells expressing wild-type receptor (Fig. 8b), providing further evidence that this residue is important for P2X7 function. To further verify that residue N187 is important for receptor activity, and that our observations with an alanine substitution at this position are not restricted to the specific chemistry (hydrophobicity) of alanine, we also mutated P2X7 residue N187 to the more polar residue histidine, as well as to a residue structurally related to the asparagine normally located at this site, i.e., glutamine. As shown in Figs. 8c and 8d, mutation of P2X7 N187 to His or Gln also results in diminished BzATP-induced ERK1/2 phosphorylation, thereby supporting the specific importance of asparagine/glycosylation at this site.
Fig. 8. Mutation of P2X7 N187 results in decreased BzATP-induced ERK1/2 activation and pore formation.
A) HEK293 cells were transfected with the indicated amounts of pCMV-Myc vector control, P2X7 WT/pCMV-Myc or P2X7 N187A/pCMV-Myc. After 24 h, the cells were treated with 250 μM BzATP for 5 min, and cell lysates were prepared and immunoblotted with anti-Myc, anti-pERK1/2 and anti-ERK1/2 antibodies. These data are representative of two independent experiments. B) COS7 were transfected with pCMV-Myc, P2X7 WT/pCMV-Myc, P2X7 N187A/pCMV-Myc or P2X7 N202A/pCMV-Myc. After 24 h, a YO-PRO-1 dye uptake was performed. Similar data were obtained in two separate experiments. C) HEK293 cells were transfected with pCMV-Myc vector control, P2X7 WT/pCMV-Myc or P2X7 N187H/pCMV-Myc. After 24 h, the cells were treated with 250 μM BzATP for 5 min, and cell lysates were prepared and immunoblotted with anti-Myc, anti-pERK1/2 and anti-ERK1/2 antibodies. These data are representative of three independent experiments. D) HEK293 cells were transfected with pCMV-Myc vector control, P2X7 WT/pCMV-Myc or P2X7 N187Q/pCMV-Myc. After 24 h, the cells were treated with 250 μM BzATP for 5 min, and cell lysates were prepared and immunoblotted with anti-Myc, anti-pERK1/2 and anti-ERK1/2 antibodies. These data are representative of three independent experiments.
Wild-type and mutant P2X7 display similar localization patterns
To delineate the mechanism by which P2X7 N187A exhibits attenuated activity, we considered the possibilities that it may be mislocalized or not efficiently trafficked to the plasma membrane. To test these ideas, we expressed wild-type P2X7 and the Asn to Ala mutants in COS7 cells and examined their overall localization patterns by immunofluorescence. As shown in Fig. 9, wild-type P2X7 and the glycosylation mutants, including N187A, all display a perinuclear, endoplasmic reticulum-like pattern. Even though P2X7 N187A has diminished activity, it appears to localize in a pattern comparable to wild-type P2X7 and to the other Asn to Ala mutants. These data support the notion that the diminished activity of P2X7 N187A is not likely the result of gross mislocalization of the receptor.
P2X7 N187A cell surface expression is altered
Because we did not detect any major differences in the localization between P2X7 wild-type and N187A in fixed cells as assessed by immunostaining, we used a live cell proteinase K digestion assay to analyze the plasma membrane expression of P2X7 over a given period of time and to test the hypothesis that P2X7 N187A does not traffic as efficiently to the plasma membrane as wild-type P2X7. When the receptor is expressed on the surface of live cells, the extracellular domain is digested with proteinase K, liberating the C-terminal tail. In this assay, we treated cells with protease for 90 min, allowing the enzyme to digest proteins that become exposed on the cell surface at any point during the incubation period. A key advantage of this approach is that it represents an accumulation of events that occur over 90 min vs the single “snapshot” in time represented by the immunostaining approach. We predicted that treatment of live cells with proteinase K would result in the cleavage of full-length wild-type P2X7, but not the N187A mutant, yielding a fragment containing only the intracellular domain. As depicted in Fig. 1, human P2X7 contains an intracellular C-terminal domain that contains more than 200 amino acids. Digestion of HEK293 cells stably expressing wild-type P2X7 or N202A but not the N187A receptor resulted in the formation of ~35 kDa proteins that are immunoreactive with an anti-P2X7 antibody that was raised against the C-terminus of the protein (Fig. 10). These immunoreactive proteins are likely the C-terminal tail of full-length plasma membrane-localized P2X7. The predicted size of the C-terminal tail, including the second transmembrane domain, is ~30 kDa. The ~35 kDa proteins were also immunoreactive with a different anti-P2X7 antibody that was also raised against the intracellular domain of the protein (data not shown). The samples were immunoblotted with anti-ERK1/2 and anti-β-tubulin antibodies to demonstrate the protease did not digest intracellular proteins. These data are evidence that P2X7 N187A does not efficiently traffic to the cell surface, providing a plausible mechanism for why this mutant displays attenuated activity.
DISCUSSION
We have presented the first evidence that the nucleotide receptor P2X7 appears glycosylated on five asparagine residues and that residue N187, which is an amino acid that is conserved among six of the seven P2X family members, is important for receptor-stimulated function/signaling. Mutation of residues N213, N241 and N284 also results in a modest attenuation of BzATP-induced ERK1/2 phosphorylation, but not to the same extent as mutation of N187. In addition, we have provided evidence that the P2X7 N187A point mutant displays reduced activity, at least in part, because it does not properly traffic to the plasma membrane when compared to wild-type receptor.
There are several potential explanations as to how mutation of P2X7 N187 attenuates receptor function. One possibility is that the proper folding and assembly of P2X7 monomers into oligomers requires glycosylation at this site (4). Cell surface-bound proteins that are not folded properly in the ER are often degraded during the unfolded protein response (28). This process may contribute to the observations that it was often difficult to express P2X7 N187A at levels that were similar to wild-type and that P2X7 Asn to Ala double mutants containing N187A were generally expressed at lower levels than the single mutants (Fig. 8 and data not shown). The PredictProtein program was used to determine where predicted secondary structures in human P2X7 are located. This analysis predicted that residue N187 is located in a short loop between an alpha helix and a beta sheet. Thus, it is possible that mutation at this residue disrupts these secondary structures, altering tertiary protein structure.
Another possible explanation for why the P2X7 N187 point mutants exhibit decreased activity is that glycosylation at this residue is required for P2X7 trafficking to the plasma membrane. Assessing P2X7 cell surface expression at a given time point has been challenging in the cell lines commonly used for P2X7 analysis. It has been reported by others that P2X7 is naturally highly represented in intracellular compartments in several cell types (29). Similarly, we have noted that endogenous P2X7 in RAW 264.7 macrophages and over-expressed P2X7 in COS7 cells appears to be predominantly localized at intracellular sites, as assessed by immunofluorescent staining (Figs. 4 and 9). It is plausible that the trafficking of P2X7 to and from the cell surface occurs rapidly in cells naturally expressing the receptor as well as in those transfected with exogenous receptor. Therefore, visualizing P2X7 localization at the plasma membrane at any single time point is likely to be limited by the possibility that only a small percentage of the receptor is present at the surface at any given time. In agreement with this concept, we have found that P2X7 levels on the cell surface appear low using other methods, including flow cytometry (data not shown) and EndoH-resistance assays (Figs. 3 and 5). Accordingly, the live cell proteinase K digestion assay described herein appears to be a valuable tool for examining the plasma membrane localization of P2X7 over one hour or more. This assay has been useful in identifying P2X7 mutants that exhibit a major defect in trafficking to the cell membrane.
Interestingly, a recent report by Roger et al. has shown that two P2X7 SNPs located near N187, E186K and L191P, exhibit decreased ion channel and pore activity, providing further evidence this extracellular region of the receptor is critical for function (17). The authors propose the E186K and L191P mutations affect ATP binding because they are in close proximity to the ATP-binding amino acids. As discussed above, our data support two alternative explanations for why P2X7 exhibits attenuated activity when residues in that region are mutated. We propose that the region of the receptor encompassing residue N187 is important for protein folding and/or trafficking to the cell surface and that any ATP binding issues may be secondary to the trafficking defect. Nonetheless, this report and that of Roger et al. demonstrate that the region surrounding N187 is important for P2X7 function, and this information should help in advancing our understanding of how naturally-occurring genetic variations in P2X7 lead to altered physiology.
As discussed previously, we and others have provided evidence that residue N284 may also be an important determinant of P2X7 function. For example, we have shown herein that mutation of N284 to Ala in the P2X7 receptor results in a modest decrease in BzATP-induced ERK1/2 activation (Fig. 7). Interestingly, Young et al. have demonstrated that introduction of Asn at the equivalent residue in mouse P2X7 (D284), which leads to increased receptor glycosylation, results in increased sensitivity to BzATP- and ATP-induced responses, and that mutation of the preceding amino acid in rat P2X7 (T283) results in attenuated channel and pore activities (27,30). This threonine residue is conserved among the human, mouse and rat sequences (Fig. 2, 27,30). It is possible that we only observed a modest decrease in P2X7 N284A activity in response to BzATP because this region of the protein may be less critical for BzATP interaction with the receptor when compared to other P2X7 ligands such as ATP. In support of this idea, introduction of Asn at mouse P2X7 D284 promoted a larger sensitivity to ATP than BzATP (27).
In summary, this work highlights the importance of N-linked glycosylation in the regulation of the immunomodulatory critical protein P2X7. We have demonstrated that both mutation of the conserved glycosylation site N187 and treatment of cells with the glycosylation synthesis inhibitor tunicamycin results in attenuated receptor activity, supporting the idea that N-linked glycosylation is essential for P2X7 function.
Acknowledgments
We thank Dr. Charles Heise (UT Southwestern Medical Center) and Dr. James Keck (University of Wisconsin-Madison) for critical comments about this manuscript, and we thank Dr. Beatriz Quinchia-Rios and Lance Rodenkirch for technical assistance.
This work was supported by National Institutes of Health (NIH) grants 1 U19 AI070503, 2 R01 HL069116, and 1 P01 HL0885940 to PJB, a postdoctoral fellowship to LYL from The Hartwell Foundation, and a Trewartha undergraduate research grant to ZW.
ABBREVIATIONS
- ERK1/2
extracellular signal-regulated kinases 1/2
- CREB
cyclic-AMP response element-binding protein
- p90RSK
p90 ribosomal S6 kinase
- MAPK
mitogen activated protein kinase
- ATF
activating transcription factor
- BzATP
2′(3′)-O-(4-benzoylbenzoyl)-ATP
- PVDF
polyvinylidene fluoride
- EGFR
epidermal growth factor receptor
- ER
endoplasmic reticulum
- PDI
protein disulfide isomerase
- TM
transmembrane
References
- 1.Burnstock G. Physiology and pathophysiology of purinergic neurotransmission. Physiol Rev. 2007;87:659–797. doi: 10.1152/physrev.00043.2006. [DOI] [PubMed] [Google Scholar]
- 2.Myrtek D, Idzko M. Chemotactic activity of extracellular nucleotides on human immune cells. Purinergic Signal. 2007;3:5–11. doi: 10.1007/s11302-006-9032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubyak GR. Signal transduction by P2-purinergic receptors for extracellular ATP. Am J Respir Cell Mol Biol. 1991;4:295–300. doi: 10.1165/ajrcmb/4.4.295. [DOI] [PubMed] [Google Scholar]
- 4.North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 5.Romagnoli R, Baraldi PG, Cruz-Lopez O, Lopez-Cara C, Preti D, Borea PA, Gessi S. The P2X7 receptor as a therapeutic target. Expert Opin Ther Targets. 2008;12:647–661. doi: 10.1517/14728222.12.5.647. [DOI] [PubMed] [Google Scholar]
- 6.Ferrari D, Pizzirani C, Adinolfi E, Lemoli RM, Curti A, Idzko M, Panther E, Di Virgilio F. The P2X7 receptor: a key player in IL-1 processing. J Immunol. 2006;176:3877–3883. doi: 10.4049/jimmunol.176.7.3877. [DOI] [PubMed] [Google Scholar]
- 7.Lenertz LY, Gavala ML, Hill LM, Bertics PJ. Cell signaling via the P2X(7) nucleotide receptor: linkage to ROS production, gene transcription, and receptor trafficking. Purinergic Signal. 2009;5:175–87. doi: 10.1007/s11302-009-9133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gavala ML, Pfeiffer ZA, Bertics PJ. The nucleotide receptor P2RX7 mediates ATP-induced CREB activation in human and murine monocytic cells. J Leukoc Biol. 2008;84:1159–1171. doi: 10.1189/jlb.0907612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford MD, Soltoff SP. P2X7 receptors activate protein kinase D and p42/p44 mitogen-activated protein kinase (MAPK) downstream of protein kinase C. Biochem J. 2002;366:745–755. doi: 10.1042/BJ20020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noguchi T, Ishii K, Fukutomi H, Naguro I, Matsuzawa A, Takeda K, Ichijo H. Requirement of reactive oxygen species-dependent activation of ASK1-p38 MAPK pathway for extracellular ATP-induced apoptosis in macrophage. J Biol Chem. 2008;283:7657–7665. doi: 10.1074/jbc.M708402200. [DOI] [PubMed] [Google Scholar]
- 11.Potucek YD, Crain JM, Watters JJ. Purinergic receptors modulate MAP kinases and transcription factors that control microglial inflammatory gene expression. Neurochem Int. 2006;49:204–214. doi: 10.1016/j.neuint.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 12.Dell’Antonio G, Quattrini A, Cin ED, Fulgenzi A, Ferrero ME. Relief of inflammatory pain in rats by local use of the selective P2X7 ATP receptor inhibitor, oxidized ATP. Arthritis Rheum. 2002;46:3378–3385. doi: 10.1002/art.10678. [DOI] [PubMed] [Google Scholar]
- 13.Dell’Antonio G, Quattrini A, Dal Cin E, Fulgenzi A, Ferrero ME. Antinociceptive effect of a new P(2Z)/P2X7 antagonist, oxidized ATP, in arthritic rats. Neurosci Lett. 2002;327:87–90. doi: 10.1016/s0304-3940(02)00385-3. [DOI] [PubMed] [Google Scholar]
- 14.Denlinger LC, Shi L, Guadarrama A, Schell K, Green D, Morrin A, Hogan K, Sorkness RL, Busse WW, Gern JE. Attenuated P2X7 pore function as a risk factor for virus-induced loss of asthma control. Am J Respir Crit Care Med. 2009;179:265–270. doi: 10.1164/rccm.200802-293OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malhotra JD, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal. 2007;9:2277–2293. doi: 10.1089/ars.2007.1782. [DOI] [PubMed] [Google Scholar]
- 16.Zhao YY, Takahashi M, Gu JG, Miyoshi E, Matsumoto A, Kitazume S, Taniguchi N. Functional roles of N-glycans in cell signaling and cell adhesion in cancer. Cancer Sci. 2008;99:1304–1310. doi: 10.1111/j.1349-7006.2008.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roger S, Mei ZZ, Baldwin JM, Dong L, Bradley H, Baldwin SA, Surprenant A, Jiang LH. Single nucleotide polymorphisms that were identified in affective mood disorders affect ATP-activated P2X(7) receptor functions. J Psychiatr Res. 2009 doi: 10.1016/j.jpsychires.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Pfeiffer ZA, Guerra AN, Hill LM, Gavala ML, Prabhu U, Aga M, Hall DJ, Bertics PJ. Nucleotide receptor signaling in murine macrophages is linked to reactive oxygen species generation. Free Radic Biol Med. 2007;42:1506–1516. doi: 10.1016/j.freeradbiomed.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerra AN, Fisette PL, Pfeiffer ZA, Quinchia-Rios BH, Prabhu U, Aga M, Denlinger LC, Guadarrama AG, Abozeid S, Sommer JA, Proctor RA, Bertics PJ. Purinergic receptor regulation of LPS-induced signaling and pathophysiology. J Endotoxin Res. 2003;9:256–263. doi: 10.1179/096805103225001468. [DOI] [PubMed] [Google Scholar]
- 20.Denlinger LC, Coursin DB, Schell K, Angelini G, Green DN, Guadarrama AG, Halsey J, Prabhu U, Hogan KJ, Bertics PJ. Human P2X7 pore function predicts allele linkage disequilibrium. Clin Chem. 2006;52:995–1004. doi: 10.1373/clinchem.2005.065425. [DOI] [PubMed] [Google Scholar]
- 21.Denlinger LC, Angelini G, Schell K, Green DN, Guadarrama AG, Prabhu U, Coursin DB, Bertics PJ, Hogan K. Detection of human P2X7 nucleotide receptor polymorphisms by a novel monocyte pore assay predictive of alterations in lipopolysaccharide-induced cytokine production. J Immunol. 2005;174:4424–4431. doi: 10.4049/jimmunol.174.7.4424. [DOI] [PubMed] [Google Scholar]
- 22.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 23.Humphreys BD, Dubyak GR. Modulation of P2X7 nucleotide receptor expression by pro- and anti-inflammatory stimuli in THP-1 monocytes. J Leukoc Biol. 1998;64:265–273. doi: 10.1002/jlb.64.2.265. [DOI] [PubMed] [Google Scholar]
- 24.Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal Biochem. 1989;180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- 25.Gamou S, Shimizu N. Glycosylation of the epidermal growth factor receptor and its relationship to membrane transport and ligand binding. J Biochem. 1988;104:388–396. doi: 10.1093/oxfordjournals.jbchem.a122478. [DOI] [PubMed] [Google Scholar]
- 26.Di Jeso B, Pereira R, Consiglio E, Formisano S, Satrustegui J, Sandoval IV. Demonstration of a Ca2+ requirement for thyroglobulin dimerization and export to the golgi complex. Eur J Biochem. 1998;252:583–590. doi: 10.1046/j.1432-1327.1998.2520583.x. [DOI] [PubMed] [Google Scholar]
- 27.Young MT, Pelegrin P, Surprenant A. Amino acid residues in the P2X7 receptor that mediate differential sensitivity to ATP and BzATP. Mol Pharmacol. 2007;71:92–100. doi: 10.1124/mol.106.030163. [DOI] [PubMed] [Google Scholar]
- 28.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 29.Gu BJ, Zhang WY, Bendall LJ, Chessell IP, Buell GN, Wiley JS. Expression of P2X(7) purinoceptors on human lymphocytes and monocytes: evidence for nonfunctional P2X(7) receptors. Am J Physiol Cell Physiol. 2000;279:C1189–1197. doi: 10.1152/ajpcell.2000.279.4.C1189. [DOI] [PubMed] [Google Scholar]
- 30.Young MT, Pelegrin P, Surprenant A. Identification of Thr283 as a key determinant of P2X7 receptor function. Br J Pharmacol. 2006;149:261–268. doi: 10.1038/sj.bjp.0706880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]