To the Editor: A recent paper in Nature Methods describes the use of fluorescence-activated cell sorting (FACS) is to sort Caenorhabditis elegans embryos1. Here we report FACS-based method to sort live C. elegans larvae, which permits us to rapidly collect large quantities of live genotyped worms from a mixed population. Using GFP-marked balancer chromosomes (Fig. 1a), and live-animal FACS laFACS, we routinely collected >100,000 genotyped animals in less than one hour. To test laFACS, we combined it with large-scale RNA interference (RNAi) screening and identified genetic interactors of mel-28, an important regulator of nuclear envelope and chromatin functions2–5.
Figure 1.
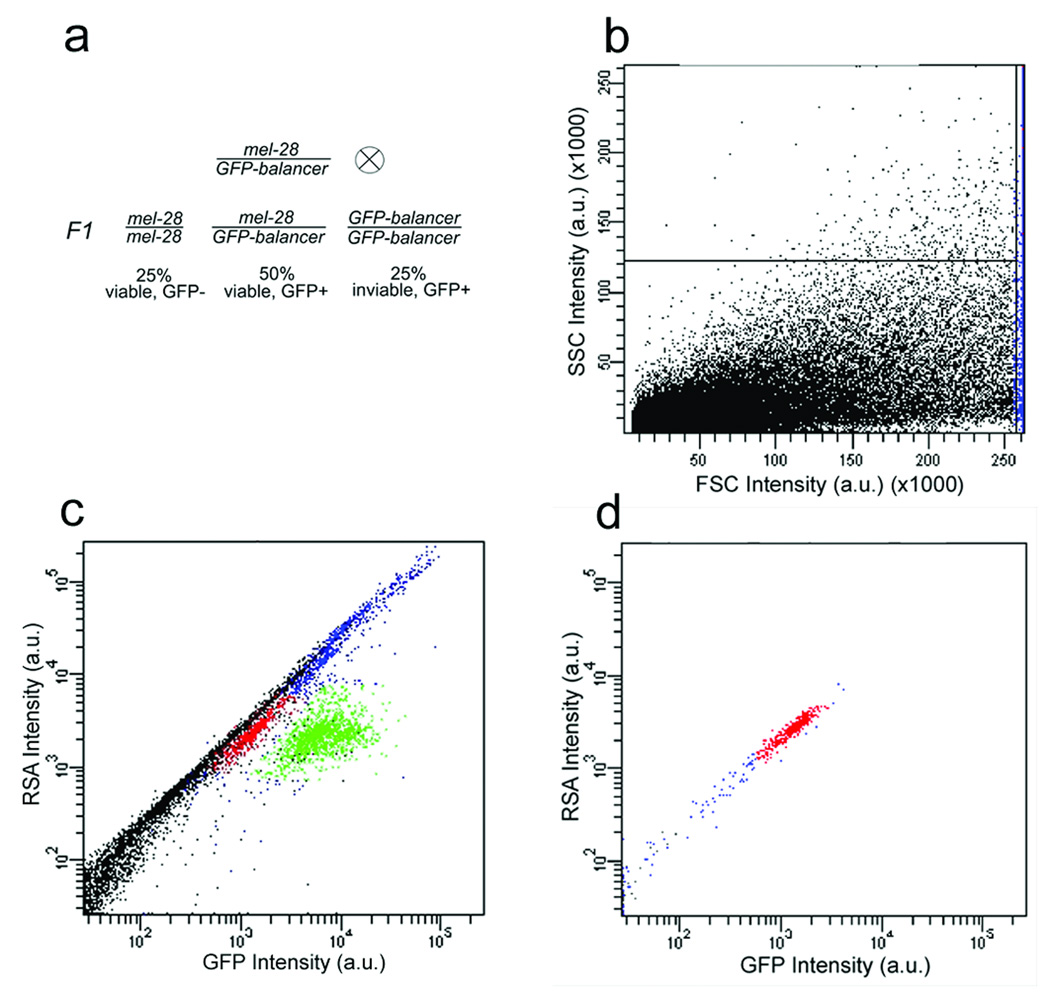
laFACS of GFP-negative L1 larvae. (a) Genetic scheme. A mel-28 mutant allele is kept over a balancer chromosome containing a GFP marker and a recessive lethal allele. GFP-negative progeny (F1) are mel-28 homozygotes and grow up to produce only dead embryos. (b) A dot-plot of forward scatter (FSC) versus side scatter (SSC) signals. The gate on FSC separates the larvae (blue) from debris (black). (c) A dot-plot of red-spectrum autofluorescence (RSA) versus GFP signals. GFP-positive mel-28 heterozygotes (green) are defined and used to set a gate for the GFP-negative larvae (red) to be sorted. (d) A dot-plot of the RSA versus GFP signals post-sort.
Although a FACS machine is designed to sort single cells, a few modifications enabled sorting of ~0.25 mm long L1 C. elegans. First, we used a reduced drop-drive frequency of ~16.4 kHz to keep the larvae undamaged. We also used a 100-µm nozzle and set a gate to capture events with a high forward-scatter signal (Fig. 1b), confining our collections to larger objects (Supplementary Methods). After worm sorting, the same FACS machine could be used for several other applications, including sorting of yeast, Drosophila melanogaster cells, mammalian cells, and plant protoplasts. The worm sort had no impact on these applications, and the worm-specific FACS modifications were easily reversed to accommodate single-cell applications.
We used laFACS to collect mel-28 (maternal-effect-lethal-28) homozygous worms from a mixed population. mel-28 homozygous hermaphrodites derived from heterozygous mothers appear phenotypically indistinguishable from wild-type animals until they mature and produce only inviable progeny. This selection is usually performed manually and thus is not amenable to large-scale applications. We first generated a GFP-marked strain in which the mel-28(t1684) mutation is balanced over a chromosome bearing lag-2::GFP6 (Fig. 1a). We sorted L1 worms, collecting GFP-negative mel-28(t1684) homozygotes. We selected GFP-negative larvae on the basis of the ratio of green (GFP fluorescence; 530/30 nm) to red (red-spectrum autofluorescence; 610/20 nm) signal (Fig. 1c,d). We separated GFP-positive and GFP-negative worms (Supplementary Figure 1). Typically, from a population of 600,000 larvae we retrieved ~130,000 healthy animals after one sort. This first-pass population was ~95-98% homozygous (n > 1,000, verified by microscopy). A second sort recovered an essentially 100% pure population of about 100,000 homozygous larvae (n > 20,000, verified by microscopy and by genetic analysis). This scheme can be easily adapted for the majority of C. elegans genes using available balancers.
We used the collected homozygous mel-28 animals to perform an RNAi-based synthetic interaction screen using clones representing chromosome I genes7. The ability to easily collect large amounts of mel-28 homozygous animals allowed us to perform the RNAi repeatedly (up to 16 times). From over 2,000 genes tested, 12 showed synthetic phenotypes with mel-28: npp-2, npp-4, npp-12, npp-14, npp-17, his-67, his-68, exos-3, pas-5, phi-56, rpa-0, and rpl-30 (Supplementary Fig. 2 and Supplementary Table 1). These results agreed well with mel-28’s role in coordinating chromatin and nuclear envelope functions2,3. Obtaining large quantities of pure mutant populations could also be useful for chemical screens, microarrays, or biochemical assays, expanding the arsenal of high-throughput tools available in C. elegans.
Supplementary Material
Acknowledgments
This work was partly funded by the NICHD (R01HD046236) and NHGRI (U01 HG004276) to FP, the NIH (R01GM078279-01) to KDB, and by the NSF (0827858) to AGF. Nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
References Cited
- 1.Stoeckius M, et al. Nat Methods. 2009 [Google Scholar]
- 2.Fernandez AG, Piano F. Curr Biol. 2006;16:1757–1763. doi: 10.1016/j.cub.2006.07.071. [DOI] [PubMed] [Google Scholar]
- 3.Galy V, Askjaer P, Franz C, Lopez-Iglesias C, Mattaj IW. Curr Biol. 2006;16:1748–1756. doi: 10.1016/j.cub.2006.06.067. [DOI] [PubMed] [Google Scholar]
- 4.Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. Proc Natl Acad Sci U S A. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franz C, et al. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgley MK, Baillie DL, Riddle DL, Rose AM. WormBook. (The C. elegans research community, 2006) [Google Scholar]
- 7.Kamath RS, et al. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.