Abstract
The voltage dependent anion channel (VDAC) forms a channel for metabolites and nutrients in the outer membrane of mitochondria, and it is also involved in apoptotic pathways. Here, we report sequence-specific NMR assignments for the isoform 1 of human VDAC reconstituted in lauryldimethylamine oxide (LDAO) detergent micelles. The assignments were deposited in the BMRB data base with accession number 16381.
Keywords: Voltage dependent anion channel, Voltage gating, β-Barrel, Protein refolding, Membrane protein structure, Multi-dimensional decomposition (MDD), TROSY, Selective labeling, 4D-NOESYs, Non-uniform sampling (NUS)
Biological context
The eukaryotic integral membrane protein VDAC is the most abundant protein of the outer mitochondrial membrane (Colombini 2004). In humans, VDAC has three highly homologous isoforms. We have recently determined the three-dimensional structure of VDAC-1 reconstituted in LDAO micelles by solution NMR (Hiller et al. 2008). The structure of human VDAC-1 in detergent micelles has also been determined by a hybrid NMR/X-ray approach (Bayrhuber et al. 2008), and the X-ray structure of mouse VDAC-1 was determined in a bicelle environment (Ujwal et al. 2008). VDAC forms a large, 19-stranded β-barrel, providing a pore for metabolite diffusion across the outer mitochondrial membrane.
Still, many open questions remain about the functions and interactions of VDAC. Electrophysiological experiments show that at low membrane potentials VDAC is in an open state, but it switches to the closed state at membrane potentials of about 30 mV (Colombini 1979). However, the mechanistic details of this voltage gating process are still unclear. Additional interest in this protein comes from its involvement in mitochondrial apoptotic pathways (Kroemer et al. 2007; Malia and Wagner 2007). VDAC can interact with anti-apoptotic proteins from the Bcl-2 family, prohibiting the opening of the mitochondrial exit channel that leads to cell death (Shimizu et al. 1999). The isoform VDAC-1 interacts directly with the antiapoptotic protein Bcl-xL, whereas the isoform VDAC-2 interacts with Bak (Shimizu et al. 1999; Malia and Wagner 2007). Other interaction partners of VDAC include the proteins hexokinase and the metabolites NADH and cholesterol (Colombini 2004; Hiller et al. 2008).
Methods and experiments
Expression, purification and refolding
VDAC-1 with a C-terminal His-tag (VDAC-1(1–283)-Leu-Glu-His6) was expressed in BL21(DE3) cells using M9 minimal medium and induction with 1 mM IPTG at 37°C for 4 h. After cell lysis by sonication, the VDAC-1 containing inclusion bodies were isolated and purified under denaturing conditions in 8 M urea on a Ni-agarose column. Purified VDAC-1 was precipitated and redissolved in 6 M GuHCl solution. Subsequently, it was refolded at 4°C by dropwise dilution into refolding buffer containing 1% (43 mM) lauryldimethylamine oxide (LDAO). Cation exchange chromatography was used to further purify the sample and to isolate well-folded protein. The final sample conditions were 25 mM Na·PO4, 5 mM DTT, pH 6.8, 300–500 mM LDAO, 0.5–1 mM VDAC-1.
Isotope labeling schemes
The following samples were used for the backbone assignment: [U-99%-2H, 13C, 15N]-VDAC-1 in [U-99%-2H]-LDAO, [U-99%-2H,13C,15N; 99%-1Hδ-IL; 99%-1Hγ-V]-VDAC-1 in [U-99%-2H]-LDAO (Goto et al. 1999), [U-99%-2H, 15N; 99%-1Hδ,13Cδ-IL; 99%-1Hγ,13Cγ-V]-VDAC-1 in [U-99%-2H]-LDAO (Goto et al. 1999), [U-99%-2H; 99%- 15N-Xxx]-VDAC-1 in LDAO, where Xxx = Ala, Asn, Asp, Cys, Met, Gly, Ile, Leu, Lys, Phe, Thr, Tyr, Val (13 samples). [U-99%-2H, 15N; 99%-13C–Yyy]-VDAC-1, with Yyy = Ala, Ile, Leu, Lys, Phe, Tyr, Val, Pro (8 samples). [U-99%-2H]-LDAO was purchased from FB Reagents, Cambridge, MA (www.fbreagents.com). All other isotopes were purchased from Cambridge Isotopes, Andover, MA. For the preparation of the two methyl-labeled samples, the 13C-labelled precursors α-ketoisovalerate and α-ketobutyrate were purchased in fully protonated form. For the substitution of 2H for 1H at the 3 positions an in-house protocol was used, which as a side reaction affected also methyl protons of α-ketoisovalerate.
NMR chemical shift assignments
All experiments were carried out at 30°C on Bruker and Varian spectrometers equipped with cryogenic probes, operating at field strengths of 600, 750 and 900 MHz. The following experiments were recorded: 2D [15N,1H]-TROSY (Pervushin et al. 1997), 3D TROSY-HNCA, 3D TROSY-HNCO, 3D TROSY-HNCOCA, 3D TROSY-HNCACB, 3D ct-TROSY-HNCA, 3D [1H,1H]-NOESY-15N-TROSY, 3D [1H,1H]-NOESY-13C-HMQC, 2D [13C,1H]-HMQC, 3D HMCM[CG]CBCA-COSY (Tugarinov and Kay 2003), 4D NUS-Co-MDD-13C-HMQC-[1H,1H]-NOESY-13C-HMQC, 4D NUS-Co-MDD-15N-HMQC-[1H,1H]-NOESY-13CHMQC (Hiller et al. 2009). The Co-MDD spectra were processed simultaneously, using the mddNMR software. All other spectra were processed with prosa. All data was analyzed using cara and xeasy.
Assignments and data deposition
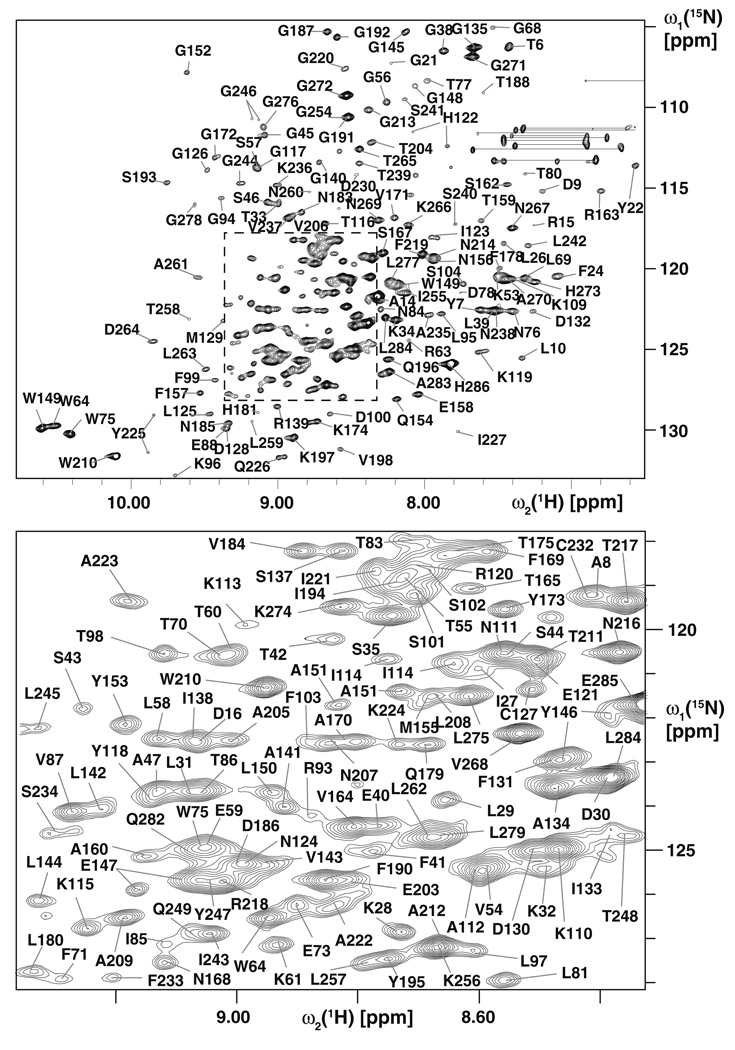
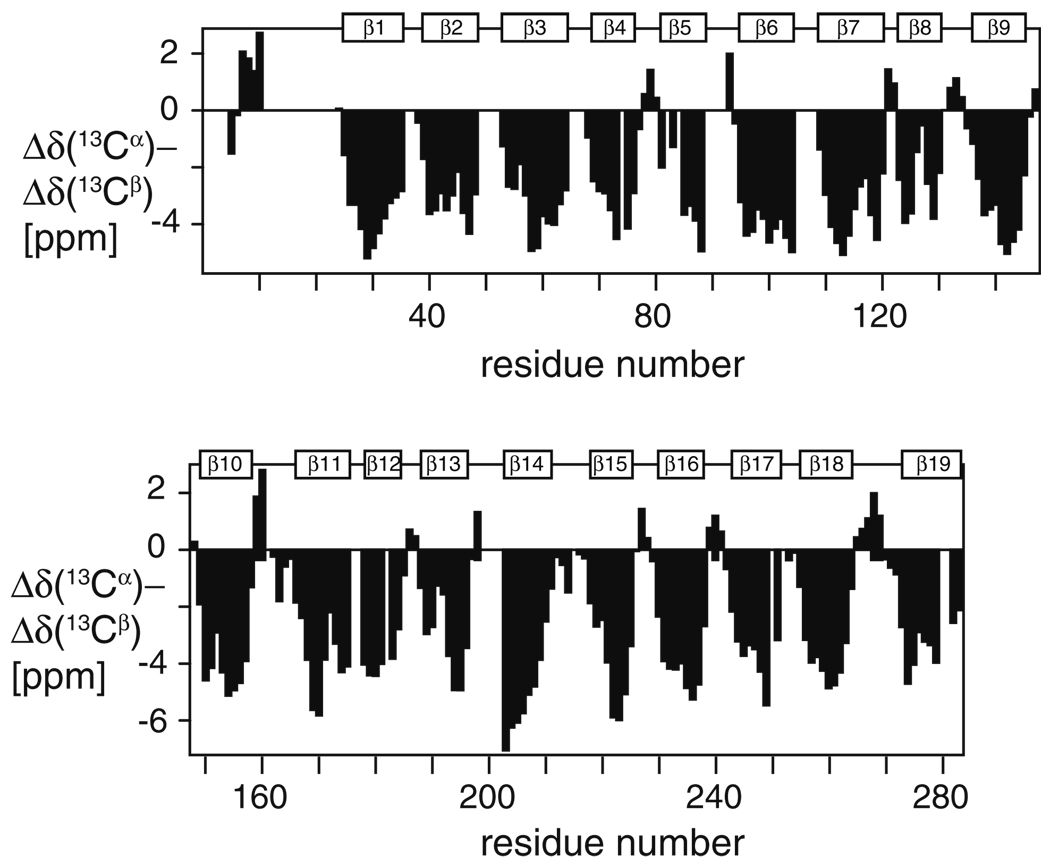
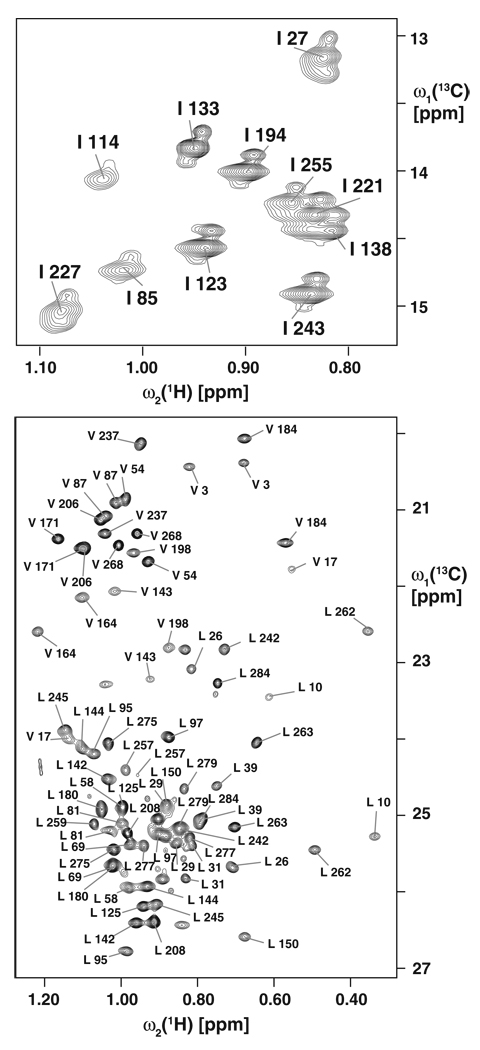
VDAC-1 consists of 283 residues. The protein exhibits a well-dispersed 2D [15N,1H]-TROSY spectrum (Fig. 1). High-field triple-resonance TROSY-type experiments and selectively labeled samples enabled sequence-specific resonance assignment of the protein backbone resonances HN, N, Cα and Cβ of 82% of the residues. Nineteen β-strands are formed within residues 25–283 and a short α-helix is located at the N-terminus containing residues 6–10, as indicated by Cα and Cβ secondary chemical shifts (Fig. 2). The missing assignments can be mostly rationalized by intermediate time scale dynamics, since they are mainly located in loops and in the polypeptide segment 11–25, which forms a dynamic linker. In addition to the backbone, 96% of the methyl groups of isoleucine, leucine and valine residues could be assigned using the HMCM[CG]CBCA-COSY experiment (Tugarinov and Kay 2003), and the 4D NUS-Co-MDD NOESYs (Hiller et al. 2009) (Fig. 3). These side-chain assignments were essential for defining the overall structure of the protein and the location of the N-terminal helix with respect to the β-barrel. Furthermore, they provide valuable sensors for the detection of protein–protein interactions on the hydrophobic face of the channel.
Fig. 1.
Amide resonance assignments of VDAC-1. A 2D [15N,1H]-TROSY spectrum of [U-2H,15N]-VDAC-1 in [U-99%-2H]-LDAO micelles is shown, collected on a Bruker 900 MHz spectrometer at 30°C. Sequence specific resonance assignments are indicated. Pairs of glutamine and asparagine side chain resonances are connected by horizontal lines. The spectral region in dashed lines in the top panel is shown enlarged in the bottom panel. Several residues exhibit two cross peaks, indicating the presence of two conformations, such as for the loop K119–I123
Fig. 2.
Secondary chemical shifts of VDAC-1 in LDAO micelles. Deviations from random coil chemical shifts are plotted versus the amino acid sequence, after multiplication with a 1:2:1 weighting function for residues i − 1: i: i + 1. The location of the β-strands is indicated by white boxes β1–β19. The secondary chemical shifts were calculated relative to average values from the BMRB data base
Fig. 3.
Isoleucine, leucine and valine methyl group resonance assignments of VDAC-1. A 2D [13C,1H]-TROSY spectrum of [U-99%-2H,13C,15N; 99%-1Hδ-IL; 99%-1Hγ-V]-VDAC-1 in [U-99%-2H]-LDAO micelles is shown, collected on a Bruker 900 MHz spectrometer at 30°C. Sequence-specific resonance assignments are indicated. The observation of three peaks per isoleucine methyl group arises from the unintentional use of an α-ketobutyrate precursor with mixed CH3, CH2D, CD2H and CD3 labeling (see “Methods and experiments”)
The chemical shifts were referenced relative to DSS for 1H and indirectly for 13C and 15N following the IUPAC conventions. The sequence-specific resonance assignments of the H–N TROSY components and the C–H HMQC components were deposited in the BMRB data base with accession number 16381. The atomic coordinates of VDAC-1 in LDAO micelles have been deposited at the PDB data base with accession code 2k4t.
Acknowledgments
This work was supported by the United States National Institute of Health by the roadmap grant GM075879 and the grants GM066360, GM47467 and EB 002026 for purchase, operation and maintenance of the instruments. S.H. was supported in part by the Swiss National Science Foundation.
References
- Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K. Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombini M. A candidate for the permeability pathway of the outer mitochondrial membrane. Nature. 1979;279:643–645. doi: 10.1038/279643a0. [DOI] [PubMed] [Google Scholar]
- Colombini M. VDAC: The channel at the interface between mitochondria and the cytosol. Mol Cell Biochem. 2004;256:107–115. doi: 10.1023/b:mcbi.0000009862.17396.8d. [DOI] [PubMed] [Google Scholar]
- Goto NK, Gardner KH, Mueller GA, Willis RC, Kay LE. A robust and cost-effective method for the production of Val, Leu, Ile (δ1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol NMR. 1999;13:369–374. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller S, Ibraghimov I, Wagner G, Orekhov VY. Coupled decomposition of four-dimensional NOESY spectra. J Am Chem Soc. 2009;131:12970–12978. doi: 10.1021/ja902012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- Malia TJ, Wagner G. NMR structural investigation of the mitochondrial outer membrane protein VDAC and its interaction with antiapoptotic Bcl-xL. Biochemistry. 2007;46:514–525. doi: 10.1021/bi061577h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Narita M, Tsujimoto Y. Bcl-2 family proteins regulate the release of apoptogenic cytochrome c by the mitochondrial channel VDAC. Nature. 1999;399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Kay LE. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- Ujwal R, Cascio D, Colletier JP, Faham S, Zhang J, Toro L, Ping P, Abramson J. The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci USA. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]