Abstract
The mammalian heart is formed from distinct sets of first (FHF) and second (SHF) heart field progenitors. Although multipotent progenitors have been previously shown to give rise to cardiomyocytes, smooth muscle, and endothelial cells, the mechanism governing the generation of large numbers of differentiated progeny remains poorly understood. Herein, we have employed a two-colored fluorescent reporter system to isolate FHF and SHF progenitors from developing mouse embryos and embryonic stem cells. Genome wide profiling of coding and non-coding transcripts revealed distinct molecular signatures of these progenitor populations. We further identify a committed ventricular progenitor cell in the Islet 1 lineage that is capable of in vitro expansion, differentiation, and assembly into functional ventricular muscle tissue. These results represent a novel approach combining tissue-engineering with stem cell biology for the generation of functional ventricular tissue.
The mammalian heart is composed of a diversified set of muscle and non-muscle cells that arise from multipotent progenitors in the FHF and SHF (1, 2). Defining the precise progenitor identity and the pathways that lead to ventricular myogenesis is critical for understanding cardiogenesis, with major implications for regenerative cardiovascular medicine.
Accordingly, we generated a transgenic mouse with the red florescent protein dsRed under the control of an Isl1-dependent SHF specific enhancer of the transcription factor Mef2c (3, 4). We then bred this mouse line with the previously described transgenic mouse line in which eGFP is under the control of the cardiac specific Nkx2.5 enhancer (5, 6). Fluorescence microscopy of double transgenic embryos on embryonic day (ED) 9.5 revealed that the entire primitive heart tube was eGFP+, but only the right ventricle (RV) and the outflow tract (OFT) were also dsRed+. Further, the pharyngeal mesoderm (PM) which contributes to the RV and OFT (7, 8) was dsRed+ but eGFP- (Figure 1A–1C). To delineate the local expression of eGFP and dsRed in the developing heart, we performed immunohistochemistry on ED9.5 embryos and found that eGFP+/dsRed+ cells (R+G+) were restricted to the RV and OFT, dsRed-/eGFP+ cells (R-G+) to the left ventricle (LV) and inflow tract (IFT), and dsRed+ cells (R+G-) to the pharyngeal mesoderm (Figure 1D).
Figure 1. ED9.5 SHF-dsRed/Nkx2.5-eGFP transgenic embryos.
A-C. Whole mount fluorescence microscopy of double transgenic ED9.5 embryos. The LV and IFT are eGFP+, the RV and OT are dsRed+ and eGFP+, and the PM is dsRed+. Right lateral (A) and (B), and left lateral (C) views are shown. Scale bar 500µm (A) and 100µm (B and C).
D. Immunofluorescence labeling of ED9.5 double transgenic mouse embryo shows that cells in the RV and OFT are R+G+, in the PM are R+G-, and in the LV and IFT are R-G+. Section levels are shown in cartoon. eGFP is shown in green, dsRed in Red, and overlay in yellow. Scale bar 50µm.
Embryonic stem cell lines (ESC) can differentiate in vitro and recapitulate many of the in vivo developmental programs providing an attractive model system for lineage commitment. We therefore generated multiple ES lines that harbor both the Nkx2.5-eGFP and the SHF-dsRed reporters (Figure S1A). Fluorescence microscopy of chimera embryos generated from these ESC lines revealed faithful recapitulation of marker expression (Figure S1B and S1C). In vitro differentiation by embryoid body (EB) formation resulted in discrete populations of R+G+, R+G-, and R-G+ cells by EB day 6 (Figure S1D).
To isolate ESC-derived FHF and SHF progenitor cells, we dissociated day 6 EBs into single cell suspension and FACS purified four distinct populations of cells: R+G+, R+G-, R-G+, and unlabeled (R-G-) (Figure 2A and S2A). We then performed DNA microarray analysis on coding and non-coding RNA from ES cell-progenitors. Hierarchical clustering (9) showed distinct reproducible expression patterns for the different cardiac progenitor subsets of mRNAs as well as microRNAs (miRNAs) (Figure S3–S5, and Table S1). Next, we FACS purified ED9.5 embryonic progenitors (Figure 2B and S2B). Real time PCR (qPCR) analysis on 100 mRNAs and 10 miRNAs revealed that ESC and embryonic derived progenitors displayed similar but non-identical patterns of expression (Figure 2C and S6). Significantly, RNA samples from embryonic progenitors were isolated immediately after FACS sorting to avoid in vitro expansion artifacts. mRNAs and miRNAs implicated in cardiac development and disease were enriched in the colored cells compared with unlabeled cells. Isl1, a marker for the SHF was appropriately enriched only in the R+G+ and the R+G- populations whereas Tbx5, a marker of the FHF (10, 11), was appropriately enriched only in the R-G+ population. Interestingly, the R+G+ cells appeared to be the most myogenic population based on the expression of myocardial markers such as cardiac Troponins, cardiogenic transcription factors, and BMP signaling molecules. Further, the R+G- population of the PM expressed high levels of Snai2, a transcription factor regulating epithelial to mesenchymal transition (EMT) and necessary for cell-migration (12, 13), raising the possibility that SHF/PM progenitors undergo EMT prior to migrating into the developing heart. In addition, miRNA199a/b were preferentially expressed in the R+G- population and miRNA200a/b in the R-G+ population and can therefore be considered markers for the SHF and FHF respectively (Figure S7).
Figure 2. Isolation of cardiac progenitors.
A&B. Representative flow cytometry plots of double transgenic day 6 EBs (A) and ED9.5 embryos (B) showing 4 populations of cells: R+G+, R+G-, RG+, and R-G-.
C. qPCR analysis of mRNA and miRNA isolated from embryonic progenitors. Values are normalized against unlabeled control. Standard deviation (SD) shown. Differences between groups were highly statistically significant, see Table S2 and S3.
D. Hoechst staining and FACS-based cell cycle analysis of undifferentiated ESC, EB day6 progenitors, and differentiated progeny.
E. ESC derived cardiac progenitors were cultured 5 days. DAPI and Ki67 staining was performed to quantify total cell number and proportion of cycling cells (Ki67+ cells/total cells). SD shown (n=4).
A hallmark of progenitor cells is their capacity for expansion prior to differentiation. Accordingly, we performed Hoechst staining and FACS-based cell cycle analysis on undifferentiated ESC, EB day 6 cardiac progenitors, and their differentiated progeny. Undifferentiated ESC and EB day 6 progenitors had approximately 40–60% of cells in S or G2 phase but the differentiated progeny had less than 10% of cells in S or G2 phase (Figure 2D). To validate these results, we isolated EB day 6 progenitors and allowed them to expand in vitro for an additional 5 days. Immunostaining with Ki67, a marker for actively cycling cells, showed that 24 hours after isolation most cells were actively cycling but this waned over five days. Conversely, total cell number increased by approximately four-fold (Figure 2E). Furthermore the expression of the progenitor markers Isl1 or Tbx5 was maximal at the time of progenitor isolation but waned with further differentiation. In contrast Troponin T expression continued to increase with differentiation (Figure S8). Thus, the progenitor populations have a real but limited in vitro expansion potential. The drop off in expansion is concomitant with differentiation and loss of progenitor marker expression suggesting that an endogenous clock may limit the proliferative capacity of cardiac progenitors.
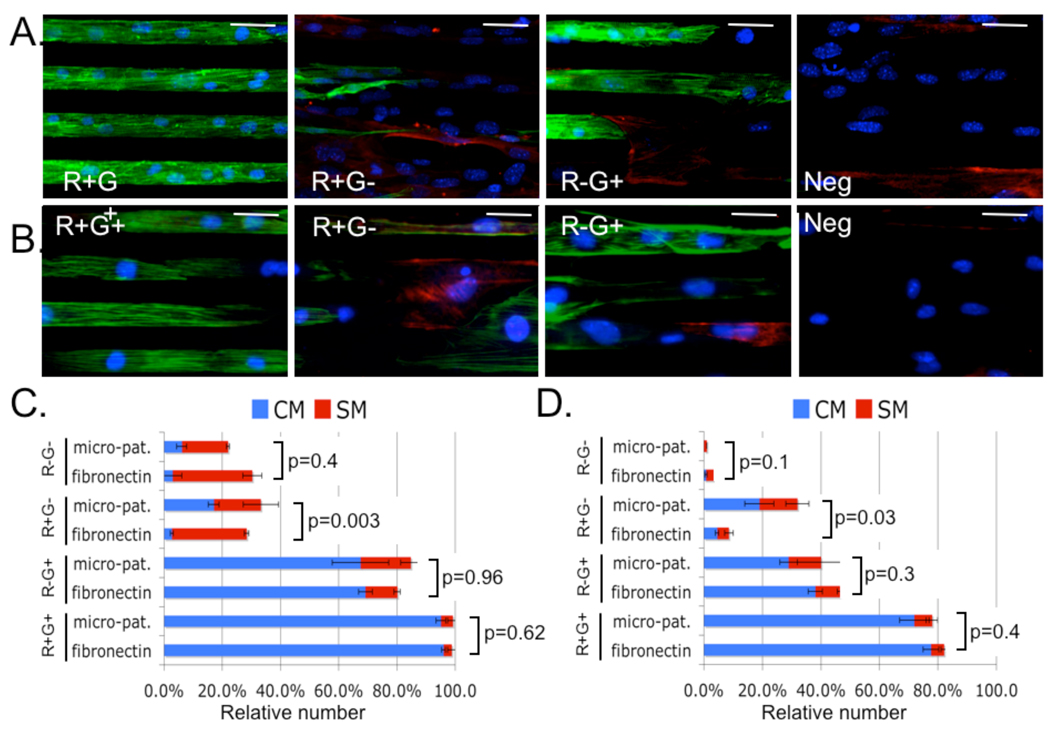
To examine progenitor myogenic potential, we cultured embryonic and ES-derived progenitors on either fibronectin coated slides or micropatterns of 20µm wide lines of fibronectin alternating with 20µm wide lines of Pluronic F127 (a surfactant that blocks cell adhesion). After 5 days of in vitro expansion and differentiation, we performed immunofluorescence staining for sarcomeric α-actinin and smooth muscle Myosin Heavy Chain (sm-MHC). Plating embryonic and ESC-derived R+G+ cells on micropatterned surfaces resulted in anisotropic tissue consisting of longitudinally aligned myocardial fibers (Figure 3A and 3B). In contrast, plating the progenitor populations on un-patterned slides resulted in isotropic unaligned tissue (Figure S9). Cell counting showed that embryonic and ESC derived R+G+ progenitors gave rise to an overwhelming majority of cardiomyocytes independent of surface culture conditions. In contrast, the R+G- and the R-G+ populations gave rise to a more heterogeneous population of both smooth muscle and cardiomyocytes (Figure 3C and 3D). At this point, it remains unclear whether these cells represent homogenous populations of multipotent progenitors or heterogeneous populations of unipotent progenitors. Interestingly, culturing R+G- (but not other) progenitors on micro-patterned surfaces resulted in a statistically significant increase in the proportion of cardiac myocytes suggesting that this population’s myogenic potential may be modulated by micro-environmental geometric cues. Single cell patch clamp experiments demonstrated that R+G+ progenitors differentiated into ventricular cardiac myocytes with typical four-phase action potential (AP), whereas R+G- and R-G+ progenitors differentiated into more heterogeneous cell types (Figures 4A, 4B, S10, and Table S4). Further, R+G+ cardiomyocytes showed sodium channel dependency, consistent with ventricular APs (Figure S11).
Figure 3. Differentiation potential of cardiac progenitors.
Embryonic and ESC-derived cardiac progenitors were cultured on fibronectin coated slides (fibronectin) or micro-patterns for five days.
A&B. Representative immunofluorescence microscopy of Embryonic (A) or ESC-derived (B) progenitors cultured on micropatterns are shown; nuclei (blue), smMHC (red) and sarcomeric α-actinin (green). Scale bar 40µm.
C&D. Cell counting was used to quantify the relative number of CM (sarcomeric α-actinin positive) or SM (smMHC positive) derived from embryonic (C) or ESC (D) progenitors. R+G+ populations resulted in the most CM (p<0.001). No significant differences were observed in SM differentiation (p=0.38-1.0). P-values for the differences in CM differentiation are displayed.
Figure 4. Engineered ventricular tissue from R+G+ progenitors.
A. R+G+ (n=12), R+G- (n=5), and R-G+ (n=5) progenitors were allowed to differentiate and single cell patch clamp recordings were performed. AP morphology was assessed for typical four-phase ventricular action potential.
B. Representative spontaneous AP from R+G+ derived cardiomyocytes.
C. ESC-derived R+G+ progenitors were used to generate MTF as described in Supplementary Materials and (14). Field stimulation (10 V, 10 ms pulse-width) induced 0.5 Hz cyclical contraction and rhythmic MTF bending. See Movie S1.
Given these results we used the R+G+ progenitors to engineer 2-dimensional cardiac tissue into a muscular thin film (MTF) as described in Supplementary Materials and (14). The MTF beat spontaneously at a rate of approximately 20 contractions per minute and could be paced by field stimulation at 0.5 and 1.0 Hz. To measure contractility, the MTF was fixed as a cantilever on one end and the contracting cardiomyocytes bent the MTF towards the cell-side during systole (Figure 4C and supplementary Movie S1). During diastole, the elastic PDMS film provided the antagonistic force that returned the MTF back to the relaxed position. The change in radius of curvature is inversely proportional to cardiomyocyte stress generation and was measured at ~5 kPa for the progenitor-derived cardiac tissue at peak systole (Figure S12), similar to MTFs engineered from neonatal rat ventricular cardiomyocytes (14).
The development of an in vivo multicolor reporter system in embryos and corresponding ES cell lines has now allowed for the purification of distinct subsets of heart field progenitors from the earliest stages of cardiogenesis. While Islet-1 primarily marks the SHF (4, 15–17), there have been no distinct markers for the FHF lineages that contribute to the left ventricle. We here document distinct transcriptional signatures for the FHF and SHF lineages including the expression of unique subsets of microRNAs. The profiles are sufficiently distinct to suggest that they have non-overlapping identities and the identification of independent FHF markers should allow a rigorous analysis of their role in development and disease.
A critical step in cardiogenesis is the formation and expansion of the ventricular myocyte lineage necessary for normal cardiac contractile function. The discovery and purification from embryos and corresponding ESC lines of committed ventricular progenitors (CVPs) uncovers a novel mechanistic pathway for organogenesis through the expansion and assembly of CVPs into fully functional ventricular muscle tissue. Thus, directed differentiation from multipotent islet progenitors to a specific differentiated progeny occurs via the formation of transient committed intermediate progenitors that are destined to become specific cell types. This finding suggests a general paradigm for the conversion of multipotent islet progenitors to other differentiated cell types, such as endothelial or conduction system cells (Figure 4D). Furthermore, recent work has now identified multiple Isl1 intermediate progenitor populations in human embryonic hearts and human ESC (18) suggesting that it may be possible to isolate self-expanding human ventricular progenitors.
Advanced heart failure is a major, unmet clinical need, arising from a loss of viable and/or fully functional cardiac muscle cells (19). Currently, a number of clinical trials have been designed to augment the function of cardiac muscle via cell transplantation. To date, while there have been encouraging early suggestions of a small therapeutic benefit, there has not been evidence for robust regeneration of heart muscle tissue (20, 21) underscoring the need for new approaches. A central challenge for cell-based therapy has been the identification of an optimal cell type to drive robust cardiac myogenesis. The ideal heart progenitor cell would be derived from a renewable cell source in sufficient quantities to drive clinically relevant levels of cardiac myogenesis. In addition, it would be critical to direct the differentiation of progenitor cells into functional ventricular myocytes, instead of related lineages such as smooth muscle cells. In this regard, the discovery of a unique subset of ESC-derived CVPs fulfills many of the above criteria. The ability to generate fully functional ventricular MTF should allow the direct chemical screening of novel molecular entities for therapeutic endpoints that can only be measured on intact muscle tissue, including force development and conduction velocity. With recent advances in the generation of induced pluripotent stem cells (iPS) (22–24), it should now be possible to isolate patient and disease specific cardiac progenitors. The combination of tissue engineering technology with stem cell biology therefore represents a new approach for the development of human models of human disease and as a platform for drug discovery and design.
Supplementary Material
References
- 1.Buckingham M, Meilhac S, Zaffran S. Nat Rev Genet. 2005 Nov;6:826. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 2.Martin-Puig S, Wang Z, Chien KR. Cell Stem Cell. 2008;2:320. doi: 10.1016/j.stem.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Dodou E, Xu SM, Black BL. Mech Dev. 2003 Sep;120:1021. doi: 10.1016/s0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- 4.Qyang Y, et al. Cell Stem Cell. 2007;1:165. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 5.Lien CL, et al. Development. 1999 Jan;126:75. doi: 10.1242/dev.126.1.75. [DOI] [PubMed] [Google Scholar]
- 6.Wu SM, et al. Cell. 2006 Dec 15;127:1137. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 7.Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Development. 2004 Aug;131:3931. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 8.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. Dev Biol. 2005 Nov 1;287:134. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 9.Reich M, et al. Nature Genetics. 2006;38:500. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 10.Bruneau BG, et al. Cell. 2001 Sep 21;106:709. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 11.Mori AD, et al. Dev Biol. 2006 Sep 15;297:566. doi: 10.1016/j.ydbio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 12.Barrallo-Gimeno A, Nieto MA. Development. 2005;132:3151. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 13.Blanco MJ, et al. Development. 2007;134:4073. doi: 10.1242/dev.006858. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg AW, et al. Science. 2007 Sep 7;317:1366. doi: 10.1126/science.1146885. [DOI] [PubMed] [Google Scholar]
- 15.Laugwitz KL, et al. Nature. 2005 Feb 10;433:647. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moretti A, et al. Cell. 2006 Dec 15;127:1151. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 17.Cai CL, et al. Dev Cell. 2003 Dec;5:877. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bu L, et al. Nature. 2009 Jul 2;460:113. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- 19.Jessup M, Brozena S. N Engl J Med. 2003 May 15;348:2007. doi: 10.1056/NEJMra021498. [DOI] [PubMed] [Google Scholar]
- 20.Menasche P. Semin Thorac Cardiovasc Surg. 2008 Summer;20:131. doi: 10.1053/j.semtcvs.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Pouly J, et al. J Thorac Cardiovasc Surg. 2008 Mar;135:673. doi: 10.1016/j.jtcvs.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K, et al. Cell. 2007 Nov 30;131:861. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Yu J, et al. Science. 2007 Dec 21;318:1917. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 24.Hanna J, et al. Cell. 2008 Apr 18;133:250. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.We thank L. Prickett and K. Folz-Donahue for expert help with flow cytometry. Financial support was provided by the de Gunzburg Family Foundation to I.J.D.; the NIH Fellowship support T32 HL002807 to I.J.D. and M.C.; The Netherlands Organization for Scientific Research Rubicon 825-07-011 to P.v.d.M. and the and the Leducq Foundation to K.R.C.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.