Abstract
Runx genes encode a family of proteins defined by the highly conserved Runt DNA-binding domain. Studies in several organisms have shown that these transcription factors regulate multiple aspects of embryonic development and are responsible for the pathogenesis of several human diseases. Here we report the cloning and expression of Runx3 during Xenopus development and compare its expression pattern to other Runx family members, Runx1 and Runx2, and to Cbfβ, the obligatory binding-partner of Runx proteins. Using in situ hybridization in the whole embryo and on sections we show that Runx3 is co-expressed with Runx1 in the hematopoietic lineage and in Rohon-Beard sensory neurons. In contrast Runx3 and Runx2 are co-expressed in craniofacial cartilage elements. Runx3 shows also unique expression domains in a number of derivatives of the neurogenic placodes, including the ganglia of the anteroposterior and middle lateral line nerves, and ganglia of the trigeminal, glossopharyngeal, facial and vagal nerves. These observations suggest a critical role for Runx3 in the development of cranial sensory neurons, while in other tissues its co-expression with Runx1 or Runx2 may signify functional redundancy between these family members.
Keywords: Xenopus, Runx1, Runx2, Runx3, Cbfβ, Islet1, Pax3, Placode, Cartilage, Craniofacial, Rohon-Beard neuron, Cranial nerve, Trigeminal, Profundal, Vagal, Glossopharyngeal, Lateral line
1. Results and Discussion
Runx3, also known as Aml2/Cbfα3/Pebp2αC, belongs to the Runx family of transcription factors, which include Runx1 (Aml1/Cbfα2/Pebp2αB) and Runx2 (Aml3/Cbfα1/Pebp2αA). This class of molecules is defined by the presence of the runt domain, a highly conserved DNA binding and protein-protein interaction domain. Runx proteins bind DNA in association with the non-DNA-binding partner, Cbfβ, that confers high-affinity DNA binding and stability to the complex. This heterodimeric complex has the ability to activate or repress transcription of key regulators of cell growth, proliferation, survival and differentiation (reviewed in Blyth et al., 2005; Ito, 2008).
The function of the three mammalian Runx genes has been studied using gene-targeting technology in the mouse. Runx1 is primarily required for hematopoiesis and is essential for the generation of hematopoietic stem cells (Okuda et al., 1996; Wang et al., 1996; North et al., 1999). RUNX1 is a frequent target of gene rearrangements and mutations in a spectrum of human leukemias including acute myeloid leukemia (Speck and Gilliland, 2002). Runx2 is required for bone development and is associated with human cleidocranial dysplasia, a skeletal disorder characterized by bone and dental abnormalities (Komori et al., 1997; Otto et al., 1997; Mundlos et al., 1997). Runx3 is expressed in a broader range of tissues compared to the other two Runx genes, and has been implicated in development of axonal projections of a subpopulation of neurons in the dorsal root ganglia (Levanon et al., 2002; Inoue et al., 2002) and in the formation of the gastrointestinal tract (Li et al., 2002). There is also evidence that Runx3 functions as a tumor suppressor in gastric cancers (Li et al., 2002; Brenner et al., 2004).
The expression and function of Runx genes has been relatively well conserved during evolution. For example, in zebrafish and Xenopus Runx1 is expressed in blood progenitors, and controls hematopoietic stem cell specification (Tracey et al., 1998; Kalev-Zylinska et al., 2002; Burns et al., 2005). Runx2 in fish and frogs is detected in developing skeletal elements where it regulates chondrogenesis (Flores et al., 2004; 2006; Kerney et al., 2007). Zebrafish runx3 is expressed in hematopoietic, neuronal and cartilaginous tissues and its function has been studied in the context of hematopoiesis (Kalev-Zylinska et al., 2003) and chondrocyte differentiation (Flores et al., 2006). Here we report the cloning of Xenopus Runx3 and provide a comprehensive analysis of Runx3 expression during Xenopus embryogenesis, comparing its expression pattern to that of Runx1, Runx2 and Cbfβ.
1.1. Cloning of Xenopus laevis Runx3
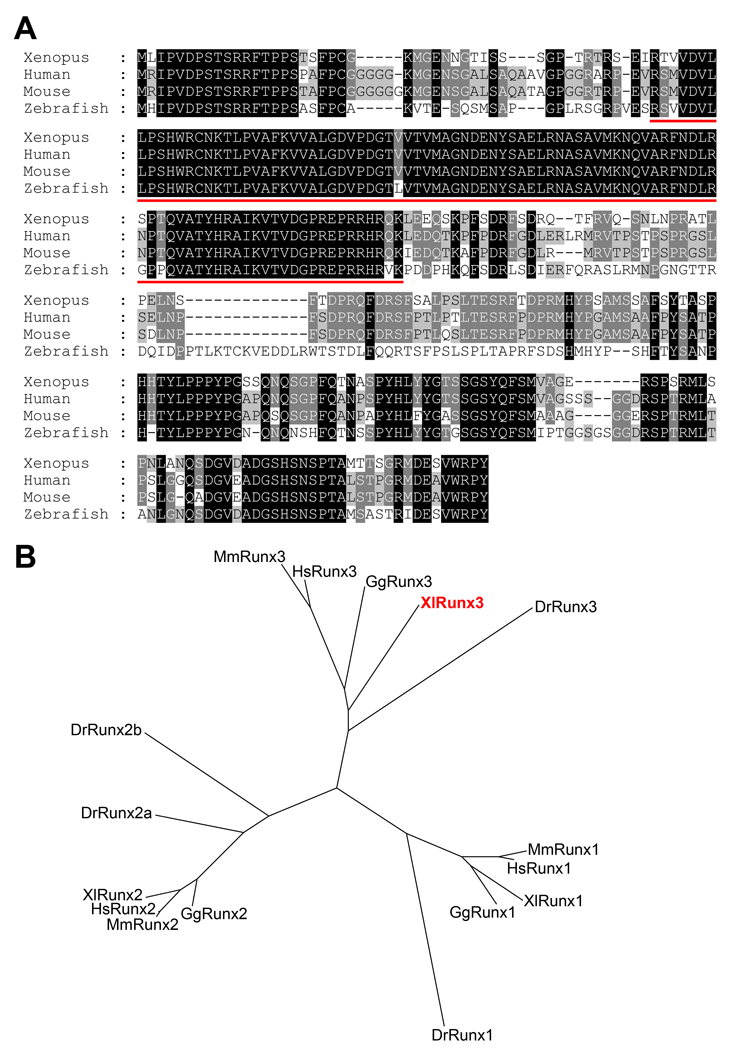
A PCR product corresponding to a partial sequence of the Xenopus laevis Runx3 open reading frame was amplified from stage 30 embryonic cDNA using primers designed based on Xenopus tropicalis Runx3 sequence. The complete coding region of Xenopus laevis Runx3 was subsequently established by two rounds of PCR using stage 41 embryonic cDNA and based on the sequence of Xenopus tropicalis Runx3. Xenopus laevis Runx3 possesses an open reading frame encoding 393 amino acids (Fig. 1A). At the amino acid level, Xenopus laevis Runx3 shares 76% identity with human RUNX3 (Levanon et al., 1994), 74% identity with mouse Runx3 (Wijmenga et al., 1995) and 65% identity with zebrafish runx3 (Kataoka et al., 2000; Burns et al., 2002). When compared to Xenopus laevis Runx1/Xaml1 (Tracey et al., 1998) and Xenopus laevis Runx2 (Kerney et al., 2007), the overall amino acid identity drops to 55% and 56%, respectively. Assignment of Xenopus laevis Runx3 sequence to the Runx family was based on phylogenetic tree analysis of the predicted amino acid sequences compared to that of selected vertebrate species (Fig 1B). This analysis indicates that Xenopus laevis Runx3 represents an ortholog of mammalian Runx3.
Figure 1. Sequence and structure comparison of Runx3 proteins across species.
(A) The predicted amino acid sequences from Xenopus laevis, human, mouse and zebrafish Runx3 were aligned using ClustalW. Conserved amino acids in all four species or in at least two species are highlighted in black and grey, respectively. The Runt domain, signature motif of this class of molecules, is underlined in red. (B) Phylogenetic tree analysis of Runx proteins from Xenopus laevis (Xl), human (Hs), mouse (Mm), chicken (Gg) and zebrafish (Dr). Accession numbers for the source sequences are indicated in materials and methods.
1.2. Temporal expression of Runx3 and other Runx family members
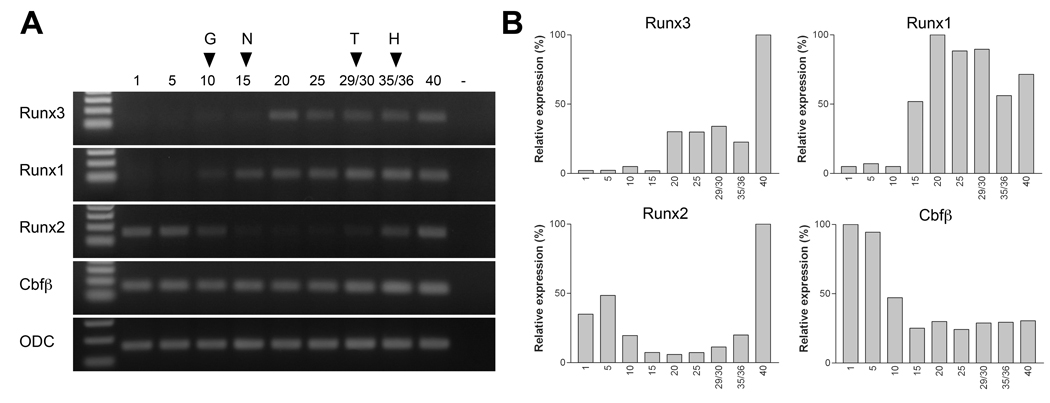
To determine the temporal expression of Runx3 we performed RT-PCR of embryonic RNA at different stages of development. We also compared Runx3 expression profile to that of Runx1, Runx2 and Cbfβ (Fig 2A). These results were independently confirmed by real-time RT-PCR (Fig 2B). Runx3 is first detected around stage 20, while Runx1 transcription is initiated before stage 10, consistent with previous work (Tracey et al., 1998). Runx1 and Runx3 transcripts persist at least up to stage 40, the latest stage analyzed in this study. Runx2 and Cbfβ are both maternally expressed, and while Cbfβ is expressed at all stages examined, Runx2 exhibits a dramatic down regulation between stage 10 (gastrula) and stage 29/30 (tailbud). The biphasic temporal expression profile of Runx2 has not been previously described in Xenopus (Kerney et al., 2007), presumably because we have analyzed a broader range of embryonic stages here than in previous work. In zebrafish runx2b type2 follow a very similar expression profile, with the maternal component detected in the early zygote, followed by a reduction in expression levels by 6 hpf, and a phase of upregulation around 18 hpf, corresponding to the zygotic expression (Flores et al., 2008).
Figure 2. Temporal expression of Runx3, Runx1, Runx2 and Cbfβ during embryogenesis.
(A) RT-PCR analysis of the developmental expression of Runx3, Runx1, Runx2 and Cbfβ. Stages are according to Nieuwkoop and Faber (1967). Ornithine decarboxylase (ODC) is shown as a loading control. G, onset of gastrulation; N, mid-neurula stage; T, tailbud stage; H, hatching stage. (B) Real-time RT-PCR analysis of Runx3, Runx1, Runx2 and Cbfβ. Each value has been normalized to the level of ODC.
1.3. Comparative analysis of Runx3, Runx1, Runx2 and Cbfβ expression
1.3.1 Whole-mount in situ hybridization analysis
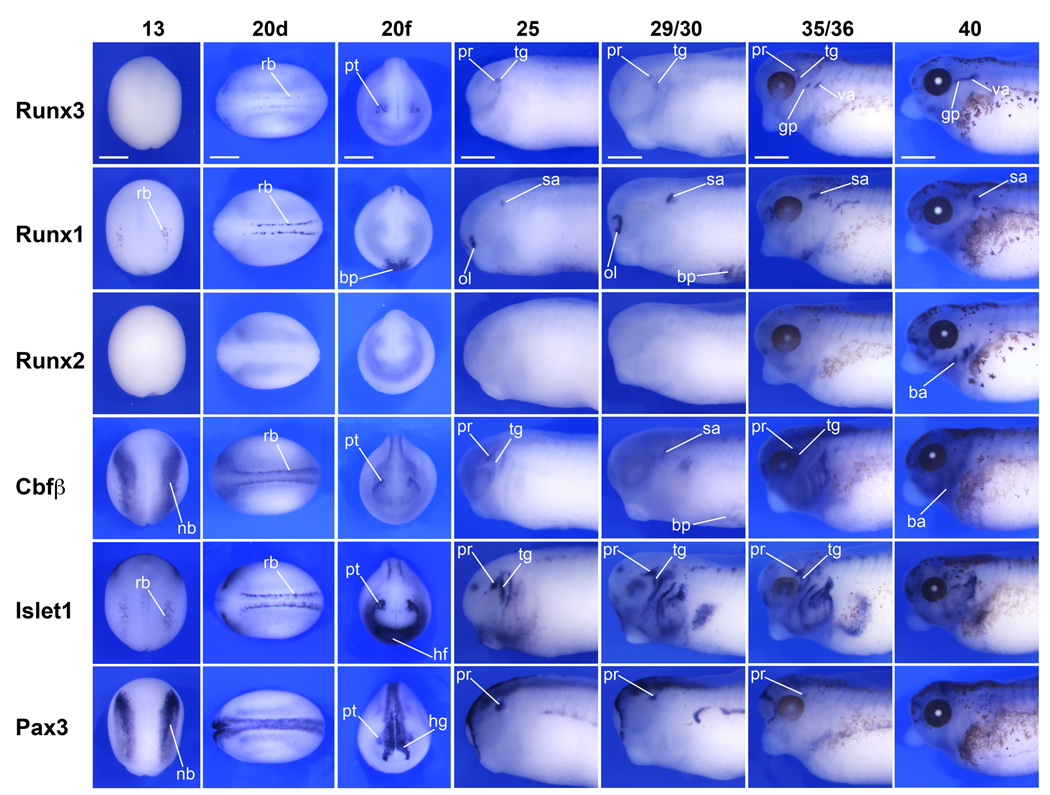
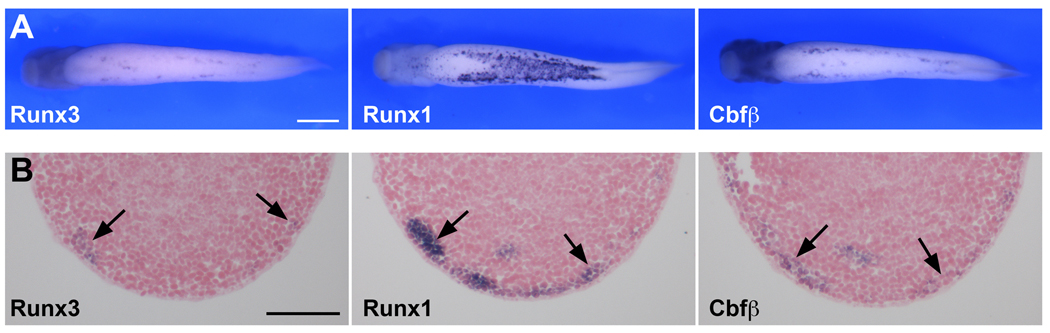
To determine the expression pattern of Runx3, we performed whole-mount in situ hybridization on embryos at different stages of development (Fig 3; Fig 4A). Antisense and control sense probes were analyzed to established specificity of the signal for each gene (control sense hybridization, not shown). Runx3 transcripts are first detected at stage 20 in two domains, the developing primary sensory (Rohon-Beard) neurons in the dorsal spinal cord and the profundal-trigeminal placode, which appear as bilateral clusters of cells posterior to the eye primordia (Fig 3). At the same stage Runx1 is also expressed in Rohon-Beard sensory neurons, as well as in the ventrally located blood progenitors (Fig 3). Runx1 can be detected in both of these domains as early as stage 13 (Fig 3 and not shown), as previously reported (Tracey et al., 1998). Runx3 is also detected in the hematopoietic progenitors but not until stage 31, in a pattern that overlaps with Runx1 and Cbfβ at least in the lateral most aspect of the lateral plate mesoderm (Fig 4). The apparent difference in the expression of these genes in the hematopoietic lineage may reflect differences in the expression level of these molecules, Runx1 being more strongly expressed than Runx3 and Cbfβ at the ventral midline (Fig 4). The obligatory partner of all Runx proteins, Cbfβ, is detected in the same three domains as Runx3 and Runx1 (Fig 3 and Fig 4). Cbfβ expression in blood progenitors is first detected at stage 22 (not shown) several hours after Runx1 onset of expression in this lineage (Fig 3). Cbfβ expression at stage 13 is expressed at the neural plate border in a pattern reminiscent to that of Pax3, and more broadly than the expression domain of Runx1 in Rohon-Beard sensory neurons. At later stages, stage 25 and up, Runx3 expression appears to be largely confined to the head region and in a number of cranial nerves including the ganglia of the trigeminal, vagal and glossopharyngeal nerves, where it overlaps with Cbfβ (Fig 3). Runx2 is first detected around stage 35/36 in the developing craniofacial structures as previously described (Kerney et al., 2007). To confirm the tissue-specific expression analysis of the Runx genes, we also performed in situ hybridization for Pax3 and Islet1 in staged matched embryos (Fig 3). These two genes share common expression domains with Runx genes in the profundal-trigeminal placode (Pax3 and Islet1), Rohon-Beard sensory neurons and cranial ganglia (Islet1). In zebrafish Runx3 is expressed in the same four domains: hematopoietic lineage, trigeminal ganglia, Rohon-Beard neurons and cartilaginous tissues in the head (Kalev-Zylinska et al., 2003). While Runx3 has been reported to be expressed in the mouse gastrointestinal tract (Li et al., 2002), and in zebrafish intestinal bulb (Kalev-Zylinska et al., 2003), we could not detect Runx3 expression in the developing gut at the stages examined, though we cannot exclude that it is expressed in this lineage later in development.
Figure 3. Comparative analysis of the expression of Runx and Cbfβ genes by whole-mount in situ hybridization.
Runx3 is first detected at stage 20 in two domains, the developing primary sensory (Rohon-Beard) neurons and the trigeminal-profundal placode. At the same stage Cbfβ is also co-expressed in both domains, while Runx1 is restricted to Rohon-Beard neurons. At later stages, stage 25 and up, Runx3 expression appears to be largely confined to the head region and in a number of cranial nerves including the ganglion of the trigeminal, vagal and glossopharyngeal nerves. To confirm the tissue-specific expression of Runx genes, we also performed in situ hybridization for Pax3 and Islet1 in staged matched embryos. Pax3 and Islet1 share common expression domains with Runx genes in the profundal-trigeminal placode (Pax3 and Islet1), Rohon-Beard sensory neurons and cranial ganglia (Islet1). Stages are according to Nieuwkoop and Faber (1967). Stage 13, dorsal view anterior to top. Stage 20d, dorsal view anterior to the left. Stage 20f, frontal view dorsal to top. Stage 25–40, lateral view of the head region, anterior to the left and dorsal to top. ba, branchial arches region; bp, blood progenitors; hf, heart field; hg, hatching gland; gp, glossopharyngeal; nb, neural plate border; ol, olfactory placode; pr, profundal ganglia; rb, Rohon-Beard neurons; sa, statoaccoustic ganglia; tg, trigeminal ganglia; pt, profundal-trigeminal placode; va, vagal nerve. The scale bars in the upper row represent 500 µm.
Figure 4. Comparative analysis of Runx3, Runx1 and Cbfβ in blood progenitors.
(A) Whole-mount in situ hybridization of stage 31 embryos viewed from the ventral side, anterior to the left, highlights the expression of Runx3, Runx1 and Cbfβ in blood progenitors. In these cells Runx1 appears to be expressed at higher level than Runx3 and Cbfβ. The scale bar represents 500 µm. (B) In situ hybridization on adjacent sections of a stage 31 embryo shows that Runx3, Runx1 and Cbfβ expression domain overlap at least in the lateral most aspect of the lateral plate mesoderm (arrows). The scale bar represents 100 µm
Next we undertook a detailed analysis of Runx3 expression using in situ hybridization on sections at stage 20 and 40, focusing on the early expression of Runx3 in the profundal-trigeminal placode and Rohon-Beard sensory neurons (Fig 5), and its later expression in the branchial arches (Fig 6) and derivatives of the neurogenic placodes (Fig 7).
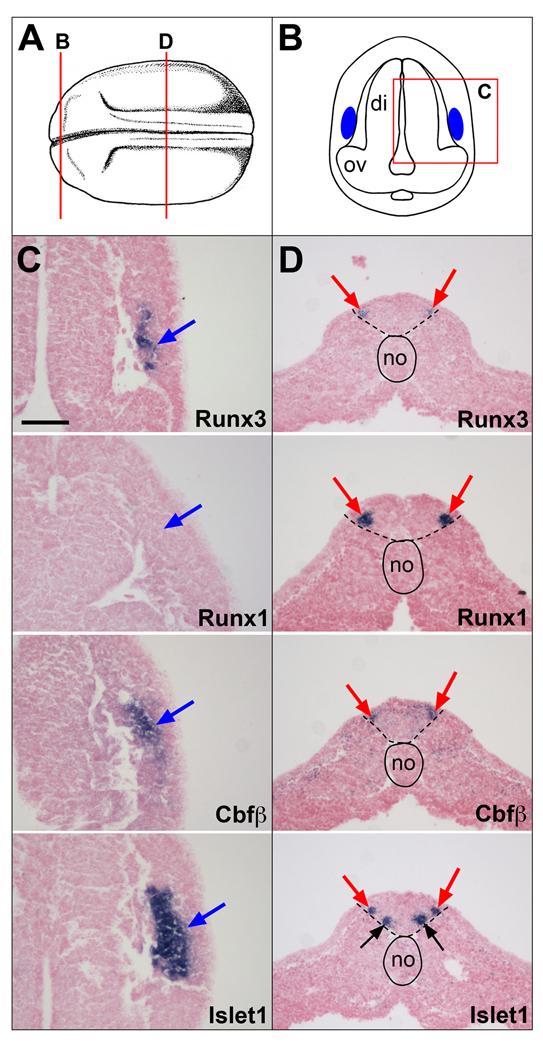
Figure 5. Comparative analysis of Runx3, Runx1, Cbfβ and Islet1 expression in the profundal-trigeminal placode and Rohon-Beard sensory neurons at stage 20.
(A) Schematic representation of a stage 20 embryo view from the dorsal side, anterior to the left. Modified from Nieuwkoop and Faber (1967). (B) Schematic representation of a transverse section at the level of the diencephalon, corresponding to the line labeled “B” in panel (A). Dorsal to top. The blue areas indicate the position of the developing profundal-trigeminal placode. The boxed area labeled “C” is the region of the section shown in subsequent panels (C). (C)Runx3, Cbfβ and Islet1 are coexpressed in the profundal-trigeminal placode (blue arrows), while Runx1 is not detected in this tissue. (D) Transverse sections in the trunk region corresponding to the line labeled “D” in panel (A). All four genes are coexpressed in Rohon-Beard sensory neurons (red arrows). Islet1 is also expressed in the population of ventral interneurons (black arrows). The notochord is outlined with a solid line, while the position of the neural tube is underlined with dashed lines. The probes are indicated in the lower right corner of each panel. di, diencephalon; no, notochord; ov, optic vesicle. The scale bar in panel C represents 100 µm.
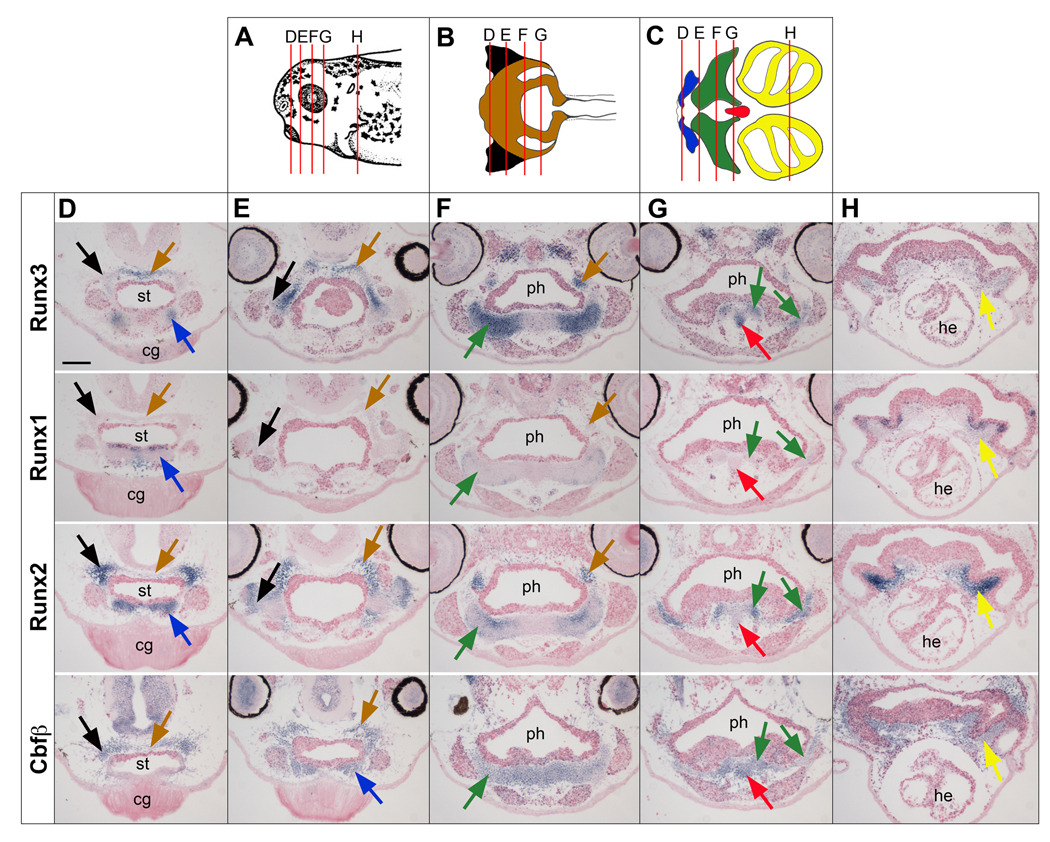
Figure 6. Comparative analysis of Runx3, Runx1, Runx2 and Cbfβ expression in the craniofacial cartilage at stage 40.
(A) Schematic representation of the head of a stage 40 embryo viewed from the lateral side, dorsal to top, anterior to the left. Modified from Nieuwkoop and Faber (1967). (B, C) Schematic representations of the ventral (B) and dorsal (C) cranial cartilage elements. Modified from Sadaghiani and Thiebaud (1987). The cartilage elements are color-coded: ethmoid-trabecular (brown), quadrate (black), Meckel’s (blue), cerathoyal (green), basihyal (red) and branchial (yellow). In panels (A), (B) and (C) the lines labeled “D”, “E”, “F”, “G” and “H” indicate the level of the transverse sections shown in the subsequent panels. cg, cement gland; he, heart; ph, pharynx; st, stomodeum. The scale bar in panel D represents 100 µm.
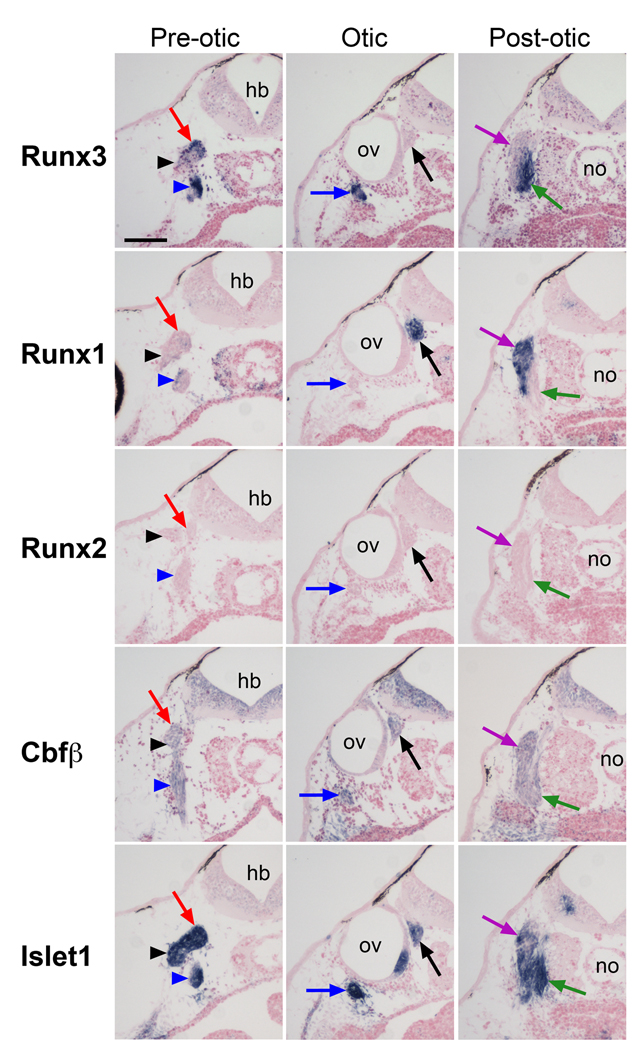
Figure 7. Comparative analysis of Runx3, Runx1, Runx2, Cbfβ and Islet1 expression in neurogenic placode derivatives at stage 40.
Transverse sections were performed at three levels along the antero-posterior axis; the pre-otic, the otic, and post-otic regions. Each column shows sections from approximately the same level. Trigeminal ganglion (red arrows), ganglion of anterodorsal lateral line nerve (black arrowheads), ganglia of facial and anteroventral lateral line nerves (blue arrowheads), ganglia of glossopharyngeal and middle lateral line nerves (blue arrows), statoaccoustic ganglion (black arrows), ganglion of vagal nerve (green arrows), ganglion of posterior lateral line nerve (purple arrows). hb, hindbrain; no, notochord; ov, otic vesicle; The scale bar in the upper left panel represents 100 µm.
1.3.2 Runx3 expression in profundal-trigeminal placode and Rohon-Beard sensory neurons at stage 20
The profundal and trigeminal placodes are initially fused into a single profundal-trigeminal complex located immediately dorsal and posterior to the developing eye. These placodes give rise to neurons of the profundal and trigeminal nerves, respectively (Schlosser and Northcutt, 2000). To confirm the expression of Runx3 in this domain we also analyzed Islet1 expression in staged matched embryos. We found that Runx3 and Islet1 are co-expressed in this placode, however unlike Islet1, which is expressed in the entire placodal domain, Runx3 and Cbfβ are detected only a sub-region of this developing placode (Fig 5 A-C).
Lower vertebrates develop a unique set of primary sensory neurons located in the dorsal spinal cord. These cells, known as Rohon-Beard sensory neurons, innervate the skin and are critical to mediate the escape response to touch during larval stages (Roberts, 2000). Later in development, these neurons undergo apoptosis and are replaced by the neural crest-derived dorsal root ganglia neurons. Runx3 is detected in Rohon-Beard sensory neurons and this expression appears to overlap with that of Islet1, Runx1 and Cbfβ in these cells (Fig 5D). Islet1 is also detected in a second row of primary neurons that will give rise to interneurons, located ventral to the row of Rohon-Beard neurons (Fig 5D). To confirm the co-expression of these genes in Rohon-Beard sensory neurons we performed in situ hybridization on adjacent sections of the same embryo alternating in situ probes. We found that while Runx3 is only expressed in a subset of Rohon-Beard neurons (Fig 1S), it is co-expressed in these cells with Cbfβ, Runx1 and Islet1 (Fig 1S). Runx1, Islet1 and Cbfβ appear to be co-expressed in the entire population of Rohon-Beard neurons (Fig 1S), although each one with slightly different onset of expression (not shown).
1.3.3 Runx3 expression in craniofacial cartilage at stage 40
Whole-mount in situ hybridization indicates that Runx3 is expressed in the branchial arches starting at stage 35/36 and is co-expressed in this domain with Runx2 and Cbfβ. To determine the specific cartilage elements in which Runx3 is expressed, we performed in situ hybridization on serial sections of stage 40 embryos. Cartilage elements (Fig 6A-C) were identified based on the nomenclature of Sadaghiani and Thiebaud (1987). With the exception of the basihyal Runx2 is detected in all cartilage elements (Fig 6D-H). Runx3 on the other hand is expressed in the basihyal cartilage in addition to the Meckel’s, ceratoyal, and ethmoid-trabecular cartilages (Fig 6D-H). Runx1 is detected in a few scattered cells within the ceratohyal cartilage (Fig 6F) and in the pharyngeal endoderm overlying the developing branchial cartilages (Fig 6H). This is in contrast to what has been reported in Zebrafish where runx3 is expressed in the pharyngeal endoderm (Kalev-Zylinska et al., 2003; Flores et al., 2006). As expected, Cbfβ is uniformly expressed in all cartilage elements (Fig 6D-H). Co-expression of Runx2 and Runx3 in the developing cartilage is conserved in zebrafish, chick and mouse embryos (Flores et al., 2006; Stricker et al., 2002).
1.3.4 Runx3 expression in neurogenic placode derivatives at stage 40
Neurogenic placodes in Xenopus laevis comprise the olfactory placode, the profundal and trigeminal placodes, a series of epibranchial placodes, two hypobranchial placodes, the otic placode, and five lateral line placodes (Schlosser and Northcutt, 2000). To precisely evaluate the expression of Runx3 in the derivatives of these placodes we also analyzed the expression of Islet1 a gene expressed in all cranial ganglia and nerves (Brade et al., 2007). We performed sections at three different levels along the anteroposterior axis focusing on the pre-otic, otic and post-otic regions.
In the pre-otic region Runx3 is expressed in the ganglia of the trigeminal, anterodorsal lateral line nerves and the fused ganglia of the facial and anteroventral lateral line nerves, though at a lower level in the anterodorsal lateral line ganglion (Fig 7; pre-otic). Ventral to the otic vesicle, Runx3 is detected in the fused ganglia of the glossopharyngeal and middle lateral line nerves (Fig 7; otic). In this region Runx1 is expressed in the statoaccoustic ganglia associated with the otic vesicle, where it is coexpressed with Islet1 (Fig 7; otic). Interestingly, posterior to the otic vesicle, in the fused ganglia of the vagal and posterior lateral line nerves, Runx1 and Runx3 have complementary and non-overlapping expression domains. Runx1 and Runx3 are restricted to the ganglion of the posterior lateral line nerve and to the ganglion of the vagal nerve, respectively (Fig 7; post-otic). Runx2 is not detected in any of these ganglia, and Cbfβ is ubiquitously expressed in all of them (Fig 7).
In summary, Runx3 is co-expressed with Runx1 in blood progenitors and Rohon-Beard neurons and with Runx2 in craniofacial cartilage (Table 1). Runx3 also has a unique expression domain in a number of derivatives of the neurogenic placodes not shared with other Runx family members, including ganglia of the anteroposterior and middle lateral line nerves, and ganglia of the trigeminal, glossopharyngeal, facial and vagal nerves (Table 1). Our data suggest that Runx3 is an important player in the regulation of cranial sensory neurons development.
Table 1.
Summary of Runx1, Runx2, Runx3, and Cbfβ tissues expression during Xenopus development
| Runx1 | Runx2 | Runx3 | Cbfβ | |
|---|---|---|---|---|
| Blood progenitors | + | − | + | + |
| Rohon-Beard sensory neurons | + | − | + | + |
| Profundal-trigeminal placode | − | − | + | + |
| Olfactory placode | + | − | − | + |
| Trigeminal ganglion | − | − | + | + |
| Ganglion of anterodorsal lateral line nerve | − | − | +/− | + |
| Ganglia of facial and anteroventral lateral line nerves | − | − | + | + |
| Statoaccoustic ganglion | + | − | − | + |
| Ganglia of glossopharyngeal and middle lateral line nerves | − | − | + | + |
| Ganglion of posterior lateral line nerve | + | − | − | + |
| Ganglion of vagal nerve | − | − | + | + |
| Meckel’s cartilage | − | + | + | + |
| Ceratoyal cartilage | +/− | + | + | + |
| Basihyal cartilage | − | − | + | + |
| Branchial cartilage | − | + | − | + |
| Quadrate cartilage | − | + | − | + |
| Ethmoid-trabecular cartilage | − | + | + | + |
“+”, gene strongly expressed in the corresponding tissue; “+/−”, weakly expressed; “−”, undetected.
2. Experimental Procedures
2.1. Cloning of Xenopus Runx3
A partial sequence of the Xenopus laevis Runx3 ORF was amplified using PCR from stage 30 cDNA using a set of primers designed against two conserved regions of Runx3 in several species (F: TGCGGAAAGATGGGCGAGAA; R: TCCATTCTTCCAC TAGTGGT) and based on the sequence of Xenopus tropicalis Runx3 (Ensembl ID, ENSXETT00000002249). The resulting 1100 bp PCR product was purified, subcloned into pGEMTeasy (Promega) and sequenced. This construct is referred as pGEMT-XRunx3. Based on Xenopus tropicalis sequence we designed a second set of primers outside Runx3 ORF (F: AACACCCTGCTGTTGTAATG; R: GGCTGTATTCACAAAG TCTC) to amplify the entire coding region of Xenopus laevis Runx3. This product was predicted to include 17 bp of 5'UTR and 20 bp of 3'UTR. The PCR was performed using Taq polymerase and stage 41 cDNA as template. The resulting 1224 bp product was subcloned into pGEMTeasy and multiple clones were sequenced to confirm the sequence of Xenopus laevis Runx3. Based on this information, we designed a third set of nested primers (F: ATGCTCATTCCCGTAGACCC; R: TCAATAGGGTCTCCAAA CTG) to amplify the Xenopus laevis Runx3 ORF using Pfu polymerase and stage 41 cDNA. The PCR product encoding Xenopus laevis Runx3 ORF was subcloned and sequenced. The sequence has been submitted to GeneBank (accession # GU725438).
2.2. Molecular Phylogeny
Multiple sequence alignment of the deduced amino acid sequences of Runx3 was performed using ClustalW (Thompson et al., 1994), and phylogenetic tree was constructed by using the neighbor-joining method (Pearson et al., 1999) through the EMBL–EBI interface (http://www.ebi.ac.uk/Tools/clustalw/). The sequences used in Fig. 1 are human RUNX1 (NP_001001890), mouse Runx1 (NP_001104493), chicken Runx1 (Ensembl ID, ENSGALP00000029722), Xenopus laevis Runx1 (AAC41269), zebrafish runx1 (NP_571678), human RUNX2 (NP_004339), mouse Runx2 (NP_001139392), chicken Runx2 (NP_989459), Xenopus laevis Runx2 (ABM05616), zebrafish runx2a (NP_998023), zebrafish runx2b (NP_998027), human RUNX3 (NP_004341), mouse Runx3 (EDL29993), chicken Runx3 (XP_001232978) and zebrafish runx3 (NP_571679).
2.3. RT-PCR analysis
Embryos were staged according to Nieuwkoop and Faber (1967). Total RNA was extracted from embryos at indicated stages using an RNeasy micro RNA isolation kit (Qiagen). To avoid contamination from genomic DNA, the RNA samples were digested with RNase-free DNase I before RT-PCR was performed. RT-PCR experiments were performed using the One Step RT-PCR kit (Qiagen) according to the manufacturer’s instructions using the following primer sets: Runx1 (F: ACTCTGAGTCCGGGGAAGAT; R: CCATATTCCGGTCTGTGCTT; 30 cycles), Runx2 (F: GCTTCCTGCTATCTCCGA TG; R: GGAGGGCTGTACGTGAATGT; 33 cycles), Runx3 (F: CACACTGGCCAACAC AAATC; R: TACGAGGGTCGGTAAACCT G; 30 cycles), Cbfβ (F: GAACGACAAGCA CGTTTTCA; R: CTCCCGTTCAAAGTCCACAT; 26 cycles) and ODC (F: ACATGGC ATTCTCCCTGAAG; R: TGGTCCCAAGGCTAAAGTTG; 25 cycles). For real-time RT-PCR the reaction was performed using the same primer sets and the QuantiTect SYBR Green RT-PCR kit (QIAGEN) on a LightCycler (Roche Diagnostics). The reaction mixture consisted of 10 µl of QuantiTect SYBR Green RT-PCR Master Mix, 500 nM forward and reverse primers, 0.2 µl of RT, and 60 ng of template RNA in a total volume of 20 µl. The cycling conditions were as follow: denaturation at 95°C (3 sec.), annealing at 55°C (4 sec.), and extension at 72°C (12 sec.). By optimizing primers and reaction conditions, a single specific product was amplified as confirmed by melting curve analysis. Each reaction included a control without template and a standard curve of serial dilutions (in 10-fold increments) of test RNAs. In each case, ornithine decarboxylase (ODC) was used as an internal reference (data not shown). Each bar on the histograms has been normalized to the level of ODC.
2.4. In situ hybridization
Sense and antisense DIG-labeled probes (Genius kit, Roche) were synthesized using template cDNA encoding Runx1/Xaml1 (Tracey et al., 1998), Runx2 (Kerney et al., 2007), Pax3 (Bang et al., 1997), Islet1 (Brade et al., 2007), Cbfβ (Sport6-Cbfβ; Open Biosystems) and Runx3 (pGEMT-XRunx3). Whole-mount in situ hybridization was performed as described (Harland, 1991). In some cases to ease visualization of the staining embryos were cleared in a mixture of benzyl alcohol and benzyl benzoate (1v:2v). For in situ hybridization on sections, embryos at stage 20 and 40 were fixed in MEMFA for 1 hour, embedded in Paraplast and 12 µm sections hybridized with the appropriate probes as described (Henry et al. 1996). Sections were then briefly counter stained with eosin.
Supplementary Material
Acknowledgements
We thank Drs. Anne Bang, Ryan Kerney, Peter Klein and Petra Pandur for reagents and Dr. Trish Labosky for comments on the manuscript. This work was supported by a grant from the National Institutes of Health to J-P S-J (RO1-DC07175).
References
- Bang AG, Papalopulu N, Kintner C, Goulding MD. Expression of Pax-3 is initiatedin the early neural plate by posteriorizing signals produced by the organizer and by posterior non-axial mesoderm. Development. 1997;124:2075–2085. doi: 10.1242/dev.124.10.2075. [DOI] [PubMed] [Google Scholar]
- Blyth K, Cameron ER, Neil JC. The RUNX genes: Gain or loss of function in cancer. Nat. Rev. Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- Brade T, Gessert S, Kühl M, Pandur P. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev. Biol. 2007;311:297–310. doi: 10.1016/j.ydbio.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Brenner O, Levanon D, Negreanu V, Golubkov O, Fainaru O, Woolf E, Groner Y. Loss of Runx3 function in leukocytes is associated with spontaneously developed colitis and gastric mucosal hyperplasia. Proc. Natl. Acad. Sci. USA. 2004;101:16016–16021. doi: 10.1073/pnas.0407180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CE, DeBlasio T, Zhou Y, Zhang J, Zon L, Nimer SD. Isolation and characterization of runxa and runxb, zebrafish members of the runt family of transcriptional regulators. Exp. Hematol. 2002;30:1381–1389. doi: 10.1016/s0301-472x(02)00955-4. [DOI] [PubMed] [Google Scholar]
- Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 2005;19:2331–2342. doi: 10.1101/gad.1337005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores MV, Tsang VW, Hu W, Kalev-Zylinska M, Postlethwait J, Crosier P, Crosier K, Fisher S. Duplicate zebrafish runx2 orthologues are expressed in developing skeletal elements. Gene Expr. Patterns. 2004;4:573–581. doi: 10.1016/j.modgep.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Flores MV, Lam EYN, Crosier P, Crosier K, Fisher S. A Hierarchy of Runx transcription factors modulate the onset of chondrogenesis in craniofacial endochondral bones in zebrafish. Dev. Dyn. 2006;235:3166–3176. doi: 10.1002/dvdy.20957. [DOI] [PubMed] [Google Scholar]
- Flores MV, Lam EYN, Crosier P, Crosier K. Osteogenic transcription factor Runx2 is a maternal determinant of dorsoventral patterning in zebrafish. Nature Cell Biol. 2008;10:346–352. doi: 10.1038/ncb1697. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Meth Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Henry GL, Brivanlou IH, Kessler DS, Hemmati-Brivanlou A, Melton DA. TGF-beta signals and a pattern in Xenopus laevis endodermal development. Development. 1996;122:1007–1015. doi: 10.1242/dev.122.3.1007. [DOI] [PubMed] [Google Scholar]
- Hong C-S, Saint-Jeannet J-P. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol. Biol. Cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Ozaki S, Shiga T, Ito K, Masuda T, Okado N, Iseda T, Kawaguchi S, Ogawa M, Bae SC, Yamashita N, Itohara S, et al. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat. Neurosci. 2002;5:946–954. doi: 10.1038/nn925. [DOI] [PubMed] [Google Scholar]
- Ito K. RUNX Genes in Development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv. Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- Kalev-Zylinska ML, Horsfield JA, Flores MV, Postlethwait JH, Vitas MR, Baas AM, Crosier PS, Crosier KE. Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development. 2002;129:2015–2030. doi: 10.1242/dev.129.8.2015. [DOI] [PubMed] [Google Scholar]
- Kalev-Zylinska ML, Horsfield JA, Flores MV, Postlethwait JH, Chau JY, Cattin PM, Vitas MR, Crosier PS, Crosier KE. Runx3 is required for hematopoietic development in zebrafish. Dev. Dyn. 2003;228:323–336. doi: 10.1002/dvdy.10388. [DOI] [PubMed] [Google Scholar]
- Kataoka H, Ochi M, Enomoto K, Yamaguchi A. Cloning and embryonic expression patterns of the zebrafish Runt domain genes, runxa and runxb. Mech. Dev. 2000;98:139–143. doi: 10.1016/s0925-4773(00)00445-7. [DOI] [PubMed] [Google Scholar]
- Kerney R, Gross JB, Hanken J. Runx2 is essential for larval hyobranchial cartilage formation in Xenopus laevis. Dev. Dyn. 2007;236:1650–1662. doi: 10.1002/dvdy.21175. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, Kremer E, Otto F, Brenner O, Lev-Tov A, Groner Y. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21:3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, Kim HM, Kim WJ, Yamamoto H, Yamashita N, Yano T, Ikeda T, Itohara S, Inazawa J, Abe T, Hagiwara A, Yamagishi H, Ooe A, Kaneda A, Sugimura T, Ushijima T, Bae SC, Ito Y. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) Amsterdam, The Netherlands: North Holland Publishing Company; 1967. [Google Scholar]
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marin-Padilla M, Speck NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Robins G, Zhang T. Generalized neighbor-joining: more reliable phylogenetic tree reconstruction. Mol. Biol. Evol. 1999;16:806–816. doi: 10.1093/oxfordjournals.molbev.a026165. [DOI] [PubMed] [Google Scholar]
- Roberts A. Early functional organization of spinal neurons in developing lower vertebrates. Brain Res. Bull. 2000;53:585–593. doi: 10.1016/s0361-9230(00)00392-0. [DOI] [PubMed] [Google Scholar]
- Sadaghiani B, Thiebaud CH. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev. Biol. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Northcutt RG. Development of neurogenic placodes in Xenopus laevis. J. Comp. Neur. 2000;418:121–146. [PubMed] [Google Scholar]
- Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- Stricker S, Fundele R, Vortkamp A, Mundlos S. Role of Runx genes in chondrocyte differentiation. Dev Biol. 2002;245:95–108. doi: 10.1006/dbio.2002.0640. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WD, Pepling ME, Marko EH, Thomsen GH, Gerben JP. A Xenopus homologue of aml-1 reveals unexpected patterning mechanisms leading to the formation of embryonic blood. Development. 1998;125:1371–1380. doi: 10.1242/dev.125.8.1371. [DOI] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijmenga C, Speck NA, Dracopoli NC, Hofker MH, Liu P, Collins FS. Identification of a new murine runt domain-containing gene, Cbfa3,and localization of the human homolog, CBFA3, to chromosome 1p35-pter. Genomics. 1995;26:611–614. doi: 10.1016/0888-7543(95)80185-o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.