Abstract
Web services have become widely used in bioinformatics analysis, but there exist incompatibilities in interfaces and data types, which prevent users from making full use of a combination of these services. Therefore, we have developed the TogoWS service to provide an integrated interface with advanced features. In the TogoWS REST (REpresentative State Transfer) API (application programming interface), we introduce a unified access method for major database resources through intuitive URIs that can be used to search, retrieve, parse and convert the database entries. The TogoWS SOAP API resolves compatibility issues found on the server and client-side SOAP implementations. The TogoWS service is freely available at: http://togows.dbcls.jp/.
INTRODUCTION
In recent years, major bioinformatics centers have begun providing SOAP-based (http://www.w3.org/2002/ws/) Web services that enable users to use these database resources with client programs in an automated manner. These include the E-Utilities service (1) provided by the National Center for Biotechnology Information (NCBI), Web services provided by the European Bioinformatics Institute (EBI) (2,3), the Web API for Bioinformatics (WABI) from the DNA Data Bank of Japan (DDBJ) (4–7), the Protein Data Bank Japan’s (PDBj) Web services (8) and the KEGG API service from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (9). Thanks to these services, users can easily perform various bioinformatics tasks through their choice of client software and can reproduce each procedure as a workflow.
However, when it comes to using these services in combination, there are several limitations (10) to their interoperability and technological implementation: (i) there are no common ontologies for operations and objects in these Web services, resulting in inconsistent naming conventions and data types; (ii) this incompatibility of data types requires format conversion of objects to use the output of one service as the input to the next service; (iii) there are several services that require specific SOAP features that are not always supported in the available SOAP libraries, even for several major programming languages; and (iv) the client developer needs to be aware of fail-safe mechanisms, such as temporary downtime of the server or the network, as well as environmental restrictions such as the maximum size of exchanged data.
To overcome these limitations [especially for (i) and (ii)], the BioMoby project (11,12) was begun to provide a central registry of operations and objects used in public Web services, along with ontologies. In this way, a number of BioMoby-compliant services were developed, and the BioMoby client can find the service that is appropriate for the type of object. The main problem here is that most major bioinformatics service providers are not compatible with the BioMoby standard, possibly because it requires a considerable amount of server-side effort. Furthermore, it is also difficult to enforce a set of standard data formats for interoperability among these providers.
To help resolve these problems, we organized DBCLS BioHackathons in 2008 (http://hackathon.dbcls.jp/) and 2009 (http://hackathon2.dbcls.jp), international workshops focusing on Web services, drawing participants from many backgrounds, including Web service providers, developers of the Open Bio* libraries and client applications as well as database creators in emerging fields such as glycoinformatics and interactomics. One interesting topic in the BioHackathon was the attempt to resolve the current limitations in interoperability among existing Web services. For this purpose, a workflow was proposed that pipelines services provided by DDBJ, PDBj and KEGG to find homologs using BLAST and annotate them with structural and pathway information. When this workflow is run in the Taverna environment (13), we again encountered the essential need for data format conversion. The Open Bio* libraries (14), including BioPerl (15), BioRuby (http://bioruby.org), BioPython (16) and BioJava (17), provide parsers for major database entry and software output formats such as the BLAST report. However, users are required to install these libraries and to write code to use their functionality.
Building upon discussions from the BioHackathon, we began to develop TogoWS, an integrated Web service (‘togo’ is a Japanese word for ‘integration’) that provides uniform access to database resources, parsers for database entries and converters among major data formats. Bioinformatics Web services can be categorized into data-retrieval services and analysis services. Although both types of services can be exposed using either the REST (18) or the SOAP architecture, REST is better suited for data-retrieval services and SOAP is more suitable for analysis services because the former can be easily mapped to resource URIs and the latter usually requires a long execution time or complex parameters.
In our survey, we discovered that most existing Web services (data not shown) are designed to search and retrieve database entries maintained at each institution. Therefore, in TogoWS, we designed a REST-based Web service for accessing database resources in a unified manner, with intuitive URI notation for searching, retrieving, parsing and converting the database entries. Moreover, we developed a unified SOAP-based Web service in TogoWS that proxies analysis services provided by Japanese institutions to resolve several incompatibilities found in these services. Supplemental documents and source code in major programming languages (Perl, Ruby, Python and Java) are also provided.
TogoWS REST API
The TogoWS REST service provides intuitive APIs to search, retrieve, parse and convert the database entries. In the following sections, we will describe these interfaces and the internal architecture of the REST service.
Database search
TogoWS provides a uniform query interface for various databases. The result of the database search can be considered a resource that is relevant to the query string. Therefore, we map each database name (DATABASE) and query string (QUERY_STRING) to a URI by the following convention:
A list of currently available databases can be obtained by accessing the following URI without a database name:
As an example, a search against the UniProt database using the phrase ‘lung cancer’ can be represented as follows:
The returned text contains matched entry IDs, one per line (Figure 1a). The QUERY_STRING can be a simple keyword or a URI-encoded string containing a structured query with logical operations. The given query is translated by the TogoWS server and then sent to the corresponding service.
Figure 1.
Examples of the TogoWS URIs and their outputs.
Hit count and pagination
A database search often returns a long list of hits. To make our search service scalable, we introduced a method for counting and pagination. To count the number of hits, simply add ‘/count’ to the end of the query URI:
Then, the user can retrieve any subset of the hits by indicating OFFSET and LIMIT numbers in the following format:
For example, to obtain 10 results starting from the 100th hit
The user can iterate over the OFFSET value, starting from 1 and incrementing it by LIMIT until all hits have been retrieved.
Entry retrieval
Each database entry can be identified by a database name and a unique identifier; therefore, it can be easily represented as a unique URI. In the TogoWS REST API, we mapped database names and entry IDs to URIs by the following convention:
where the ‘/entry’ prefix indicates a REST action to retrieve the resource specified by DATABASE and ENTRY_ID, which represent the name of the database and the entry ID string, respectively.
For example, the URI to retrieve a KEGG GENES database entry ‘sec:YDR074W’ can be represented as follows, and it will return the flatfile entry as a text string, without any decoration:
Multiple entries can be retrieved at once by concatenating entry IDs with commas. Therefore, PubMed entries ‘18077471′ and ‘19151099′ can be retrieved at a time by accessing the following URI:
A list of currently available databases can be obtained by accessing the following URI without a database name:
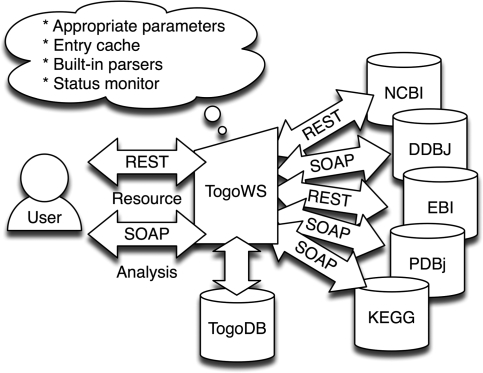
To obtain actual database entries, TogoWS internally uses existing SOAP or REST interfaces provided by each database (Figure 2). Since the TogoWS acts as a proxy to various data sources, the user does not need to worry about the internals of the SOAP messages or complex CGI parameters that each database usually requires for access. The TogoWS server also caches the retrieved entries for a period of time to avoid overloading the original servers.
Figure 2.
Schematic overview of the TogoWS service.
Entry field extraction
A unique feature of the TogoWS REST API is that it comes with built-in parsers for various database formats. Without this, the user will need to install a bioinformatics library such as BioPerl, BioPython, BioRuby or BioJava and to write a program to extract the desired information from the retrieved entries. This requirement has been a bottleneck to the creation of an automated workflow that consumes a list of database entries and extracts information for the next step of the analysis pipeline. To resolve this situation, we embedded BioPerl and BioRuby libraries in the TogoWS server. These bioinformatics libraries cover a wide range of biomedical databases and provide efficient parsing functionality for various database entries. We extended the TogoWS REST API to support extraction of the field contents just by adding a specific field name at the end of the URI, as follows:
where FIELD is one of the supported field names. The list of available field names differs from database to database and can be obtained by accessing the following URI:
As described in the previous section, TogoWS will retrieve specified entries from the original database. Then, the cached contents are internally processed by built-in parsers. In this manner, the user can access any field values of the given entries without programming.
For example, a name, a molecular weight and relevant enzymes of the KEGG COMPOUND entry ‘C01083′ can be extracted by the following URIs, respectively (Figure 1b–d):
Similarly, the authors and abstract of the PubMed entry ‘19151099′ can be retrieved by
where ‘au’ and ‘ab’ correspond to the AU and AB lines, respectively, of the PubMed record in MEDLINE format.
Entry format conversion
Even though a specific field of an entry can be extracted, it is often required to convert the data format for further use. With the help of built-in parsers, TogoWS provides format conversion of the entry simply by specifying the format as a URI suffix, analogous to the extension of a filename:
For example, the DDBJ entry ‘M13899′ can be converted into the FASTA, INSDC-XML and GFF formats by the following URIs, respectively:
Acceptable formats can vary according to the database and currently include XML, JSON, GFF version 3 and FASTA. In the future, RDF/XML and Turtle will also be supported. The FASTA and GFF formats are valid for nucleotide or peptide sequence databases, and the XML format is available if the original database is also provided as XML.
Format conversion can also be applied to the extracted field. The following URI returns the associated enzymes of the KEGG COMPOUND entry ‘C01083′ in JSON format (Figure 1e).
The JSON format (http://tools.ietf.org/html/rfc4627) is particularly useful when this service is used in a Web application that retrieves relevant information on the fly via an AJAX method.
A list of available format names differs from database to database and can be obtained by accessing the following URI:
Data format conversion
TogoWS also provides format-to-format conversion functionality. Unlike the methods described above, this method uses the HTTP POST protocol instead of HTTP GET. The end-point URI of the data format conversion service uses the following convention:
For example, to convert a BLAST result to GFF format, simply POST the BLAST report string to the following URI:
Figure 3 shows a sample Ruby program demonstrating how to read a BLAST output stored in the file ‘blast_result.txt’ and convert its contents into GFF format:
Figure 3.
Ruby program to convert a BLAST output into GFF format.
Currently, GenBank, EMBL, UniProt, BLAST, FASTA, PSL, Sim4, HMMER, Exonerate and Wise formats are supported as source data types. This service is intended to be used in the workflow management software, in which the pipeline is often bottlenecked by incompatible data formats. TogoWS fills this kind of gap without requiring the user to install additional software on the local computer.
TogoWS SOAP API
The other half of TogoWS is a SOAP-based proxy service for Japanese bioinformatics resources, including DDBJ, PDBj and KEGG. In contrast to the REST service, SOAP is suitable for services requiring long execution time, returning structured objects, or expecting complex parameters in the query. The SOAP specification itself is an open standard and is independent of the programming languages. However, its implementation in each programming language tends to be incomplete because of the complexity of the specification. Because of this, there appear to be several technical incompatibilities in each service. We have been collaboratively working with some of these institutions to resolve the issues; however, there still remain problems that require modifications to their service specifications. These problems include the use of a MIME attachment for returning the results, the use of an HTTP cookie for stateful transactions and different designs for asynchronous transactions, features that are not always supported by the SOAP library of choice.
Integrated WSDL file
Instead of asking all service providers to modify their services, we developed the TogoWS SOAP API, which proxies their services and thus hides the incompatibilities and differences between them. All services across these servers (DDBJ, PDBj and KEGG) are integrated into only one WSDL file,
so that the user can use all 368 operations that were originally spread among 26 WSDL files. Our service has been tested in several major programming languages (Perl, Python, Ruby and Java), so the user can use each service in the preferred language without difficulty. This approach also eliminates a burden from the service providers because they do not themselves need to test or improve the language compatibility of their services.
Sample code and documents
The TogoWS SOAP service comes with comprehensive sample code covering all operations of the DDBJ, PDBj and KEGG services written in four programming languages (Perl, Python, Ruby and Java). The user can freely examine and download the code from the following database and use them as references for further development.
Web services often lack documentation, forcing users to consult the WSDL file to learn what kind of operations are available, what data types are used for input and output, etc. However, this is not an effortless task, as the WSDL file was not designed to be read by a human. To remedy this problem, we have created a list of Web service operations from existing bioinformatics Web services worldwide:
This list contains information extracted from the WSDL files, such as the description and input/output data types for 4172 operations, including services integrated in the TogoWS SOAP API. In addition, we also assigned a functional classification to each operation.
Server status monitor
Web services are often used by computer programs in a pipeline. However, it is often difficult to detect temporary error caused by server-side problems. We have monitored the availability of all operations in DDBJ, PDBj and KEGG over the past 2 years. The result is stored and summarized in the TogoWS status report:
Since the monitoring is performed every day, these records may help the user determine whether the source of the problem is the local configuration or the remote server. The record also contains statistical information such as output size and response time, which has helped service providers to detect unexpected errors several times.
DISCUSSION
In TogoWS, we proposed an integrated service focused on the interface and compatibility of existing bioinformatics Web services. We successfully developed a REST interface for accessing database resources with intuitive and persistent URIs. For other services, we developed a highly compatible SOAP interface supplemented by sample codes and a status monitor. These services are stable and have been used for about 2 years, but there remains room for improvement.
We will continue to increase the number of supported formats and databases in TogoWS. Most importantly, we are planning to extend the TogoWS REST API to support the Semantic Web framework. During the course of development, we will extend the TogoWS to support private datasets stored in the TogoDB database (http://togodb.dbcls.jp) in addition to the major public databases. By exporting these data in RDF format, TogoWS can contribute as a provider of Linked Data.
FUNDING
The Integrated Database Project of the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charge: Integrated Database Project in Japan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Mr. Tatsuya Nishizawa for his support in the development of the TogoWS server and the participants of the DBCLS BioHackathon (http://hackathon.dbcls.jp/), in which valuable discussions helped to clarify bottlenecks in the current Web services in bioinformatics and determined the required infrastructure to make these services interoperable.
REFERENCES
- 1.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labarga A, Valentin F, Anderson M, Lopez R. Web services at the European bioinformatics institute. Nucleic Acids Res. 2007;35:W6–W11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pillai S, Silventoinen V, Kallio K, Senger M, Sobhany S, Tate J, Velankar S, Golovin A, Henrick K, Rice P, et al. SOAP-based services provided by the European Bioinformatics Institute. Nucleic Acids Res. 2005;33:W25–W28. doi: 10.1093/nar/gki491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugawara H, Miyazaki S. Biological SOAP servers and web services provided by the public sequence data bank. Nucleic Acids Res. 2003;31:3836–3839. doi: 10.1093/nar/gkg558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazaki S, Sugawara H, Ikeo K, Gojobori T, Tateno Y. DDBJ in the stream of various biological data. Nucleic Acids Res. 2004;32:D31–D34. doi: 10.1093/nar/gkh127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugawara H, Ogasawara O, Okubo K, Gojobori T, Tateno Y. DDBJ with new system and face. Nucleic Acids Res. 2007;36:D22–D24. doi: 10.1093/nar/gkm889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon Y, Shigemoto Y, Kuwana Y, Sugawara H. Web API for biology with a workflow navigation system. Nucleic Acids Res. 2009;37:W11–W6. doi: 10.1093/nar/gkp300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Standley DM, Kinjo AR, Kinoshita K, Nakamura H. Protein structure databases with new web services for structural biology and biomedical research. Brief. Bioinform. 2008;9:276–285. doi: 10.1093/bib/bbn015. [DOI] [PubMed] [Google Scholar]
- 9.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stockinger H, Attwood T, Chohan SN, Côté R, Cudré-Mauroux P, Falquet L, Fernandes P, Finn RD, Hupponen T, Korpelainen E, et al. Experience using web services for biological sequence analysis. Brief. Bioinform. 2008;9:493–505. doi: 10.1093/bib/bbn029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkinson MD, Links M. BioMOBY: an open source biological web services proposal. Brief. Bioinform. 2002;3:331–341. doi: 10.1093/bib/3.4.331. [DOI] [PubMed] [Google Scholar]
- 12.Vandervalk BP, McCarthy EL, Wilkinson MD. Moby and Moby 2: creatures of the deep (web) Brief. Bioinform. 2009;10:114–128. doi: 10.1093/bib/bbn051. [DOI] [PubMed] [Google Scholar]
- 13.Hull D, Wolstencroft K, Stevens R, Goble C, Pocock MR, Li P, Oinn T. Taverna: a tool for building and running workflows of services. Nucleic Acids Res. 2006;34:W729–W732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stajich J, Lapp H. Open source tools and toolkits for bioinformatics: significance, and where are we? Brief. Bioinform. 2006;7:287–296. doi: 10.1093/bib/bbl026. [DOI] [PubMed] [Google Scholar]
- 15.Stajich J, Block D, Boulez K, Brenner S, Chervitz S, Dagdigian C, Fuellen G, Gilbert J, Korf I, Lapp H, et al. The Bioperl Toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cock P, Antao T, Chang J, Chapman B, Cox C, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holland R, Down T, Pocock M, Prlic A, Huen D, James K, Foisy S, Dräger A, Yates A, Heuer M, et al. BioJava: an open-source framework for bioinformatics. Bioinformatics. 2008;24:2096–2097. doi: 10.1093/bioinformatics/btn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fielding R. Architectural Styles and the Design of Network-based Software Architectures. Irvine: University of California; 2000. [Google Scholar]