Abstract
Inflammatory bowel disease (IBD), encompassing Crohn’s disease (CD) and ulcerative colitis (UC), is a chronic inflammatory disorder caused by dysregulated immune responses in a genetically predisposed individual. Recent accumulating data, including genome-wide association studies, have identified more than 50 distinct genetic loci that confer susceptibility. We highlight the role of microbial-host interaction, particularly with respect to the overlap of common genetic and pathophysiologic mechanisms of CD and UC, interleukin-22–producing natural killer cells, autophagy, and TL1A, a member of the tumor necrosis factor (TNF) family, in gut homeostasis and IBD pathogenesis. This article focuses on the recent advances in understanding of IBD from the past year, including advances in genetics and immunobiology.
Introduction
Crohn’s disease (CD) and ulcerative colitis (UC) are two disparate forms of inflammatory bowel disease (IBD) that share related characteristics, but have unique clinical distinguishing features. Genome-wide association studies (GWAS) have identified more than 50 genetic associations with CD and UC. Similar to clinical phenotypes, UC and CD share genetic susceptibility loci but differ at others, suggesting common genetic and pathophysiologic mechanisms that underlie these two disease entities. In addition to the evidence indicating that CD occurs when commensal, opportunistic, or pathogenic bacteria initiate and perpetuate a dysregulated mucosal immune response, recent UC GWAS also revealed genetic association between UC and genes important in host defense against infection, highlighting the importance of bacterial-host interaction in IBD.
The main genetic associations in IBD can be divided into genes that contribute to the innate and adaptive immune response. In the innate immune arm, the association of CD with polymorphisms in NOD2 (CARD15) and the two autophagy-related genes, ATG16L1 and IRGM, implicate defects in the recognition and handling of intracellular bacteria in the immunopathogenesis of IBD. Recent studies have shown that human and mouse gut-associated lymphoid tissues harbor a unique natural killer (NK) cell subset that produce interleukin (IL)-22, a cytokine that plays a role in host defense and autoimmune diseases, including IBD. In the adaptive immune arm, CD has been considered a condition driven by T-helper 1 (Th1) and/or Th17 whereas UC appears to be Th2 dominant [1]. However, recent genetic association studies indicate that overlapping immunogenetic pathways, especially the Th17 inflammatory axis, drive UC. Tregs, an immune-modulating subset of CD4+ T-cells, can suppress the differentiation and function of Th1 and Th2 cells. The immunopathologic concept of IBD is evolving in light of recent studies that unveiled novel effector pathways in IBD, one of which is the TL1A-DR3 pathway in microbial-host interaction and bridging innate and adaptive immune response.
GWAS and Overlapping Immunogenetic Pathways Between IBD Subtypes
GWAS examine tens to hundreds of thousands of single nucleotide polymorphisms (SNPs) across the human genome in cases and controls. Simply, the allele frequencies of these SNPs are statistically compared between cases and controls to determine any association between the SNP and the disease or condition in question. Independent confirmation of association is often performed for findings generated in a GWAS. Therefore, any confirmed SNP associations that are seen with a GWAS are likely to be in linkage disequilibrium with a true disease susceptibility allele. Several GWASs have been performed in IBD (Table 1) [2,3••,4–12].
Table 1.
Summary of design, population, and findings in eight inflammatory bowel disease genome-wide association scans and meta-analysis
| Study and population | SNPs analyzed | Sample size: cases–controls | Phenotype associated | Genes identified | Loci identified |
|---|---|---|---|---|---|
| Yamazaki [11]; Japanese and British Caucasian | 72,000 | 94–752* | CD and UC | TL1A(TNFSF15) | |
| Duerr et al. [4]; North American Caucasian | 304,000 | 946–977 | Ileal CD |
IL23R ATG16L1 PHOX2B FAM92B NCF4 |
Chromosome 10§ |
| Hampe et al. [7]; German Caucasian | 7000† | 735–368 | CD | ATG16L1 | |
| Libioulle et al. [8]; Belgian/French Caucasian | 302,000 | 547–928 | CD | IL23R | 5p13‡ |
| Wellcome Trust Case-Control Consortium [2]; British Caucasian | 469,000 | 1748–2938 | CD |
IL23R ATG16L1, IRGM NKX2-3 PTPN2 |
Chromosome 3§ 5p13‡ |
| Franke et al. [6]; German Caucasian | 92,000 | 393–399 | CD | NELL1 | 5p13‡ |
| Raelson et al. [9]; Quebec Caucasian | 164,000 | 382 trios | CD | IL23R | Chromosome 17§ |
| Fisher et al. [5]; British Caucasian | 10,886¶ | 905–1465** | UC |
ECM1 MST1 BTNL2 HLA-DRB1 |
|
| Barrett et al. [3••]; North-American, British, and Belgian/French populations | NA | 3230–4829 |
IL23R ATG16L1, IRGM MST1 PTGER4 TL1A(TNFSF15) ZNF365 NKX2-3 NOD2 PTPN2, PTPN22 ITLN1 IL12B CDKAL1 CCR6 C11orf30 LRRK2 MUC19 STAT3, JAK2 ICOSLG ORMDL3 |
5q31§ 1q24§ 1q32§ 6q21§ 7p12§ 8q24§ 10p11§ 13q14§ 21q21 |
|
| Franke et al. [12]; European population | 440,794 | 1167–777 | UC |
HLA-DRA BTNL2 IL23R ARPC2 IL10 JAK2 S100Z |
|
| Silverberg et al. [10]; European Caucasian | 280,748 | 1052–2571 | UC |
IL22 IL23R IL26 MHC PLA2G2E IFN |
1p36 1p31 6p21 12q15 |
Study performed in 94 cases and 752 controls as stage 1. Findings reproduced in 490 Japanese cases and 345 controls, 347 British inflammatory bowel disease trios, and 363 cases and 372 controls.
These were all nonsynonymous (amino acid changing) polymorphisms.
Several groups identified association with a variant in a gene desert on chromosome 5p13. By using an expression database, this region may be involved in regulation of the prostaglandin E receptor 4 gene (PTGER4).
Several studies identified association with genetic regions in which it was unclear which was the specific causative gene.
Nonsynonymous and MHC tagging SNPs.
Study performed in 905 UC cases and 1465 controls. Positive findings tested in 936 UC cases and 1470 controls.
CD—Crohn’s disease; MHC—major histocompatibility complex; NA—not applicable; SNP—single nucleotide polymorphism; UC—ulcerative colitis.
Although CD and UC differ in disease localization, endoscopic appearance, histology, and clinical course, common clinical and pathologic features are shared by these two conditions, suggesting that CD and UC share some genetic susceptibility loci but differ at others. Indeed, two recent systematic GWAS of UC revealed overlap of genetic markers between UC and CD including IL23R and JAK2 whereas other markers are specific to UC including ARPC2, PLA2G2E, and IL-22 [10,12]. Furthermore, a candidate gene experiment tagging 29 of the CD loci and testing for potential overlap with UC confirmed the overlap between the IBD subtypes [13]. This study also shows an association of JAK2 and STAT3 in UC [13]. Some of the downstream targets of the JAK-STAT pathway are the IL12/23 axis, IL23 signaling, and maturation of naïve CD4+ T cells to Th17 cells. Together, the two UC GWAS and the candidate association studies reveal a common immunogenetic pathway between CD and UC, particularly the Th17 inflammatory axis.
Similar to the CD GWAS, the two recent UC GWAS reveal genetic associations that underscore the importance of bacterial-host interaction in IBD. For example, PLA2G2E gene is in close proximity to the UC susceptibility SNP rs6426833 [10]. Evidence for PLA2G2E in bacteria-mediated inflammation lies in the fact that this family of secretory phospholipase A2 leads to the production of proinflammatory lipid mediators (eg, prostaglandins and leukotrienes) and that bacterial lipopolysaccharide can upregulate PLA2G2E expression [14]. Another genetic UC association that implicates the importance of bacterial-host interaction is at SNP rs1558744 in close proximity to IL-22 [10]. IL-22 is a member of the family of IL-10–related cytokines, which mediate host defense against infections as well as tissue inflammation in many chronic, immune-mediated disorders, including IBD [15].
Remarkably, despite the discovery of new genetic susceptibility loci with GWAS, the identified IBD markers account for only 20% of the heritable risk [3••]. Thus, a considerable amount of genetic research remains to be done to identify the risk and protective IBD genes to better elucidate IBD pathogenesis.
Innate Immunity
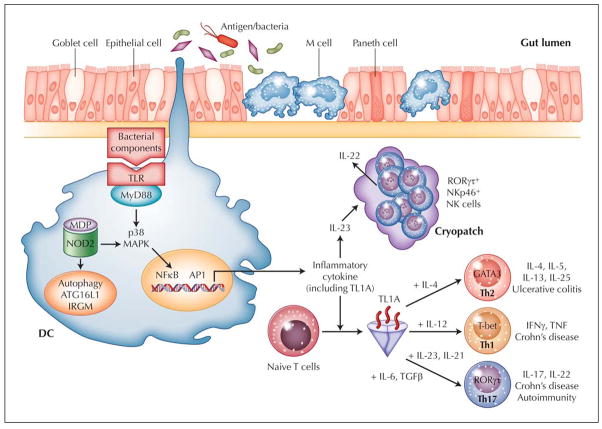
Antigen presentation to T cells by professional antigen-presenting cells (APCs) such as dendritic cells (DCs) is critical for the activation of adaptive immune responses. There are three main pathways to transport luminal antigen to lamina propria (LP). The first is mediated by microfold (M) cells through the capture of luminal antigens and their presentation to T cells. The second pathway, performed by LP DCs, involves dendritic cell extension to the intestinal lumen across epithelial cells, facilitating the capture of luminal antigens (Fig. 1). The third pathway involves the neonatal Fc receptor (FcRn), which serves as the vehicle for transporting LP immunoglobulin G (IgG) across the colonic epithelial layer into the lumen where the IgG can bind enteric bacterial antigens to form immune complexes (IC). The FcRn then recycles the antigens/IgG IC back across the colonic epithelial cells into the LP for processing by APCs such as DCs, which are capable of presenting the antigens to T cells.
Figure 1.
Working hypothesis of inflammatory bowel disease. Intestinal immune system is in close apposition to luminal antigen/bacteria, which are separated by a single layer of epithelial cells. Goblet cells contribute to the formation of the protective mucus layer, and M cells and dendritic cells (DC) sample intestinal luminal contents. Overresponse to antigens, either through Toll-like receptors (TLR), intracellular sensor NOD2, or antigen processing via autophagy, results in stimulated DC that recruit and generate T-cell (Th1, Th2, and Th17) and NK-cell subtypes. One subpopulation of RORγt+NKp46+ NK cells that resides in gut cryptopatches is found to readily produce IL-22 in response to IL-23, likely released by DC. IL-22 may play an important role in epithelial homeostasis, bacteria clearance, and tissue repair. TL1A appears to be a critical factor important in generating Th1, Th2, and Th17 cells. For each T-helper cell differentiation program, specific transcription factors and cytokine milieu are required (indicated in the figure). Terminally differentiated T-helper cells are characterized by a specific combination of effector cytokines (indicated in the figure) that orchestrate effector function of the adaptive immune system. Crohn’s disease is a predominantly Th1- and Th17-mediated process, whereas ulcerative colitis appears to be mediated by Th2 and possibly Th17.
One of the ways that host discerns foreign from self antigen is through pattern recognition receptor (PRR), which recognizes specific molecular patterns of pathogens. One group of PRRs consists of the Toll-like receptors (TLRs) with variable specificities for sensing microbial products. TLR activation can stimulate signaling cascades leading to proinflammatory cytokine production and induction of co-stimulatory signals to further augment innate and initiate effector adaptive immune responses (Fig. 1). One of the mechanisms by which sentinel APCs augment innate immune response is enhancement of the lytic potential of NK cells and their ability to produce interferon (IFN)-γ [15]. Recently, a unique NK cell subset that specializes in the production of IL-22 and is responsive to IL-23 was identified [16]. Several murine studies in the past year point to a protective role for these IL-22–producing NK cells in host defense and IBD [15]. Lastly, genetic associations between autophagy genes and CD highlight the importance of intracellular processing of bacterial components in mucosal homeostasis.
IL-22–Producing NK Cells: Protective Role in Host Defense and IBD
The recent identification of IL-22 as a UC susceptibility gene implicates this cytokine in the pathogenesis of IBD [10]. IL-22 is a member of IL-10–related cytokines and plays a role in maintaining the integrity of epithelial cell barrier in the gut from pathogens [15]. Binding of IL-22 to its receptor initiates a signal transducer and activator of transcription (STAT) 3 signaling pathway that induces the production of antimicrobial proteins including β-defensin-2, β-definsin-3, S100A7, S100A8, S100A9, RegIIIβ, RegIIIγ, and lipocalin-2, which protects the epithelial barrier against microbes [17,18]. Direct induction of antimicrobial proteins may be one of the pathways for IL-22 in regulating early defense mechanisms against extracellular bacterial infection caused by Citrobacter rodentium [18].
Several studies have identified a distinct population of NKp46+ cells that express and depend on the RORγt transcription factor for their development [16,19,20]. Interestingly, the differentiation of IL-22–producing RORγt+NKp46+ cells is dependent on the presence of gut microflora since germ-free mice have fewer IL-22–producing NK cells [19,21]. Additionally, IL-22 is produced in an IL-23–dependent way during inflammation and promoted the expression of antimicrobial proteins [16,19]. Although IL-23 is important in the differentiation of Th17 cells that produce IL-22 [22,23], several recent reports also demonstrated that IL-23 is an effective trigger of IL-22 secretion in RORγt+NKp46+ NK cells (Fig. 1) [23]. As discussed above, GWAS have identified IL-23 polymorphisms that are associated with CD and UC. It is noteworthy that IL-22 was found in the two recent GWAS to be associated with UC, suggesting that the IL-23–IL-22 axis is involved in the pathogenesis of UC.
Several recent studies implicated a protective role for IL-22–producing NK cells in IBD. Adoptively transferring CD4+CD45RBhi T cells from wild-type (WT) or IL-22–deficient mice into Rag1−/− or Rag1−/−IL22−/− mice showed that the absence of exogenous and endogenous non-T non-B cell sources of IL-22 led to worsened murine colitis than with lack of either exogenous T-cell or endogenous sources of IL-22 [24]. NK cells were identified as the likely population that produced IL-22, the absence of which led to worsened murine colitis [24]. The protective role of IL-22 in mucosal inflammation further demonstrated that IL-22 protected against intestinal C. rodentium infection in Rag2−/− mice [18]. Because Rag mice lack T-cell sources of IL-22, the protective role of IL-22 in this model may be from NK cells. Furthermore, using anti-NK1.1 antibody to deplete NK cells resulted in heightened susceptibility to of this type of infection [16,21]. These results implicate that RORγt+NKp46+ NK cells provide an innate source of IL-22 that has a protective role in host defense and mucosal inflammation (Fig. 1).
Defects in Autophagy Associated With Crohn’s Disease
Autophagy (from the Greek “auto,” oneself and “phagy,” to eat) is a biologic process of membrane trafficking in which autophagosomes engulf organelles and cytosolic macromolecules, followed by fusion of the autophagosome with a lysosome to form an autolysosome in which sequestered material is degraded. The degraded content can then be loaded onto human leukocyte antigen (HLA) class II molecules or to compartments where recognition by TLR may occur. Autophagy is important for cellular homeostatic functions including structural remodeling, nutrient and energy generation, degradation of damaged or long-lived cytoplasmic components, and protection against invading microorganisms. Its protective role in infectious disease is an important part of the innate immune armamentarium and links to adaptive immunity by delivery of foreign antigens necessary for immune recognition. Several GWAS identified ATG16L1 and IRGM as new CD susceptibility genes [3••,25–27].
The ATG16L1 gene product is comprised of an N-terminal ATG16 domain thought to be essential for interaction with other autophagy proteins such as ATG5 and ATG12; a coiled-coil domains postulated to mediate homodimeric interactions; and seven C-terminal WD repeats thought to create stable platforms that can coordinate the formation of multimeric protein complex, which is essential for the formation of the autophagosome [1]. ATG16L1 is broadly expressed by intestinal epithelial cells, APCs, CD4+, CD8+, and CD19+ primary human T cells [1].
The role of ATG16L1 in mucosal inflammation is being studied in mice with targeted genetic manipulations. Recent data using mice hypomorphic for ATG16L1 showed that the defect appears to be confined to gut paneth cells [28]. The investigators showed that ATG16L1 hypomorphic paneth cells exhibited abnormalities in the granule exocytosis pathway characterized by defective antimicrobial peptide secretion [28]. Importantly, similar reduction in secreted antimicrobial peptides was observed in biopsies from a CD patient homozygous for the ATG16L1 risk allele [28]. This suggests that the human ATG16L1 risk allele has reduced autophagy function, leading to reduced secretion of antimicrobial peptides by paneth cells.
In another study, ATG16L1 deficiency in mice macrophages produced high amounts of inflammatory cytokines IL-1β and IL-18 when these macrophages were exposed to bacterial lipopolysaccharide [29]. The increased production of IL-1β and IL-18 is due to Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF)-dependent activation of caspase-1; where caspase 1 is thought to cleave pro–IL-1β to yield active cytokine [29]. Chimeric ATG16L1-deficient mice are highly susceptible to dextran sulphate sodium-induced (DSS) acute colitis, which can be rescued by injection of neutralizing antibodies to IL-1β and IL-18 [29]. These findings indicate a role of autophagy in regulating the production of proinflammatory cytokines and in controlling bacterial endotoxin–mediated gut inflammation.
Given the important role of autophagy in the control of microbial-induced inflammatory immune response, mutations affecting important factors involved in autophagy may result in immune dysregulation seen in IBD. The association between CD and a nonsynonymous amino acid change—a threonine to alanine substitution at position 300 (T300A)—is found in several GWAS [2,7,25]. The role of the T300A risk-associated variant was studied in a model of intracellular pathogen infection using Salmonella enterica serovar typhimurium [30]. The investigators reported that the T300A substitution resulted in a variant of ATG16L1 that is less stable and has reduced capability to mediate antibacterial autophagy [30].
IRGM is a member of a family of genes encoding interferon-inducible guanosine triphosphatases that are involved in pathogen clearance. IRGM is expressed in several tissues including colon, small intestine, peripheral blood leukocytes, and monocytes that have been found to have an important role in eliminating the intracellular pathogens Toxoplasma gondii and Listeria monocytogenes [26]. Furthermore, in gene-knockdown experiments, IRGM has been shown to induce autophagy to control the intracellular mycobacterial [31] and Salmonella typhimurium load [32].
Although multiple IRGM SNPs were found to be associated with CD [2], sequencing of the IRGM coding region did not detect any causal amino acid–changing SNPs [26]. A recent report found a 20-kb deletion polymorphism upstream of the IRGM gene to be strongly associated with CD-associated SNP [32]. Therefore, the genetic variants conferring susceptibility to CD may lie in regulatory sequences that affect either IRGM expression, transcript splicing, or the rate of protein translation. Taken together, the associations of SNPs in ATG16L1 and IRGM with CD indicate that proteins involved in autophagic machinery are an important biologic pathway in chronic inflammatory diseases of the digestive tract. One possible hypothesis is that alterations in an individual’s intracellular processing of bacteria impact how the innate immune system interacts with gut microflora and may contribute to the pathogenesis of CD. Further understanding of the relevant autophagic processes, including elucidation of the molecular mechanisms by which ATG16L1 and IRGM variants contribute to CD susceptibility, will have significant impact on our understanding of IBD.
Impaired Clearance of Microbes in IBD
One of the etiologic theories on the pathogenesis of IBD is impaired clearance of foreign material leading to a sustained activation of monocytes and a compensatory induction of adaptive immune response [33]. The compensatory activation of adaptive immune response is temporally different from the initial acute inflammatory response and leads to perpetuation of the chronic inflammatory response contributing to IBD. The immunodeficiency phenotype is also consistent with the finding of CARD15 as a CD susceptibility gene [34] and further discussed below. CARD15 encodes NOD2, which is a general sensor of bacteria that recognizes muramyl dipeptide (MDP), a component of both gram-positive and gram-negative bacteria [34]. The main outcomes of NOD2 activation is the induction of nuclear factor (NF)-κB and the mitogen-activated protein kinase (MAPK) pathway resulting in the production of proinflammatory mediators [34]. Peripheral-blood mononuclear cells isolated from CD patients who carry the risk CARD15 allele show a defect in IL-1β secretion [35]. Additionally, epithelial cells transfected with 3020insC CARD15 mutation have defective NF-κB activation upon MDP stimulation [36,37]. The defective induction of proinflammatory cytokines may result in impaired mucosal host clearance of bacteria, as seen in NOD2-deficient mice that develop worse liver infection to L. monocytogenes [38]. The findings that CD variants in the autophagy genes ATG16L1 and IRGM result in reduced clearance of bacteria also support the idea that impaired clearance of microbial organism from bowel wall causes IBD [30,32].
Recruitment of neutrophils to sites of trauma in the bowel is impaired in CD patients [33]. This finding led to the hypothesis that delayed neutrophil recruitment impairs the clearance of microbial organisms, which then leads to gut inflammation. This hypothesis was recently tested by injecting heat-killed Escherichia coli into the forearm of human CD, UC, and healthy control subjects [39•]. The investigators found accumulation of neutrophils at the site of injection as well as the clearance of injected E. coli was markedly impaired in CD patients [39•]. Together, these data support the notion that impaired clearance of antigenic material contributes to the pathogenesis of IBD.
Adaptive Immunity
Recognition of commensal-derived antigens by the adaptive immune system or its stimulation by the innate immune system plays a key role in the pathogenesis of IBD. Polymorphisms of genes involved in the Th1, Th2, and Th17 immune pathway have been shown to be associated with IBD, with CD having predominately a Th1 and/or Th17 cytokine profile and UC a Th2 and, recently, a Th17 cytokine profile.
TL1A (TNFSF15) Bridges Innate and Adaptive Immune Response in IBD
TL1A (TNFSF15), an IBD-associated gene, is a member of the tumor necrosis factor (TNF) superfamily that binds to death domain receptor 3 (DR3, TNFRSF25), and its expression is increased in inflamed tissue of colon and small bowel of CD patients and colocalizes to macrophages and T cells [1]. In particular, lamina propria, but also peripheral CD4+CCR9+ T cells, constitutively express membrane TL1A and are especially sensitive to TL1A stimulation [1]. The role of TL1A in the pathology of mucosal inflammation was recently tested by using neutralizing antibodies to TL1A where inactivating the function of TL1A leads to attenuation of inflammation in two different murine colitis models [40••].
GWAS have revealed a significant association of genetic variants of the TL1A (TNFSF15) gene with CD in a large cohort of Japanese patients, in several European cohorts [11,41], in US Jewish patients [42], and combined data from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) IBD Genetics Consortium, Belgian-French IBD Consortium, and the Wellcome Trust Case-Control Consortium (WTCCC) [3••]. TL1A is the only gene that has been associated with both Asian and Caucasian IBD. Using case-control and family-based study designs, TNFSF15 haplotype A has been found to confer significant CD risk whereas haplotype B confers protection in Asian and European populations [3••,11,43]. However, a recent report demonstrated that TL1A haplotype B is a risk haplotype in Jewish CD patients with antibody titers for the E. coli outer membrane porin C (OmpC) [44]. Moreover, CD14+ monocytes isolated from Jewish OmpC+ patients homozygous for TL1A gene haplotype B express higher levels of TL1A in response to FcγR stimulation [44]. These results show that TL1A genetic variations contribute to exacerbated induction of TL1A, which may lead to dysregulated immune response and chronic inflammation. Further work is needed to determine the exact TL1A disease-associated variants that will allow us to further explain the complicated genotype/functional correlations in these different ethnic groups.
We previously showed that TL1A is induced by the FcγR signaling pathway [45]. Recently, the potential interaction of TL1A in microbial-host interaction was illustrated by a report showing microbial organisms can induce TL1A in APCs [46]. Microbial-activated TL1A was in part mediated by TLR 1, 2, 4, and 9 signaling pathways and dependent on downstream p38 MAP kinase and NF-κB activation (Fig. 1) [46]. Negative regulators of the TL1A signaling pathway also exist and may serve to maintain gut immune homeostasis. Our group showed that TLR8 or TLR7/8 ligand (R848) can inhibit TL1A production in a dose dependent manner [47•]. Therefore, TLR8 signaling pathway may represent a novel therapeutic target in IBD.
TL1A plays an important role in modulating adaptive immune response (Fig. 1). In the Th1 effector arm, TL1A was found to augment IFN-γ production by IL-12/IL-18–stimulated CD4+/CCR9+ T cells that are specifically enriched in the intestinal immune compartment [1]. Furthermore, in autologous monocyte–T-cell co-cultures, TL1A production by monocytes potentiated IFN-γ production by CD4+ T cells [45,46]. In addition to mediating Th1 response, the role of TL1A in Th2-mediated functions and disease pathology was demonstrated in a mouse model of allergic lung inflammation where TL1A signaling is required to exert Th2 effector function in Th2-polarized CD4 cells and co-stimulate IL-13 production by activated NK T cells [48]. Additionally, antibody blockade of TL1A inhibits lung inflammation and production of Th2 cytokines such as IL-13. Together, this study implicates TL1A in augmenting Th2 effector function [48]. Using TL1A-deficient mice, another report showed that TL1A−/− dendritic cells exhibited a reduced capacity in supporting Th17 differentiation and proliferation [49]. Consistent with these data, TL1A−/− mice display decreased clinical severity in experimental autoimmune encephalomyelitis, a Th17-mediated autoimmune disease model [49]. Furthermore, IFN-γ and IL-17 production induced by IL-12 and -23 respectively, can be synergistically enhanced by TL1A [40••,49]. Interestingly, a report using DR3-deficient mice showed that TL1A/DR3 signaling is dispensable for polarization of naïve CD4+ T cells into Th1, Th2, or Th17 effector cell subtypes [50]. Instead, DR3 expression is required on T cells for immunopathology, local T-cell accumulation, and cytokine production, suggesting that TL1A/DR3 signaling is important to co-stimulate antigen-induced expansion of primed T cells in the target organ of T cell–mediated autoimmune and inflammatory diseases.
Together, TL1A/DR3 signaling appears to have pleiotropic effects that include amplifying the innate immune response, modulating adaptive immunity by augmenting Th1, Th2, and Th17 effector cell function, and T-cell accumulation and immunopathology of inflamed tissue (Fig. 1). Given its immune modulatory effects, blocking TL1A-DR3 signaling is a promising therapeutic strategy in a variety of T cell–dependent autoimmune diseases, including IBD.
Conclusions
The recent GWAS and the identification of additional susceptibility UC loci demonstrate that the pathophysiology of CD and UC involves overlapping and divergent pathways. One of the new findings is the shared immunopathophysiologic pathway between IBD subtypes, particularly the IL23/Th17 inflammatory axis. In addition, these genetic findings have further implicated both innate and adaptive immune dysregulation as risk factors for developing IBD. These immunogenic parameters are biologically divergent, which explains the heterogeneous clinical manifestations and lack of a universal therapeutic response to any single agent. The challenge for the future is to correlate IBD susceptibility loci to molecular pathways and clinical phenotypes. These multidisciplinary approaches are likely to stratify this complex disease into distinct subgroups that are more biologically homogenous with more defined pathogenic parameters and aid in the evaluation of IBD susceptibility genes.
Acknowledgments
We would like to acknowledge support from Program Project Grant, National Institutes of Health grant DK046763, and Cindy Ting for critical reading of the manuscript.
Footnotes
Disclosure
No potential conflicts of interest relevant to this article were reported.
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Shih DQ, Targan SR, McGovern D. Recent advances in IBD pathogenesis: genetics and immunobiology. Curr Gastroenterol Rep. 2008;10:568–575. doi: 10.1007/s11894-008-0104-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wellcome Trust Case-Control Consortium. Genome-wide association study of 14, 000 cases of seven common diseases and 3, 000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3••.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. To advance gene discovery, the NIDDK, the WTCCC, and the Belgian-French group pooled data to perform a meta-analysis of GWAS. They confirmed 11 previously reported IBD loci and provided genome-wide significant evidence for 21 additional loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher SA, Tremelling M, Anderson CA, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat Genet. 2008;40:710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franke A, Balschun T, Karlsen TH, et al. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713–715. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 7.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 8.Libioulle C, Louis E, Hansoul S, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raelson JV, Little RD, Ruether A, et al. Genome-wide association study for Crohn’s disease in the Quebec Founder Population identifles multiple validated disease loci. Proc Natl Acad Sci U S A. 2007;104:14747–14752. doi: 10.1073/pnas.0706645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silverberg MS, Cho JH, Rioux JD, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamazaki K, McGovern D, Ragoussis J, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn’s disease. Hum Mol Genet. 2005;14:3499–3506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 12.Franke A, Balschun T, Karlsen TH, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 13.Anderson CA, Massey DC, Barrett JC, et al. Investigation of Crohn’s disease risk loci in ulcerative colitis further defines their molecular relationship. Gastroenterology. 2009;136:523–529. doi: 10.1053/j.gastro.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami M, Yoshihara K, Shimbara S, et al. Arachidonate release and eicosanoid generation by group IIE phospholipase A(2) Biochem Biophys Res Commun. 2002;292:689–696. doi: 10.1006/bbrc.2002.6716. [DOI] [PubMed] [Google Scholar]
- 15.Colonna M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 2009;31:15–23. doi: 10.1016/j.immuni.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Cella M, Fuchs A, Vermi W, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aujla SJ, Chan YR, Zheng M, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Y, Valdez PA, Danilenko DM, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 19.Sanos SL, Bui VL, Mortha A, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luci C, Reynders A, Ivanov II, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 21.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Cua DJ, Sherlock J, Chen Y, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 23.Veldhoen M, Hocking RJ, Flavell RA, Stockinger B. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat Immunol. 2006;7:1151–1156. doi: 10.1038/ni1391. [DOI] [PubMed] [Google Scholar]
- 24.Zenewicz LA, Yancopoulos GD, Valenzuela DM, et al. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prescott NJ, Fisher SA, Franke A, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 28.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 30.Kuballa P, Huett A, Rioux JD, et al. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 32.McCarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sewell GW, Marks DJ, Segal AW. The immunopathogenesis of Crohn’s disease: a three-stage model. Curr Opin Immunol. 2009;21:506–513. doi: 10.1016/j.coi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shih DQ, Targan SR. Immunopathogenesis of inflammatory bowel disease. World J Gastroenterol. 2008;14:390–400. doi: 10.3748/wjg.14.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Heel DA, Ghosh S, Butler M, et al. Muramyl dipeptide and toll-like receptor sensitivity in NOD2-associated Crohn’s disease. Lancet. 2005;365:1794–1796. doi: 10.1016/S0140-6736(05)66582-8. [DOI] [PubMed] [Google Scholar]
- 36.Girardin SE, Boneca IG, Viala J, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 37.Inohara N, Ogura Y, Fontalba A, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi KS, Chamaillard M, Ogura Y, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 39•.Smith AM, Rahman FZ, Hayee B, et al. Disordered macrophage cytokine secretion underlies impaired acute inflammation and bacterial clearance in Crohn’s disease. J Exp Med. 2009;206:1883–1897. doi: 10.1084/jem.20091233. This study demonstrated impaired recruitment of neutrophils in CD patients that led to impaired clearance of bacteria. Macrophages cultured from CD patients were found to secrete less proinflammatory cytokines in response to E. coli stimulation, likely from abnormal routing of cytokines to the lysosomal compartments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Takedatsu H, Michelsen KS, Wei B, et al. TL1A (TNFSF15) regulates the development of chronic colitis by modulating both T-helper 1 and T-helper 17 activation. Gastroenterology. 2008;135:552–567. doi: 10.1053/j.gastro.2008.04.037. This outstanding study demonstrates that TL1A is an important modulator of chronic mucosal inflammation by enhancing Th1 and Th17 effector functions. The investigators show that neutralizing TL1A antibodies can ameliorate two murine models of colitis and provide evidence that TL1A represents an attractive, novel therapeutic target for the treatment of IBD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tremelling M, Berzuini C, Massey D, et al. Contribution of TNFSF15 gene variants to Crohn’s disease susceptibility confirmed in UK population. Inflamm Bowel Dis. 2008;14:733–737. doi: 10.1002/ibd.20399. [DOI] [PubMed] [Google Scholar]
- 42.Picornell Y, Mei L, Taylor K, et al. TNFSF15 is an ethnic-specific IBD gene. Inflamm Bowel Dis. 2007;13:1333–1338. doi: 10.1002/ibd.20223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kakuta Y, Kinouchi Y, Negoro K, et al. Association study of TNFSF15 polymorphisms in Japanese patients with inflammatory bowel disease. Gut. 2006;55:1527–1528. doi: 10.1136/gut.2006.100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michelsen KS, Thomas LS, Taylor KD, et al. IBD-associated TL1A gene (TNFSF15) haplotypes determine increased expression of TL1A protein. PLoS One. 2009;4:e4719. doi: 10.1371/journal.pone.0004719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prehn JL, Thomas LS, Landers CJ, et al. The T cell costimulator TL1A is induced by FcgammaR signaling in human monocytes and dendritic cells. J Immunol. 2007;178:4033–4038. doi: 10.4049/jimmunol.178.7.4033. [DOI] [PubMed] [Google Scholar]
- 46.Shih DQ, Kwan LY, Chavez V, et al. Microbial induction of inflammatory bowel disease associated gene TL1A (TNFSF15) in antigen presenting cells. Eur J Immunol. 2009 doi: 10.1002/eji.200839087. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Saruta M, Michelsen KS, Thomas LS, et al. TLR8-mediated activation of human monocytes inhibits TL1A expression. Eur J Immunol. 2009;39:2195–2202. doi: 10.1002/eji.200939216. This article describes the first study to identify TLR7/8 pathway as a negative regulator of the TL1A signaling pathway, suggesting that the TLR8 signaling pathway may represent a novel therapeutic target in IBD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang L, Adkins B, Deyev V, Podack ER. Essential role of TNF receptor superfamily 25 (TNFRSF25) in the development of allergic lung inflammation. J Exp Med. 2008;205:1037–1048. doi: 10.1084/jem.20072528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pappu BP, Borodovsky A, Zheng TS, et al. TL1A-DR3 interaction regulates Th17 cell function and Th17-mediated autoimmune disease. J Exp Med. 2008;205:1049–1062. doi: 10.1084/jem.20071364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meylan F, Davidson TS, Kahle E, et al. The TNF-family receptor DR3 is essential for diverse T cell-mediated inflammatory diseases. Immunity. 2008;29:79–89. doi: 10.1016/j.immuni.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]