Summary
The majority of bacterial species do not grow on synthetic media. Many non-growers require growth factors from other bacteria, but the nature of these compounds is largely unknown. We show here that previously uncultured isolates from marine sediment biofilm grow on a Petri dish in the presence of cultured organisms from the same environment. The growth factors produced by one cultured helper strain were identified as new acyl-desferrioxamine siderophores. A panel of previously uncultured isolates exhibited a range of siderophore promiscuity for growth promotion. This siderophore-based approach has enabled the culturing of organisms only distantly related to previously cultured microbes. The lack of growth in the lab for many strains from this habitat stems from an inability to autonomously produce siderophores, and the resulting chemical dependence on other microorganisms regulates community establishment in the environment.
Introduction
Sampling of diverse environments, such as soil, marine sediment, or hot springs shows that only 0.01 – 1% of cells visible under the microscope will form colonies on a Petri dish, leaving the remaining majority “uncultured”. This paradoxical result, known as “the great plate count anomaly,” is a major unsolved problem in microbiology (Butkevich, 1932; Keller and Zengler, 2004; Rappe and Giovannoni, 2003; Staley and Konopka, 1985). Surveys of 16S rRNA gene sequences directly isolated from the environment showed that the anomaly is more than a gap in the total cell count, since >99% of species are uncultured (Barer and Harwood, 1999; Colwell and Grimes, 2000; Giovannoni, 2000; Grimes et al., 2000; Stackebrandt and Embley, 2000), although the precise percentages are hotly debated (Reeder and Knight, 2009).
We recently developed a general method to grow previously uncultured bacteria (here referred to as “uncultured”) from a variety of environments (Bollmann et al., 2007; Kaeberlein et al., 2002; Nichols et al., 2008). The method grows cells in their natural environment in a diffusion chamber. Cells are taken from an environment such as marine sediment, diluted, mixed with agar, and sandwiched between two semi-permeable membranes. The chamber is then placed back in the same environment where the sample originated. Alternatively, a porous membrane allows filamentous microbes to selectively migrate into a sterile chamber, which serves as a trap (Gavrish et al., 2008). The success of these approaches relies on the ability of small molecules from the natural environment to freely diffuse in and out of the chamber, while the movement of cells is restricted. After 1–2 weeks of incubation, colonies appear in the chamber, resulting in recovery of up to 40% of the bacteria in that environment (Kaeberlein et al., 2002). In contrast, recovery by standard cultivation methods from the same environment is only 0.01–0.05% (Cifuentes et al., 2000; Keller and Zengler, 2004; Llobet-Brossa et al., 1998). Application of the method to freshwater sediment showed that the richness and variety of species cultured in the chamber was considerably greater than on a Petri dish (Bollmann et al., 2007). Growth in a natural environment was used to successfully culture Pelagibacter, the most abundant marine bacterial species that does not grow on synthetic media (Rappe et al., 2002). Soil organisms have also been cultured on the surface of a membrane placed on top of soil (Ferrari et al., 2005), and while quantitative recovery has not been reported for this technique, FISH analysis indicated microcolonies of representatives of the elusive domain TM7, for which no cultured representatives are known.
We noticed that some organisms forming colonies in the diffusion chamber can grow on a Petri dish, but only in the presence of other species from the same environment (Kaeberlein et al., 2002; Nichols et al., 2008). Interspecies symbiosis based on nutrient exchange (syntrophy) is well known in the bacterial world (McInerney et al., 2008). Bacteria are also known to communicate using an interspecies quorum sensing factor (autoinducer 2, or AI-2) that induces synthesis of proteins that are useful for a community rather than a single cell, such as toxins or polymer hydrolases (Williams et al., 2007). Uncultured bacteria, however, do not grow on rich synthetic media (such media should largely obviate the need for nutrient supply by other species), and AI-2 has not been found to act as a growth-promoting factor, raising questions about the nature of unknown growth promoting factors in microbial communities.
We suggested that uncultured bacteria only commit to division in a familiar environment, which they recognize by the presence of growth factors released by their neighbors (Kaeberlein et al., 2002). Here we use co-culture of organisms from marine sediment to determine the nature of these diffusible molecules and find that siderophores from neighboring species induce growth of uncultured marine bacteria. Siderophores are low molecular weight compounds that have a high binding affinity for insoluble Fe(III). Microorganisms release siderophores to scavenge Fe(III) and then transport the ferric form back into the cells (Crosa and Walsh, 2002; Neilands, 1995; Wandersman and Delepelaire, 2004). Soluble Fe(II) is severely limited in most aerobic environments. Siderophore biosynthesis is tightly regulated in cultured species, and the chelators are produced when available iron is low. Uncultured species would be expected to have the same ability to autonomously produce siderophores since maintaining functional genes for siderophore production consumes few resources, and an inability would severely limit their ability to grow in Fe(II)-deficient but otherwise nutrient-rich environments.
Results
Recovery of Environmental Isolates Depends on Growth-Inducing Helper Bacteria
Samples of intertidal coarse sand sediment were collected from Canoe Beach in Massachusetts Bay, near Nahant, MA. The sediment was separated from seawater and vortexed in sterile saltwater to disperse the biofilm covering the sand particles (Fig. 1). The liquid phase containing bacterial cells was serially diluted and plated on nutrient medium containing 50% sterile seawater (R2Asea). After 5 days of incubation at 30 °C, colonies of varied morphology appeared on the plates (Fig. 2A). Similar results were obtained at 24 °C. There were disproportionately more colonies appearing on densely inoculated plates compared with more dilute plates. We hypothesized that some of the cells that grew on the densely seeded plates were receiving growth factors from neighboring colonies and would not grow in isolation. To test the possible growth dependence of microorganisms on neighboring species, pairs of colonies growing within a 2 cm distance of each other were picked from high-density plates (50–200 colonies per plate) and restreaked in close proximity to each other. Each of the two isolates was streaked on one half of an R2Asea plate and cross-streaked through the center of the plate, resulting in regions of proximal, distal and overlapping inoculation (Fig. 2B). Potential uncultured isolates were identified by their diminishing growth with increasing distance from the cultured “helper” strain on the cross streak plates. In the example given in Fig. 2B, colonies of the cultured organism Micrococcus luteus KLE1011 (a marine sand sediment isolate 99.5% identical to Micrococcus luteus DSM 200030T according to 16S rRNA gene sequence) grew larger as their distance from other colonies increased. This pattern of growth is typical for cultured bacteria, with the size of colonies increasing in a less crowded, nutrient-rich environment. In contrast, colonies of the uncultured isolate Maribacter polysiphoniae KLE1104 (99.9% similar to Maribacter polysiphoniae LMG 23671T) became barely visible as the distance from the cultured helper organism increased. Approximately 100 randomly picked pairs of colonies were restreaked from the high density plates, and 10% of these pairs showed this pattern of growth induction on cross-streaked plates.
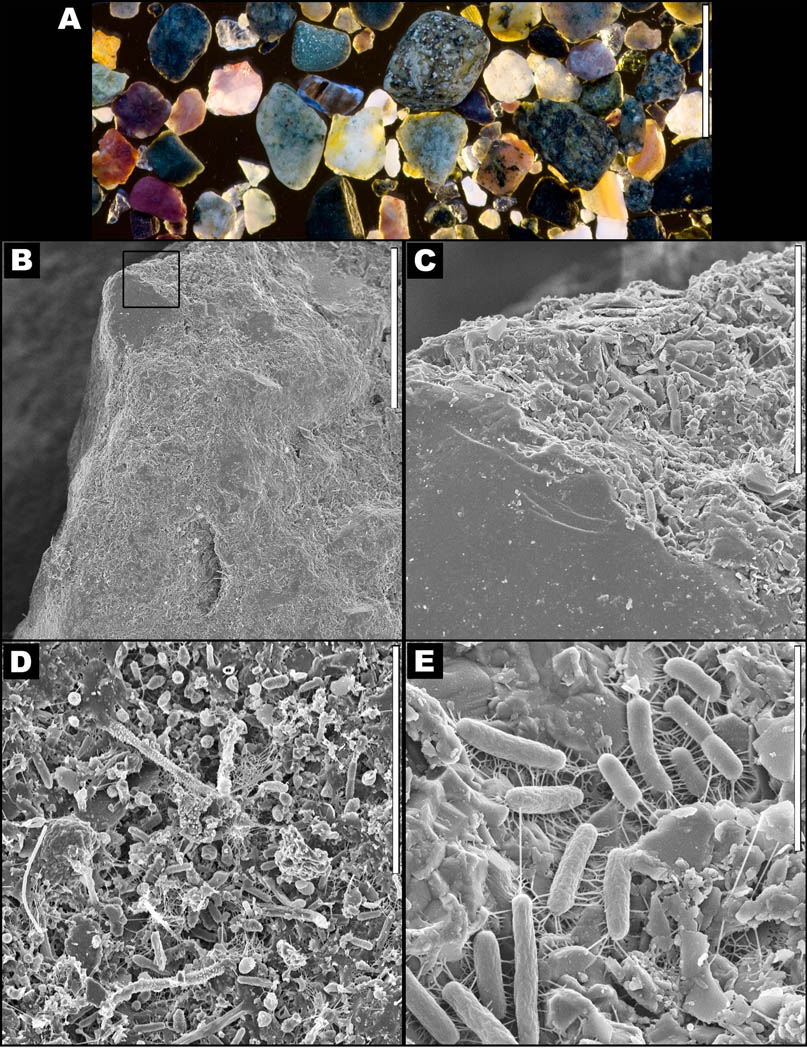
Fig. 1.
Intertidal sand grains. (A) Photograph of a collection of washed sand grains in seawater, scale bar = 3 mm. (B) Overview scanning electron micrograph (SEM) of a single sand grain, scale bar = 50 µm. (C) Higher resolution SEM of the boxed region in image B. Scale bar = 10 µm. (D) SEM image of a biofilm that can be seen attached to the surface of the sand particle, which represents the source of the bacterial cells that were resuspended by vortexing for isolation in this study. Scale bar = 10 µm. (E) SEM image of individual bacteria attached to the surface of the sand particle. Scale bar = 3 µm.
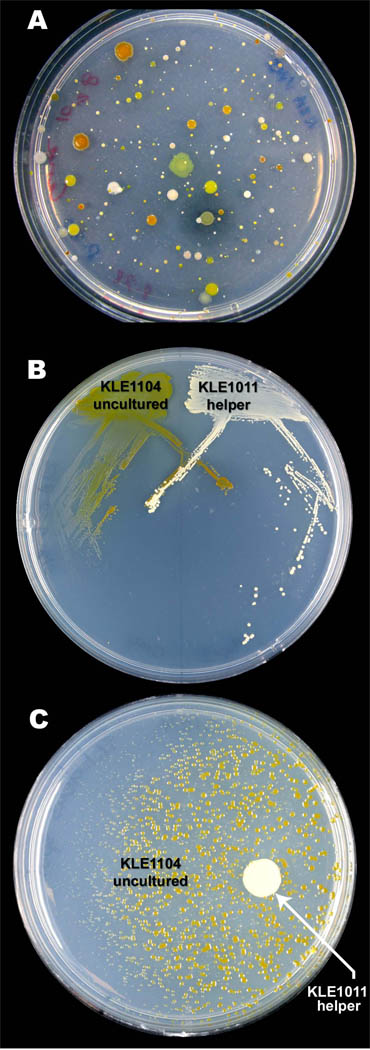
Fig. 2.
Growth of M. polysiphoniae KLE1104 is induced by M. luteus KLE1011. Bacterial cells were re-suspended from sand biofilms and plated to isolate species dependent on neighboring colonies. (A) Isolation spread plate of bacteria from sand grains. (B) Environmental helper M. luteus KLE1011 cross-streaked (right side of plate) with unculturable isolate M. polysiphoniae KLE1104 (left side of plate). Colonies of the unculturable strain are larger when in closer proximity to the helper. (C) Filtered spent supernatant of M. luteus KLE1011 induces growth of M. polysiphoniae KLE1104. In all cases tested, the uncultured phenotypes, including KLE1011, remained stable. See also Table S7.
Identification of Genes Coding for a Growth Factor
We reasoned that a small molecule growth factor could be readily identified if we found an uncultured isolate that was helped by Escherichia coli. Testing a collection of E. coli deletion strains for a lack of helping ability would then reveal its biosynthetic genes. Filtered spent supernatant of E. coli was able to induce growth of several uncultured isolates, including M. polysiphoniae KLE1104 (Fig. 3A).
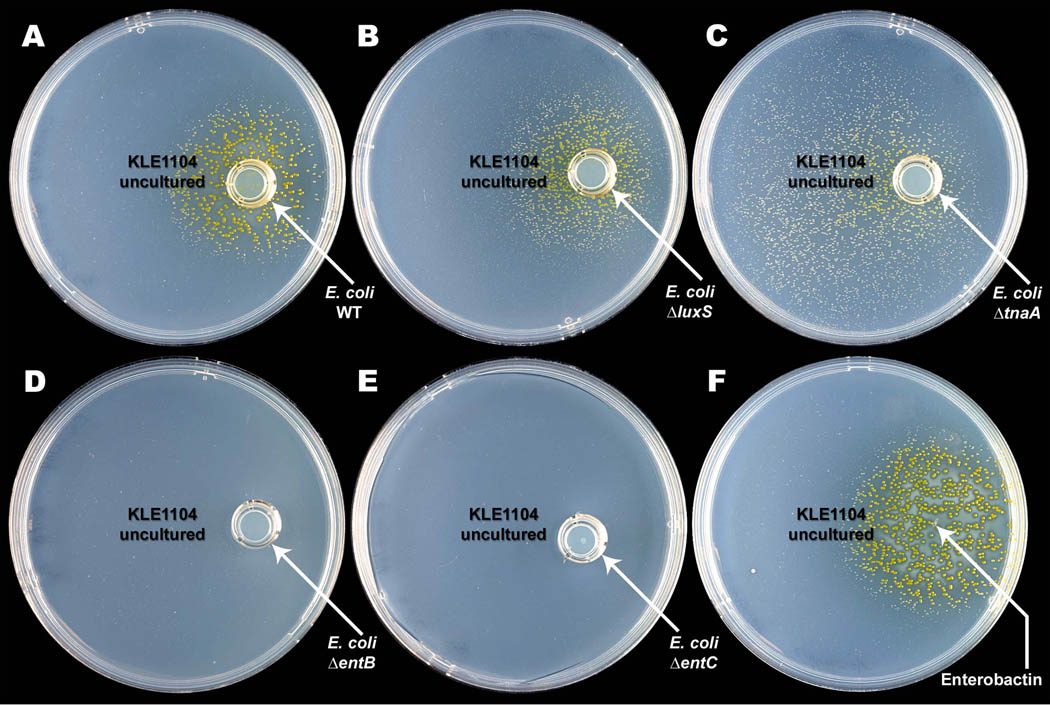
Fig. 3.
Growth induction of M. polysiphoniae KLE1104 by enterobactin from E. coli. Filtered spent supernatant of E. coli BW25113 induces growth of M. polysiphoniae KLE1104, but spent culture media from enterobactin mutant strains do not. (A) Supernatant from the parental E. coli BW25113 strain induces growth, as does supernatant from strains deficient in (B) ΔluxS and (C) ΔtnaA. There was more growth around the tnaA mutant in this experiment, but subsequent examination showed that it was due to variation in this qualitative test. Supernatant of strains deficient in enterobactin synthesis (D) ΔentB and (E) ΔentC do not induce growth of the unculturable. Growth was not observed even at high densities of KLE1104. (F) Purified enterobactin spotted on a plate evenly spread with M. polysiphoniae KLE1104 induced growth of the isolate. Colony size decreased with increasing distance from the enterobactin source and no growth of the unculturable was seen further away from the source.
E. coli has a limited repertoire of known secreted metabolites, which include the siderophore enterobactin (Braun and Braun, 2002), the universal quorum sensing factor AI-2 (Bassler and Losick, 2006; Surette et al., 1999), and the autoinducer indole (Di Martino et al., 2003). Supernatant from a strain deficient in AI-2 production (ΔluxS) and from a strain that does not secrete indole (ΔtnaA) showed normal growth induction (Fig. 3B, 3C). However, ΔentB or ΔentC, strains disabled in enterobactin synthesis were unable to induce the growth of M. polysiphoniae KLE1104 (Fig. 3D, 3E). Enterobactin spotted onto a plate with M. polysiphoniae KLE1104 strongly induced growth (Fig. 3F).
Structure Elucidation of Siderophores from M. luteus KLE1011
Spent medium from the natural helper M. luteus KLE1011 induced growth of the uncultured M. polysiphoniae KLE1104 (Fig. 2C). An iron-binding assay-guided fractionation ((chromazurol S, (Schwyn and Neilands, 1987)) was then employed to purify possible siderophores from the spent culture supernatant. Marine species survive with very low iron, 20 pM to 1 nM (Granger and Price, 1999; Johnson et al., 1997), and siderophore production in artificial media is induced under iron-limited conditions (Payne, 1994). Consequently, M. luteus KLE1011 was cultured in an iron-limited (100 pM Fe(III)) artificial seawater medium (Haygood et al., 1993). The spent culture supernatant was acidified (pH = 2) and the metabolites were purified by solid phase extraction (Amberlite XAD-2). Five different siderophores were further purified over size exclusion chromatography (LH-20) and reversed-phase (C18) high-pressure liquid chromatography (HPLC). In addition to exhibiting iron binding properties in the chromazurol S assay, both the desferric and ferric forms of each siderophore were observed by HPLC-mass spectrometry (MS) coupled with UV-visible spectroscopy analysis, confirming their role in iron acquisition (Fig. 4A).
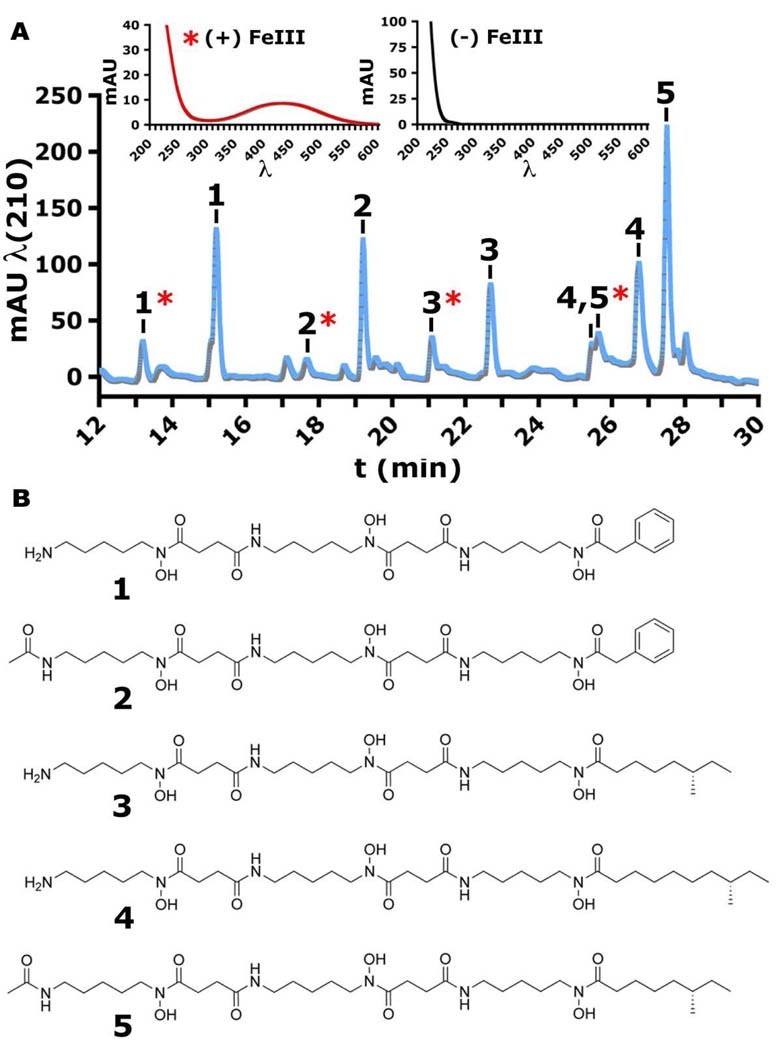
Fig. 4.
Active siderophores produced by helper strain M. luteus KLE1011. (A) Reversed-phase C18 HPLC-MS/UV-visible spectroscopic analysis. The ferric and desferric siderophores were separated over C18 HPLC, demonstrating a range of hydrophobic interactions with the nonpolar resin. The more polar ferric forms eluted faster than the desferric forms, showed masses indicative of iron complexation (MS), and exhibited characteristic iron-ligand charge transfer absorption (UV-visible). (B) Acyl-desferrioxamine siderophore structures. See also Figures S1–S2 and Tables S1–S6.
Detailed nuclear magnetic resonance (NMR) spectroscopy allowed structural elucidation of the purified siderophores (Fig. S1 and Tables S1–5). One dimensional (1H) chemical shifts and two dimensional homonuclear correlations (COSY), heteronuclear single quantum coherence correlations (HSQC), and heteronuclear multiple bond correlations (HMBC) revealed that all of the siderophores consisted of a central core with alternating N-hydroxycadaverine and succinic acid units and were of the desferrioxamine class (Challis, 2005). The metabolites differed by terminal acyl attachments. High-resolution LC Fourier-transform MS (Table S6) provided further evidence supporting their molecular formulas, and subsequent fragmentation analysis (MS/MS) confirmed the amide/hydroxamate backbone linkages, corroborating the proposed structures (Fig. S2). M. luteus KLE1011 produces a series of new acyl-desferrioxamine siderophores (Fig. 4B) with variable flanking acyl-side chains that drastically affect the central core’s aqueous solubility. Three of the siderophores (3–5) terminate with branched chain fatty acids while two (1–2) terminate with an atypical phenylacetyl moiety. Acyl-desferrioxamines 1 and 3 also were observed, beginning with a primary amine acetyl-cap (2 and 5). The five new metabolites were able to individually induce the growth of the previously uncultured M. polysiphoniae KLE1104, demonstrating that these siderophores represent the growth factors responsible for the helping activity.
As siderophores provide bacteria with a soluble source of iron, we investigated whether drastically increased quantities of soluble elemental iron could similarly allow growth of the uncultured isolates. Iron completely oxidizes to the insoluble Fe(III) state at the pH and oxygen levels of both seawater (the intertidal surface sand is continually mixed by wave action) and the laboratory media; however, by adding soluble FeSO4 after the media is autoclaved, the level of bioavailable Fe(II) is temporarily increased. Incubation of M. polysiphoniae KLE1104 over a range of Fe(II) concentrations showed that ~40 µM FeSO4 was necessary for optimal growth. Since this exceeds the reported picomolar levels of iron in the surface ocean (Johnson et al., 1997) by some six orders of magnitude, it is unlikely that uncomplexed iron is available to these organisms in the environment.
Siderophore Specificity of Uncultured Isolates
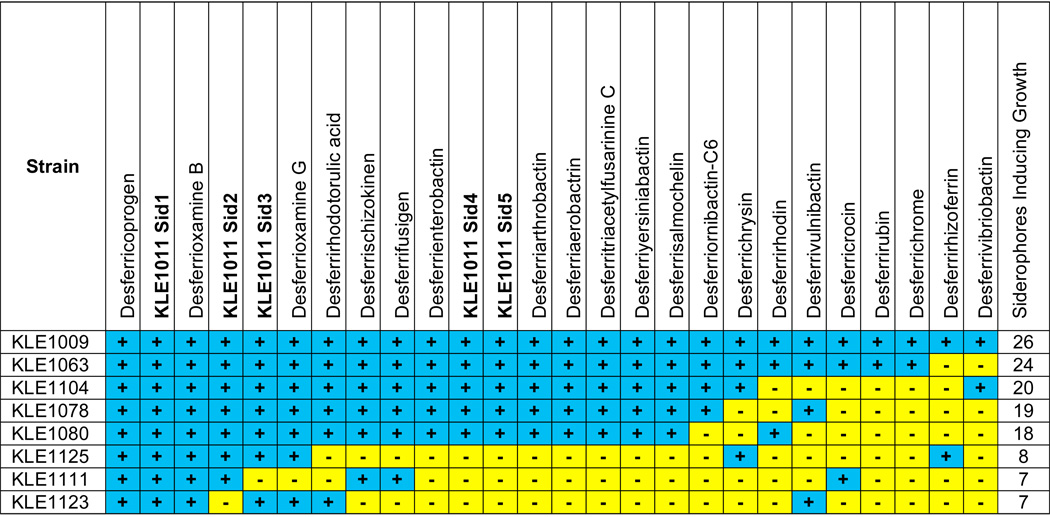
Given that siderophores from both E. coli and a neighboring species (M. luteus KLE1011) were able to induce growth of uncultured M. polysiphoniae KLE1104, it was important to determine the extent of siderophore promiscuity in growth promotion. Therefore, a screen was conducted to isolate uncultured bacteria induced by M. luteus KLE1011 and test their siderophore preferences. Environmental samples from Canoe Beach were spread on solid medium and then a culture of M. luteus KLE1011 was spotted on the plate as a potential helper. After incubation, colonies were picked and cross-streaked against M. luteus KLE1011. Plates were observed for a growth dependence pattern and candidate uncultured organisms were verified by seeding a plate spotted with the helper. A total of 185 colonies were screened of which 46 (25%) were helped by M. luteus KLE1011; the rest were independently fully cultured. Twenty of the M. luteus KLE1011-dependent isolates showed weak growth when plated alone, while the remaining 26 completely dependent isolates (14% of 185 screened) had no growth in the absence of helper. Six isolates that showed strong dependence on M. luteus KLE1011 and were not capable of producing colonies visible at 160× magnification on their own were chosen for further study. Based on 16S rRNA gene sequence comparisons, the six uncultured isolates were 96.5% to 99.3% identical to previously cultured relatives, which are typical marine bacteria (Table S7).
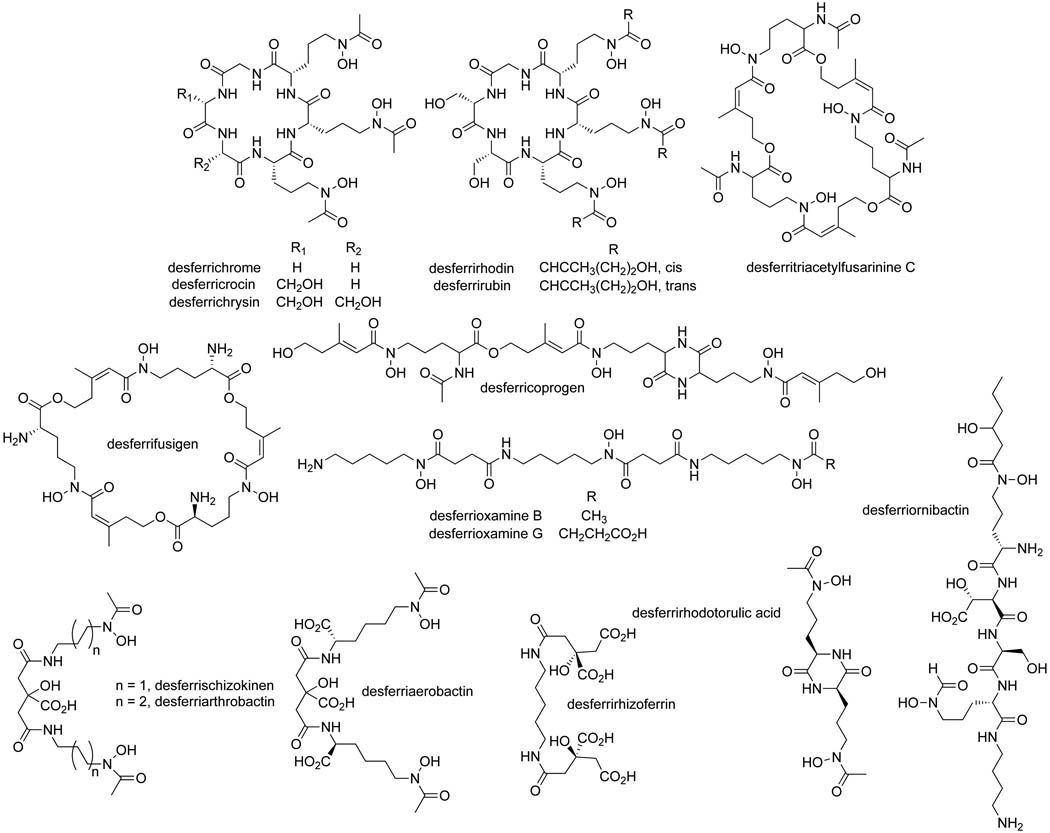
To further examine siderophore specificity in growth induction, a panel of 20 commercially available siderophores (Fig. 5) from both bacterial and fungal sources, desferrivulnibactin purified from Vibrio vulnificus and the five siderophores from M. luteus KLE1011 were tested for their ability to induce growth of the six uncultured organisms identified in the screen (Table. 1). The model uncultured M. polysiphoniae KLE1104 and one other strain (KLE1009) isolated from initial cross-streaks with environmental helpers were also tested with the siderophore panel. The siderophore panel consisted of linear and cyclic trihydroxamates, dihydroxamates, phenols/catechols, oxazolines/thiazolines, and carboxylic acid-type siderophores, providing a range of iron-binding structural types that target different siderophore receptors. Each of the eight isolates was induced by a particular set of siderophores. KLE1009 displayed wide siderophore promiscuity and was induced by all 26 siderophores tested, while other strains such as KLE1123, showed greater selectivity and only grew in the presence of 7 of the siderophores. Acyl-desferrioxamine 1 identified from the model helper M. luteus KLE1011 was capable of inducing growth of all 8 isolates tested, while the variable side chains in acyl-desferrioxamines 2–5 caused a decrease in the spectrum of growth induction. A broad range of growth induction was only seen for 2 of the commercial siderophores: the linear trihydroxamates desferrioxamine B (equivalent to the hydrophilic core of the M. luteus KLE1011 siderophores) and desferricoprogen. Isolates belonging to the division Bacteroidetes were able to utilize a wider variety than the Proteobacteria tested. The Bacteroidetes isolates were induced by an average of 22 of the siderophores, while the Proteobacteria were induced by an average of 10. Taken together, these data show a considerable variation in siderophore preferences among the uncultured isolates.
Fig. 5.
Siderophores tested for growth induction ability. The structures of 21 representative siderophores are shown, which were each tested for growth induction of 8 uncultured species. The siderophores include cyclic and linear trihydroxamates, dihydroxamates, carboxylic acid-types, phenols, catechols, oxazolines, and thiazolines that provide a variety of iron-binding structural types that would target various siderophore receptors.
Table. 1.
Growth Induction of Unculturable Isolates by Different Siderophores. Strains Maribacter sp. KLE1063, Winogradskyella sp. KLE1078, Hyphomonas sp. KLE1080, Reinekea sp. KLE1125, Simiduia sp. KLE1111 and Sulfitobacter sp. KLE1123 were isolated by screening for environmental bacteria helped by M. luteus KLE1011. Strains Cyclobacterium sp. KLE1009 and M. polysiphoniae KLE1104 were isolated from earlier cross-streak plates with environmental helpers. The five M. luteus KLE1011 siderophores and 21 representative siderophores were spotted on plates seeded with each unculturable and observed for growth induction after one week of incubation at 30 °C. Siderophores which induced growth are indicated in blue while siderophores that failed to induce growth are indicated in yellow.
 |
Isolation of Distantly Related Uncultured Microorganisms
The screening for uncultured microorganisms that require M. luteus KLE1011 produced a variety of isolates, but all of them are closely related to known cultured species. This may have resulted either from the preponderance of common microorganisms in this environment, both previously cultured and uncultured, or from the bias in the screen. Given the preference for particular siderophores, we sought to obtain uncultured isolates by an unbiased approach. For this purpose unphysiologically high concentrations of soluble Fe(II) was used as a surrogate for siderophores in an attempt to culture distant uncultured microorganisms and circumvent the natural systems that would likely require new siderophore classes.
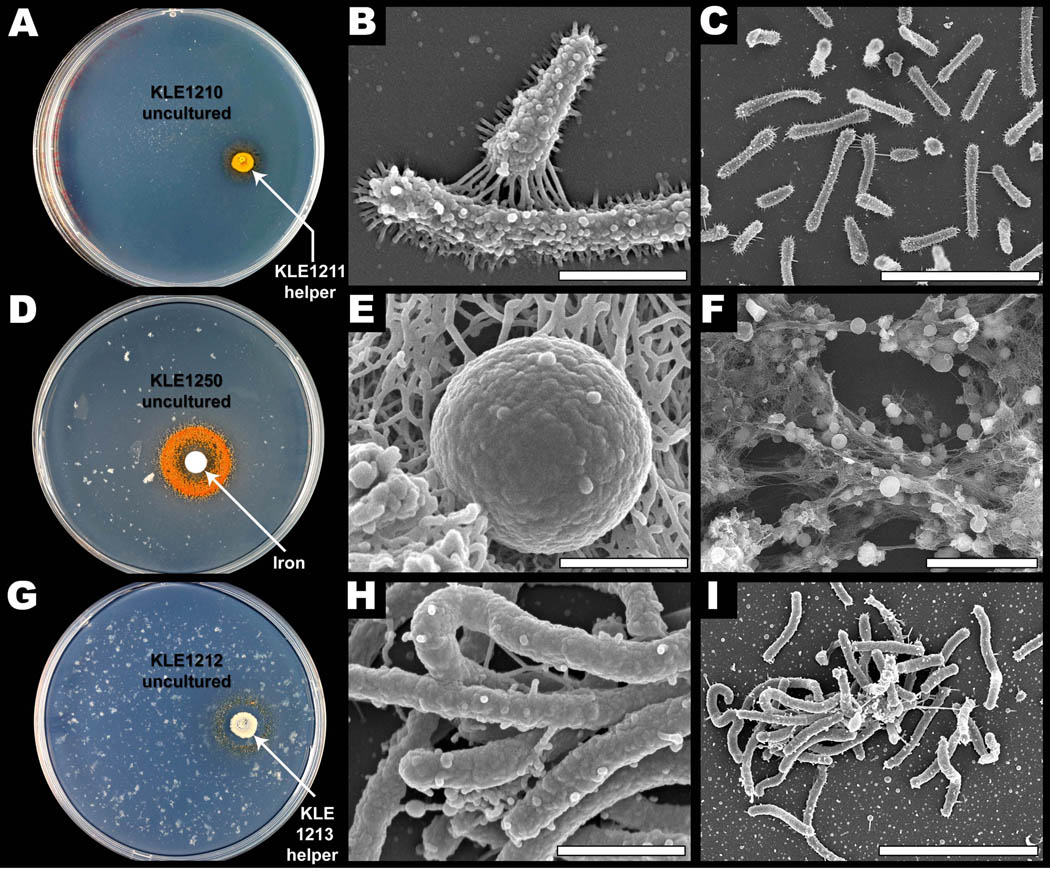
A sample from marine sediment was cultured on R2Asea with a high concentration of Fe(II), and then screened for isolates that would only grow in the presence of Fe(II). Of a total of 121 isolates, 31 showed strict dependence on Fe(II). Of a total of 121 isolates, 31 showed strict dependence on Fe(II). 16S rRNA genes were sequenced for fifteen of these, identifying 13 separate isolates and two potential duplicates. The 13 separate isolates varied in their sequence identity to typed species between 91.7% and 99.8% (Table S7), and the large proportion of strains isolated only once suggests that this environment has a considerable diversity of uncultured organisms. Importantly, three organisms in this small set of 15 were highly different from any typed strains by 16S rRNA gene sequence. One is a Verrucomicrobia sp. (KLE1210), a member of a phylum that is widespread in nature but is known mainly from uncultured isolates based on direct determination of 16S rRNA gene sequences from environmental DNA. Verrucomicrobia bacterium KLE1210 is only 91.7% identical to the closest cultured species, Rubritalea spongiae YM21-132T (Hedlund et al., 1996; Wagner and Horn, 2006) (Fig. S3).
A sample from the same environment was screened for cultured organisms able to support growth of Verrucomicrobia sp. KLE1210, as the model helper M. luteus KLE1011 was incapable of inducing growth. The intertidal sediment sample was added to the surface of ten plates seeded with Verrucomicrobia sp. KLE1210 and observed for growth promotion activity. A candidate helper was identified, isolated from the mix, and verified to induce growth of KLE1210. This led to the identification of an isolate with 97.1% 16S sequence identity to Lacinutrix mariniflava AKS432T (KLE1211), which is capable of inducing growth of KLE1210 (Fig. 6A,B,C). Assay-guided fractionation and MS analysis of spent supernatant resulted in the detection of the siderophore growth induction factor (Fig. S4). While this siderophore induces growth, none of the 26 tested siderophores from the panel induced growth, revealing the high siderophore selectivity of KLE1210.
Fig. 6.
Isolation of distantly related uncultured microorganisms. Three organisms only distantly related to typed strains were isolated on high [Fe(II)] media. (A) The cultivable Lacinutrix sp. KLE1211 (yellow spot) induces growth of the uncultured Verrucomicrobia isolate, KLE1210 (orange ring). (B,C) SEM images of Verrucomicrobia KLE1210, scale bars B,C = 1 µm, 5 µm. (D) Parvularculaceae KLE1250 (orange ring), growing near a filter disk (white spot) containing 10 µl of 1% iron sulfate. (E–F) SEM images of Parvularculaceae KLE1250, scale bars E,F = 1 µm, 10 µm. (G) Gammaproteobacterium KLE1212 (yellow ring) is induced by a cultivable species, Vibrio tasmaniensis KLE1213 (beige spot), isolated from the same sand biofilm. (H–I) SEM images of the uncultured isolate, Gammaproteobacterium KLE1212, scale bars H,I = 1 µm, 5 µm. See also Figures S3–S6.
The second distant isolate, KLE1250, belongs to a recently described alpha-proteobacterial order ‘Parvularculales’ (Lee et al., 2005). Parvularculaceae KLE1250 shows 93.7% identity by 16S rRNA gene sequence to Parvularcula bermudensis KCTC 1208T, one of only two initially described isolates (Cho and Giovannoni, 2003) that established this putative order and the only examples reported to be cultured. The apparent rarity of this order may be due to most species being uncultured, rather than an actual scarcity in the environment. Isolate KLE1250 may represent a new genus within this putative order (Fig. 6D, E, F and Fig. S5).
The third unusual isolate, KLE1212, is a member of the gamma-proteobacteria and shows only 90.4% 16S identity to the nearest typed strain, Kangiella aquimarina SW-154T (Fig. S6). The isolate was cultured by plating cells resuspended from a single sand grain on media supplemented with Fe(II) and screening for species strongly dependent on high concentrations of Fe(II). Cultured isolates from the same sand biofilm were screened for their ability to induce growth of KLE1212, resulting in the identification of a helper (KLE1213) with 99.7% 16S identity to Vibrio tasmaniensis LMG 21574T (Fig. 6G, H, I). Interestingly, none of the characterized isolates from Fe(II) plates could be induced to grow by the model helper M. luteus KLE1011, indicating both altered specificity in the growth dependence, and high diversity of uncultured species.
Discussion
Uncultured bacteria from many environments, including intertidal sediment, do not form colonies on select synthetic media and could account for up to 99.9% of the cells present in a sample (Keller and Zengler, 2004). Our results indicate that many of these bacterial strains will not commit to division unless they are provided with a small molecule growth-inducing factor from a neighboring microbe from that community.
Based on previous observations of bacteria growing only in the presence of other strains from the same environment (Kaeberlein et al., 2002; Morris et al., 2008; Nichols et al., 2008), we isolated pairs of colonies growing in close proximity on a nutrient-rich solid medium and plated them in a cross-streak to observe a helper-dependent pattern. This allowed us to isolate a number of organisms that were unable to grow in isolation, as well as the organisms that induced their growth. In addition to the environmental helper M. luteus KLE1011, wild type E. coli was able to induce the growth of a particular uncultured isolate, M. polysiphoniae KLE1104, which then allowed us to identify mutant strains lacking the helping activity. E. coli entB and entC mutants defective in enterobactin synthesis failed to induce growth of M. polysiphoniae KLE1104.
We then examined the ability of the natural helper organism M. luteus KLE1011 to produce siderophore growth factors. While marine isolates of M. luteus have been previously reported to show iron-binding activity, the identity of its siderophores has been unknown (Cabaj and Kosakowska, 2007). Filtered spent supernatant of M. luteus KLE1011 induced growth of M. polysiphoniae KLE1104. Subsequent iron-binding assay-guided fractionation followed by MS and NMR characterization lead to the isolation and characterization of M. luteus siderophores. Five new acyl-desferrioxamine siderophores were identified and each was able to induce growth of M. polysiphoniae KLE1104.
The five siderophores isolated from M. luteus KLE1011 harbor terminal hydrophobic modifications that alter their aqueous solubility. It has been proposed that marine bacteria produce siderophores with a range of hydrophobicities that could avoid significant losses to the open ocean. For example, the aquachelin- and marinobactin-class of marine siderophores have a long fatty acyl side chain of varying length to increase lipophilicity, leading to varying degrees of membrane association (Martinez et al., 2003; Martinez et al., 2000). Similarly, the acyl-desferrioxamines discovered here show a variety of hydrophobicities in contrast to their hydrophilic terrestrial counterpart, desferrioxamine B, suggesting that this is a common strategy in marine siderophore biosynthesis. Synthetic N-acyl derivatives of desferrioxamine B drastically reduced aqueous solubility and enhanced membrane permeability (Ihnat et al., 2000). M. luteus KLE1011 employs the same strategy of capping the primary amine with an acetyl group (acyl-desferrioxamines 2 and 5). Unlike the single fatty acid modification in aquachelins and marinobactins, M. luteus KLE1011 utilizes a two-component modification strategy to tailor both ends of the central core, including an atypical aromatic phenylacetyl side chain.
Using M. luteus KLE1011 as bait, a substantial number of uncultured bacteria were isolated in this study. Fourteen percent of colonies from a marine sediment sample growing in the vicinity of M. luteus KLE1011 appeared to depend on this isolate for growth. The relative ease of identifying strains dependent on M. luteus KLE1011 illustrates the commonality of siderophore dependence in the environment. Six of the uncultured strains from the screen along with two previously isolated organisms from the same environment were tested for their ability to grow in the presence of a variety of siderophores, including the five M. luteus KLE1011 acyl-desferrioxamines and 21 representative siderophores. Each uncultured isolate showed a particular pattern of siderophore dependence. Cyclobacterium sp. KLE1009 appeared the most promiscuous, and grew in the presence of all 26 siderophores tested, while Sulfitobacter sp. KLE1123 was the most restrictive and was only induced by 7 of the siderophores. The Bacteroidetes strains on average were capable of using a larger array of siderophores than the Proteobacteria.
In an unbiased screen for uncultured organisms, high levels of soluble Fe(II) were used as a surrogate for siderophores, and a small sampling of candidates resulted in the discovery of three organisms highly divergent from known species. These organisms were not induced to grow by M. luteus KLE1011, illustrating the specificity and diversity of dependent species in the sand biofilm.
Perhaps the most puzzling aspect of the “uncultured” phenomenon is that many of the uncultured microorganisms are closely related to known and well-studied cultured species by their 16S rRNA gene sequence signatures. Indeed, why would bacteria closely related to organisms that thrive on a large variety of nutrients in vitro fail to grow similarly well? We find that a common theme among uncultured bacteria in the marine sediment environment is their dependence on siderophores from neighboring species. Early studies of the function of siderophores revealed the growth promoting ability of these compounds. The lag phase of Bacillus megaterium, for example, was shown to be reduced by the addition of schizokinen, a siderophore it is capable of producing (Lankford et al., 1966). Although, the strains in these earlier studies were capable of growth in isolation in contrast to the strains isolated in this study. The results imply that the microorganisms examined in this study, which belong to unrelated groups, have lost the ability to autonomously produce siderophores or fail to upregulate production on synthetic media without assistance (Reissbrodt et al., 2000; Reissbrodt et al., 2002).
The results are surprising and counter-intuitive from what we know about siderophore production by cultured microorganisms. Many species can acquire siderophores they do not produce, a phenomenon known as ‘siderophore piracy’ (Luckey et al., 1972; Schubert et al., 1999). For example, mycobactin and carboxymycobactin produced by Mycobacterium species induce the FemIRA siderophore transport system in P. aeruginosa (Llamas et al., 2006). It appears that it is a common phenomenon for bacterial genomes to contain more siderophore uptake genes than synthesis genes. For example, genome and biochemical analyses on Pseudomonas fluorescens SBW25 showed the presence of 24 putative siderophore receptors, allowing the cell to acquire a range of heterologous pyoverdine siderophores in addition to its own (Moon et al., 2008). Siderophore synthesis is typically upregulated when available iron is low (Winkelmann, 2002), but some uncultured bacteria do not seem to follow this strategy. The cost of maintaining siderophore biosynthetic genes is probably very low, on the other hand, the cost of losing the ability to autonomously produce siderophores seems imposing - inabiliy to grow in any environment that lacks suitable siderophores. Due to the widespread nature of siderophore synthesis in aerobes (Kraemer, 2004) and the considerable level of horizontal transfer of these systems (Challis and Hopwood, 2003), it is unlikely that there is an aerobic lineage that has never possessed the ability to make them, suggesting a functional loss or altered siderophore regulation in uncultured organisms. A possible advantage to not being able to autonomously produce siderophores may be the security of initiating growth only in an environment populated by compatible neighbors. In this model, the specificity of siderophore uptake determines the range of environments suitable for growth. Growing populations are vulnerable to noxious conditions, while non-growing stationary and dormant persister cells are very resilient (Lewis, 2007), which may provide a distinct advantage for cells that only grow in familiar environments.
Interestingly, this strategy seems very similar to the germination of spores. Recently, it was reported that germination of B. subtilis spores is strongly induced by muropeptides, breakdown products of peptidoglycan derived from other bacteria (Shah et al., 2008). Apparently, the presence of nutrients is not enough, and there must be other factors indicating the presence of an environment favorable for growth.
Significance
The results of this study indicate a widespread occurrence of siderophore-dependent uncultured bacteria in marine sediment and point toward siderophores being key components in establishing microbial communities in this environment. It is likely that many uncultured microorganisms require as yet undiscovered siderophore classes for growth promotion. The diversity of uncultured organisms should vary widely based on siderophore receptor specificity, providing a general strategy to access new microorganisms. Their growth can be induced by the addition of exogenously supplied helper siderophores or artificially high concentrations of Fe(II), and these approaches could expand drug discovery efforts, as microbial metabolites have afforded some of our most useful small molecule therapeutic agents (Newman and Cragg, 2007).
Experimental Procedures
Isolation of Uncultured Bacteria and Helpers
Samples of intertidal sediment were collected from Canoe Beach in Nahant, MA. Sediment (5 g) was resuspended in 5 mL of filtered, autoclaved seawater and vortexed for 1 min. Samples were diluted and plated on solid R2Asea medium (R2A (Difco) diluted in 50% filtered, autoclaved seawater) in ten-fold dilutions to obtain plates of varying colony density. Plates were incubated at 30 °C for 5 days. Pairs of colonies growing within a 2 cm distance were cross-streaked in the pattern shown in Fig. 2B on fresh plates to screen for helper-dependent patterns. Potential uncultured and helper isolates were resuspended in sterile seawater with 15% glycerol and frozen at −80 °C. Dependence on a helper was verified by spreading a plate with 103 to 104 cells of the potential uncultured and spotting the potential helper species. Plates were incubated at 30 °C for 3–7 days and observed for growth of the potential uncultured near the helper spot. To isolate species dependent on M. luteus KLE1011, environmental samples were plated and spotted with KLE1011. After incubation for 5 days at 30 °C, colonies were screened that grew within 2 cm of KLE1011.
Isolation of Distantly Related Uncultured Bacteria
Samples of intertidal sediment were collected from Canoe Beach in Nahant, MA. R2Asea agar medium (R2A (Difco) diluted in 50% filtered, autoclaved seawater) was supplemented with 0.001% Iron(II) Sulfate after autoclaving. Sediment was resuspended in 5 mL of filtered, autoclaved seawater, vortexed for 1 min and plated. After incubation at 30 °C for 5 days, colonies were picked and restreaked on R2Asea media with and without Fe(II) supplementation to screen for isolates with Fe(II) growth dependence.
Identification of isolates by 16S rRNA gene sequencing
16S rRNA gene fragment (ca. 1400–1500 bp) was amplified using universal bacterial primers 27F and 1492R (E. coli numbering system; (Weisburg, 1991)). The identification of phylogenetic neighbors and the calculation of pairwise sequence similarity were carried out by using the EzTaxon server (http://www.eztaxon.org/; (Chun et al., 2007)). The obtained sequences were aligned with phylogenetically related type strains obtained from the Ribosomal Database Project (Cole et al., 2003). Phylogenetic tree was obtained by neighbor-joining (Saitou and Nei, 1987) method using MEGA 4 (Tamura et al., 2007). An evolutionary distance matrix for neighbor-joining method was generated according to the model of Jukes and Cantor (Jukes and Cantor, 1969). The robustness of the tree topologies was assessed by bootstrap analyses based on 1000 replications. Nucleotide sequences obtained in this study have been deposited in the GenBank database (see Table S7 for accession numbers).
Testing Spent Media of M. luteus KLE1011 and E. coli Strains
To test the helping ability of M. luteus KLE1011 spent media, 5 mL cultures of liquid R2NP media (0.5 g yeast extract, 0.5 g casamino acids, 0.5 g dextrose, 0.3 g sodium pyruvate and 0.3 g dipotassium phosphate dissolved in 50% filtered, autoclaved seawater) were inoculated from a glycerol stock and incubated at 30 °C with shaking for 2 days. Cultures were then centrifuged and filtered (0.22 µm). M. polysiphoniae KLE1104 was seeded on R2Asea plates followed by the addition of a tissue insert (Nunc) with a 0.02 µm bottom filter. 500 µL of spent media was added to the insert and plates were incubated at 30 °C for 5 days. E. coli BW25113 and the ΔluxS, ΔtnaA, ΔentB and ΔentC knockouts from the Keio Collection knockout library (Baba et al., 2006) were tested using the same protocol.
Structure elucidation of KLE1011 acyl-desferrioxamine siderophores
NMR experiments were performed on desferric siderophores in deuterated dimethylsulfoxide with a symmetrical NMR microtube susceptibility-matched with dimethylsulfoxide (Shigemi, Inc.) on a Varian INOVA 600 MHz NMR. The ferric and desfferic forms of each purified siderophore were also separated over HPLC and detected using either a linear quadrupole ion trap / Fourier transform ion cyclotron resonance hybrid MS (Thermo Electron) or a linear quadropole MS (Agilent 6130). The +1 ion of the desferric and ferric siderophores were selected for fragmentation analysis by MS/MS.
Growth Induction by Siderophores
The commercial siderophores include desferrifusigen, desferriaerobactin, desferricrocin, desferriarthrobactin, desferrischizokinen, desferrichrome, desferrirhizoferrin, desferrirhodin, desferrirhodotorulic acid, desferritriacyetylfusarinine C, desferrioxamine G, desferriornibactin-C6, desferrichrysin, desferricoprogen, desferrirubin, desferrioxamine B, desferriyersiniabactin, desferrisalmochelin, desferrivibriobactin (EMC Microcollections, Germany) and enterobactin (Genaxxon Bioscience, Germany). Vulnibactin was isolated from Vibrio vulnificus grown in iron-deplete conditions similarly to as previously described (Okujo et al., 1994). To test growth induction, 10 µL of siderophore was spotted on R2Asea plates seeded with an uncultured isolate. All commercial siderophores were tested at 0.01 mg/mL except desferrienterobactin, desferriyersiniabactin, desferrisalmochelin and desferrivibriobactin, which were tested at 1 mg/mL. Plates were incubated for 3–7 days at 30 °C and observed for growth around the spotted siderophore.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R21 AI070736 to KL and R01 CA24487 to JC. JMC is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2002-09). We thank Dr. Jessica Dawlaty for preliminary assay-guided fractionation, Drs. Wilhelm Haas and Steven Gygi (Harvard Medical School) for high-resolution MS acquisition, and William Fowle (Northeastern University) for SEM sample preparation and imaging. We are grateful to Dr. Michael Fischbach (University of California, San Francisco) for critical reading of the manuscript and helpful suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Deposition:
16S rRNA gene sequences reported in this study have been deposited in the GenBank database (accession numbers: FJ229465, FJ229461, FJ229467, FJ229459, FJ229460, FJ229466, FJ229462, GU644354, FJ229463, GU644361, GU644355, GU644363, GU644367, GU644356, GU644360, GU644358, GU644357, GU644359, GU644362, GU644368, GU644365, GU644366, GU644364, GQ262724, GU262724).
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barer MR, Harwood CR. Bacterial viability and culturability. Adv Microb Physiol. 1999;41:93–137. doi: 10.1016/s0065-2911(08)60166-6. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Bollmann A, Lewis K, Epstein SS. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl Environ Microbiol. 2007;73:6386–6390. doi: 10.1128/AEM.01309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Braun M. Iron transport and signaling in Escherichia coli. FEBS Lett. 2002;529:78–85. doi: 10.1016/s0014-5793(02)03185-x. [DOI] [PubMed] [Google Scholar]
- Butkevich VS. Zür Methodik der bakterioloschen Meeresuntersuchungen und einige Angaben über die Verteilung der Bakterien im Wasser und in den Büden des Barents Meeres. Trans Oceanogr Inst Moscow (in Russian with German summary) 1932;2:5–39. [Google Scholar]
- Cabaj A, Kosakowska A. Iron-dependent growth of and siderophore production by two heterotrophic bacteria isolated from brackish water of the southern Baltic Sea. Microbiol Res. 2007;164:570–577. doi: 10.1016/j.micres.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Challis GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem. 2005;6:601–611. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- Challis GL, Hopwood DA. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci USA. 2003;100:14555–14561. doi: 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JC, Giovannoni SJ. Parvularcula bermudensis gen. nov., sp. nov., a marine bacterium that forms a deep branch in the alpha-Proteobacteria. Int J Syst Evol Microbiol. 2003;53:1031–1036. doi: 10.1099/ijs.0.02566-0. [DOI] [PubMed] [Google Scholar]
- Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57:2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- Cifuentes A, Anton J, Benlloch S, Donnelly A, Herbert RA, Rodriguez-Valera F. Prokaryotic diversity in Zostera noltii-colonized marine sediments. Appl Environ Microbiol. 2000;66:1715–1719. doi: 10.1128/aem.66.4.1715-1719.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, et al. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell RR, Grimes DJ. Semantics and strategies. In: Colwell RR, Grimes DJ, editors. Nonculturable microorganisms in the environment. Washington DC: ASM Press; 2000. pp. 1–6. [Google Scholar]
- Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66:223-+. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino P, Fursy R, Bret L, Sundararaju B, Phillips RS. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can J Microbiol. 2003;49:443–449. doi: 10.1139/w03-056. [DOI] [PubMed] [Google Scholar]
- Ferrari BC, Binnerup SJ, Gillings M. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl Environ Microbiol. 2005;71:8714–8720. doi: 10.1128/AEM.71.12.8714-8720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrish E, Bollmann A, Epstein S, Lewis K. A trap for in situ cultivation of filamentous actinobacteria. J Microbiol Methods. 2008;72:257–262. doi: 10.1016/j.mimet.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJR. Evolution, Diversity and Molecular Ecology of Marine Prokaryotes. In: Kirchman D, editor. Microbial Ecology of the Oceans. New York: Wiley-Liss. Inc; 2000. pp. 47–84. [Google Scholar]
- Granger J, Price NM. The importance of siderophores in iron nutrition of heterotrophic marine bacteria. Limnol Oceanogr. 1999;44:541–555. [Google Scholar]
- Grimes DJ, Mills AL, Nealson KH. The importance of viable but nonculturable bacteria in biogeochemistry. In: Colwell RR, Grimes DJ, editors. Nonculturable microorganisms in the environment. Washington DC: ASM Press; 2000. pp. 209–227. [Google Scholar]
- Haygood MG, Holt PD, Butler A. Aerobactin Production by a Planktonic Marine Vibrio sp. Limnol Oceanogr. 1993;38:1091–1097. [Google Scholar]
- Hedlund BP, Gosink JJ, Staley JT. Phylogeny of Prosthecobacter, the fusiform caulobacters: members of a recently discovered division of the bacteria. Int J Syst Bacteriol. 1996;46:960–966. doi: 10.1099/00207713-46-4-960. [DOI] [PubMed] [Google Scholar]
- Ihnat PM, Vennerstrom JL, Robinson DH. Synthesis and solution properties of deferoxamine amides. J Pharm Sci. 2000;89:1525–1536. doi: 10.1002/1520-6017(200012)89:12<1525::aid-jps3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Johnson KS, Gordon RM, Coale KH. What controls dissolved iron concentrations in the world ocean? Mar Chem. 1997;57:137–161. [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. New York, NY: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Kaeberlein T, Lewis K, Epstein SS. Isolating "uncultivable" microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- Keller M, Zengler K. Tapping into microbial diversity. Nat Rev Microbiol. 2004;2:141–150. doi: 10.1038/nrmicro819. [DOI] [PubMed] [Google Scholar]
- Kraemer S. Iron oxide dissolution and solubility in the presence of siderophores. Aquatic Sciences - Research Across Boundaries. 2004;66:3–18. [Google Scholar]
- Lankford CE, Walker JR, Reeves JB, Nabbut NH, Byers BR, Jones RJ. Inoculum-dependent division lag of Bacillus cultures and its relation to an endogenous factor(s) ("schizokinen") J Bacteriol. 1966;91:1070–1079. doi: 10.1128/jb.91.3.1070-1079.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KB, Liu CT, Anzai Y, Kim H, Aono T, Oyaizu H. The hierarchical system of the 'Alphaproteobacteria': description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int J Syst Evol Microbiol. 2005;55:1907–1919. doi: 10.1099/ijs.0.63663-0. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- Llamas MA, Sparrius M, Kloet R, Jimenez CR, Vandenbroucke-Grauls C, Bitter W. The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J Bacteriol. 2006;188:1882–1891. doi: 10.1128/JB.188.5.1882-1891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llobet-Brossa E, Rossello-Mora R, Amann R. Microbial community composition of wadden sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey M, Pollack JR, Wayne R, Ames BN, Neilands JB. Iron uptake in Salmonella typhimurium: utilization of exogenous siderochromes as iron carriers. J Bacteriol. 1972;111:731–738. doi: 10.1128/jb.111.3.731-738.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JS, Carter-Franklin JN, Mann EL, Martin JD, Haygood MG, Butler A. Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium. Proc Natl Acad Sci USA. 2003;100:3754–3759. doi: 10.1073/pnas.0637444100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JS, Zhang GP, Holt PD, Jung HT, Carrano CJ, Haygood MG, Butler A. Self-assembling amphiphilic siderophores from marine bacteria. Science. 2000;287:1245–1247. doi: 10.1126/science.287.5456.1245. [DOI] [PubMed] [Google Scholar]
- McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJ, Schink B, Rohlin L, Gunsalus RP. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Annals of the New York Academy of Sciences. 2008;1125:58–72. doi: 10.1196/annals.1419.005. [DOI] [PubMed] [Google Scholar]
- Moon CD, Zhang XX, Matthijs S, Schafer M, Budzikiewicz H, Rainey PB. Genomic, genetic and structural analysis of pyoverdine-mediated iron acquisition in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25. BMC Microbiol. 2008;8:7. doi: 10.1186/1471-2180-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by "helper" heterotrophic bacteria. Appl Environ Microbiol. 2008;74:4530–4534. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilands JB. Siderophores - Structure and Function of Microbial Iron Transport Compounds. J Biol Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- Nichols D, Lewis K, Orjala J, Mo S, Ortenberg R, O'Connor P, Zhao C, Vouros P, Kaeberlein T, Epstein SS. Short peptide induces an "uncultivable" microorganism to grow in vitro. Appl Environ Microbiol. 2008;74:4889–4897. doi: 10.1128/AEM.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okujo N, Saito M, Yamamoto S, Yoshida T, Miyoshi S, Shinoda S. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals. 1994;7:109–116. doi: 10.1007/BF00140480. [DOI] [PubMed] [Google Scholar]
- Payne SM. Detection, isolation, and characterization of siderophores. Methods Enzymol. 1994;235:329–344. doi: 10.1016/0076-6879(94)35151-1. [DOI] [PubMed] [Google Scholar]
- Rappe MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- Reeder J, Knight R. The 'rare biosphere': a reality check. Nature Methods. 2009;6:636–637. doi: 10.1038/nmeth0909-636. [DOI] [PubMed] [Google Scholar]
- Reissbrodt R, Heier H, Tschape H, Kingsley RA, Williams PH. Resuscitation by ferrioxamine E of stressed Salmonella enterica serovar typhimurium from soil and water microcosms. Appl Environ Microbiol. 2000;66:4128–4130. doi: 10.1128/aem.66.9.4128-4130.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissbrodt R, Rienaecker I, Romanova JM, Freestone PP, Haigh RD, Lyte M, Tschape H, Williams PH. Resuscitation of Salmonella enterica serovar typhimurium and enterohemorrhagic Escherichia coli from the viable but nonculturable state by heat-stable enterobacterial autoinducer. Appl Environ Microbiol. 2002;68:4788–4794. doi: 10.1128/AEM.68.10.4788-4794.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schubert S, Fischer D, Heesemann J. Ferric enterochelin transport in Yersinia enterocolitica: Molecular and evolutionary aspects. J Bacteriol. 1999;181:6387–6395. doi: 10.1128/jb.181.20.6387-6395.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Shah IM, Laaberki M-H, Popham DL, Dworkin J. A Eukaryotic-like Ser/Thr Kinase Signals Bacteria to Exit Dormancy in Response to Peptidoglycan Fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Embley TM. Diversity of uncultured microorganisms in the environment. In: Colwell RR, Grimes DJ, editors. Nonculturable microorganisms in the environment. Washington DC: ASM Press; 2000. pp. 57–75. [Google Scholar]
- Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biol Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wagner M, Horn M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr Opin Biotechnol. 2006;17:241–249. doi: 10.1016/j.copbio.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- Weisburg WW, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P, Winzer K, Chan WC, Camara M. Look who's talking: communication and quorum sensing in the bacterial world. Philosophical transactions of the Royal Society of London. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelmann G. Microbial siderophore-mediated transport. Biochem Soc Trans. 2002;30:691–696. doi: 10.1042/bst0300691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.