Abstract
Short, linear motifs (SLiMs) play a critical role in many biological processes, particularly in protein–protein interactions. The Short, Linear Motif Finder (SLiMFinder) web server is a de novo motif discovery tool that identifies statistically over-represented motifs in a set of protein sequences, accounting for the evolutionary relationships between them. Motifs are returned with an intuitive P-value that greatly reduces the problem of false positives and is accessible to biologists of all disciplines. Input can be uploaded by the user or extracted directly from UniProt. Numerous masking options give the user great control over the contextual information to be included in the analyses. The SLiMFinder server combines these with user-friendly output and visualizations of motif context to allow the user to quickly gain insight into the validity of a putatively functional motif. These visualizations include alignments of motif occurrences, alignments of motifs and their homologues and a visual schematic of the top-ranked motifs. Returned motifs can also be compared with known SLiMs from the literature using CompariMotif. All results are available for download. The SLiMFinder server is available at: http://bioware.ucd.ie/slimfinder.html.
INTRODUCTION
The purpose of the Short, Linear Motif Finder (SLiMFinder) web server is to allow researchers to identify novel Short Linear Motifs (SLiMs) in a set of sequences. SLiMs, also referred to as linear motifs or minimotifs, are functional microdomains that play a central role in many diverse biological pathways (1). SLiM-mediated protein interactions include post-translational modification (including cleavage), subcellular localization and ligand binding (2). SLiMs are typically less than 10 amino acids long and have less than five defined positions, many of which will be ‘degenerate’ and incorporate some degree of flexibility in terms of the amino acid at that position. Their length and degeneracy gives them an evolutionary plasticity which is unavailable to domains meaning that they will often evolve convergently, adding new functionality to proteins (1). SLiMs hold great promise as future therapeutic targets, which makes their discovery of great interest (3,4).
Several web-based methods to discover novel instances of known SLiMs are available such as ELM (2), MnM (5) and Quasimotifinder (6). Proteins can be searched using these methods to return putatively functional sites and additional contextual information such as sequence conservation (7) and structural context (8) used to assess the likelihood of true functional significance. Because of the small, degenerate nature of SLiMs, stochastic occurrences of motifs are common and distinguishing real occurrences from random remains the greatest challenge in a priori motif discovery. This challenge is increased further still in de novo motif discovery as the motif search space is considerably greater and often it is not even known if the proteins being examined share a common SLiM or not.
The concept of over-representation as an indicator of functionality is currently the most powerful and widely used approach for discovering de novo SLiMs computationally (9–13). A set of proteins for which there is a suspected SLiM-mediated common function (e.g. targeting protein localization, mediating protein binding or acting as a recognition site for a post-translational modification) is analysed under the hypothesis that the function-mediating SLiM would occur more often than expected by chance due to the selection for the motif in these proteins. Such SLiM discovery algorithms therefore need to overcome two obstacles: (i) identify over-represented motifs; and (ii) assess whether such motifs are expected by chance. Many algorithms are very good at (i) but not (ii), i.e. they will find over-represented motifs that are genuinely present in a data set but are poor at discriminating these from false positives. This is particularly true for algorithms that do not consider the evolutionary relationships within the input proteins as results will tend to be dominated by longer regions of conservation or homology (e.g. globular domains) at the cost of SLiM detection.
Motif discovery algorithms can be broadly divided into two types: alignment-based and alignment-free. Those that look for motifs in related sequences, using alignment-based approaches, are generally optimal for discovery of very strong (or long) ‘family descriptor’ motifs [e.g. MEME (14) and PRATT (15)] or for improving definitions of known motifs [e.g. GLAM2 (16)]. More successful de novo SLiM discovery methods (10–12) are alignment-free and built on an explicit model of convergent evolution, using over-representation of motifs in ‘unrelated’ proteins. Dilimot (13) and SLiMDisc (10) combine these techniques with heuristic scoring schemes to successfully discover new functional motifs and rediscover known motifs. Neduva et al. (12) clearly demonstrated the potential of models based on convergent evolution when they applied Dilimot to discover SLiMs in multiple HPRD data sets. These algorithms still struggled to identify false-positive predictions, however.
SLiMFinder (11) extended these heuristics to estimate the statistical significance of returned motifs, through improved statistics that account for the background of randomly recurring motifs, correcting for evolutionary relationships within the data. The P-value returned by SLiMFinder greatly reduces the problem of false positives and provides a score that is intuitive and accessible to biologists of all disciplines. Additional development of this algorithm has seen further improvements in both sensitivity and specificity through use of conservation-based masking (17) and more accurate statistical models (18). The SLiMFinder server combines these new features with user-friendly input/output and visualizations of motif context to allow the user to quickly gain insight into the validity of a putatively functional motif.
THE SLiMFinder ALGORITHM
SLiMFinder (11) is a probabilistic SLiM discovery program building on the principles of the SLiMDisc algorithm (10). As input, the algorithm takes a data set of proteins with a common feature (e.g. common binding partner) that might be SLiM-mediated. These proteins can be masked to exclude under-conserved residues (17), non-disordered regions predicted using IUPred (19), low complexity regions, specific amino acids or motifs, and annotated features including domains or user-annotated regions to allow any contextual information to be included in the analyses. The TEIRESIAS raw motif discovery tool is (20) replaced by SLiMBuild (11) allowing flexible and ambiguous motifs to be returned. Motifs are built by combining dimers into longer patterns, retaining only those motifs occurring in a sufficient number of unrelated proteins. Motifs with fixed amino acid positions are identified and then combined to incorporate amino acid ambiguity and variable-length wildcards.
Statistics are implemented in the SLiMChance algorithm (11), which is based on the binomial statistics introduced by ASSET (21) [also used by Dilimot (13)] with two major extensions: (i) homologous proteins are weighted (as in SLiMDisc) to account for the dependencies introduced into the probabilistic framework by homologous proteins; and (ii) introduction of significance scores, i.e. the probability that any motif considered would reach a binomial P-value by chance is calculated and used to rank motifs. This greatly increased the stringency of de novo SLiM discovery, and substantially reduced false-positive predictions (11). The original SLiMChance algorithm incorporated some simplifying assumptions for the sake of computational efficiency, with the resulting tendency to increase returned P-values slightly [i.e. increase the chance of a false-negative result (18)]. Although full correction is not computationally efficient enough for the web server, a partial correction is available at the cost of increased run times.
THE SLIMFINDER WEB SERVER
The SLiMFinder server is available at: http://bioware.ucd.ie/slimfinder.html. The purpose of the web server is to allow researchers to identify novel SLiMs in a set of sequences. Sequences are first masked according to user specifications before recurring motifs are identified using the SLiMBuild algorithm. The SLiMChance algorithm then estimates statistical significance of recurring motifs and the most significant, filtered according to user specifications, are returned. Interactive output permits easy exploration and visualizations of motif context to allow the user to quickly gain insight into the validity of a putatively functional motif. The web server is powered by the same code as the downloadable version of SLiMFinder and details of the algorithm can be found in the previous publications (11,17,18). The novel features of the web server are described in more detail in the following sections.
Input
SLiMFinder is optimized for, and requires, at least three sequences for analysis. Two options are provided for sequence input:
1. UniProt IDs can be used to extract entries directly from the UniProtKB (22). Extracted entries are available for download by the user for additional reference/analysis. This is the preferred method of input and enables the full functionality of the web server, including conservation-based masking (17).
2. User-constructed sequences files can be uploaded or pasted directly into the input form. UniProt flat files and FASTA format sequences are accepted. UniProt entries are recommended for full feature-based masking options. Because conservation-based masking makes use of pre-computed alignments, this is not available for user-entered sequences. To use conservation-based masking of user-entered sequences, it is recommended to download SLiMFinder and run it locally.
Examples of all input formats are available in the help pages of the web server. An example data set can also be loaded directly into the input entry form for user experimentation (see example analysis).
Options
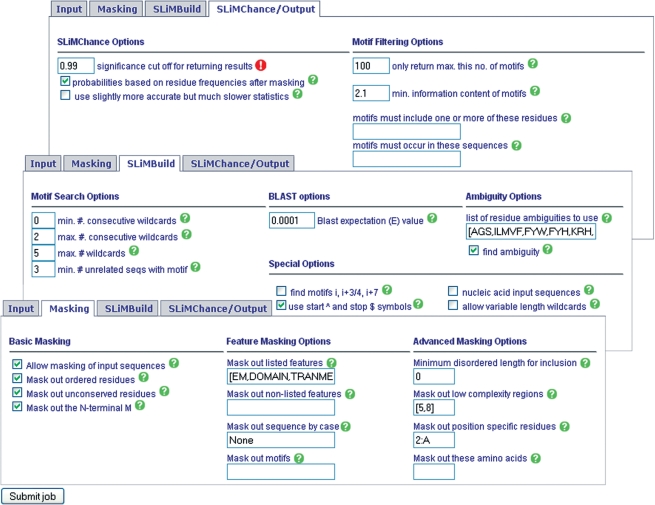
Following sequence input, the user can run SLiMFinder with default parameters, if desired. One of the strengths of SLiMFinder, however, is the ability to tailor the options to specific motif discovery needs. In particular, a large range of masking options are available to exclude (or concentrate on) specific features. Options are divided into Masking, SLiMBuild and SLiMChance/Output filtering (Figure 1). All options are explained in both the online help pages and the SLiMFinder manual; options are named for consistency and ease of transition between both web server and commandline implementations of SLiMFinder.
Figure 1.
Input options. Options are separated into sequence masking, SLiMBuild motif construction and SLiMChance/Output filtering. For clarity, all options correspond to commandline parameters of downloadable SLiMFinder program; short descriptions and commandline parameter names are given if the mouse hovers over the help buttons. All options are described in the help pages. Once options have been set/reviewed, ‘Submit job’ will move the job into the run queue.
Note that, by default, the server will return up to 100 motifs at P ≤ 0.99. This is because the default SLiMChance statistics are slightly conservative (18) and returning more motifs gives the user more control to determine what they think is interesting; for many applications a high false-positive rate might be tolerated. We urge extreme caution when interpreting motifs with Sig > 0.5 as they are most probably over-represented by chance and a much stricter cut-off (e.g. 0.05) should be used when stringency and lack of false positives is important.
Submitting jobs
Once options have been reviewed, clicking ‘Submit job’ will enter the run queue. Run times will vary according to input data size/complexity, Masking/SLiMBuild options and the current load of the server. Users can either wait for their jobs to run, or bookmark the page and return to it later.
Output
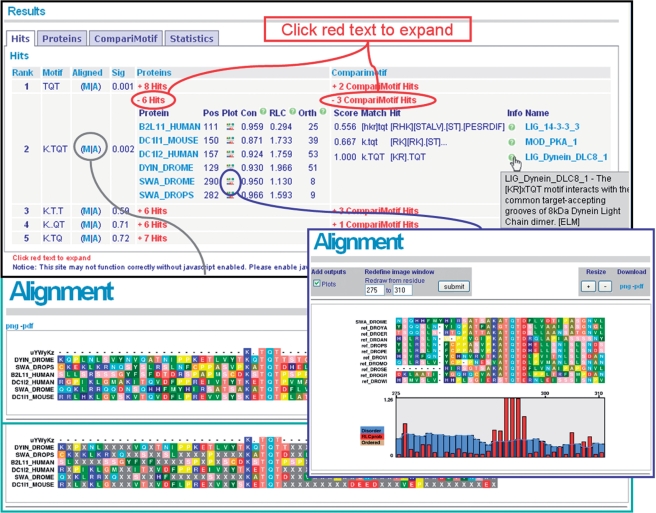
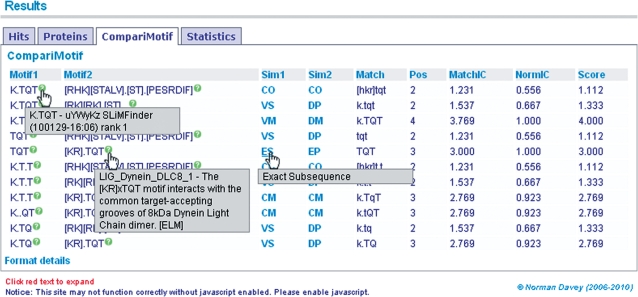
The initial results page shows summary results for returned motifs (Figure 2). The ‘Sig’ indicates the estimated significance level of each motif. Clicking the red links expands details for the proteins and CompariMotif hits, while the ‘M’ and ‘A’ alignment links will visualize the motif in the input proteins, masked and unmasked, respectively (Figure 2). Alignments for each protein and its GOPHER (9) orthologues around the motif of interest can be accessed by clicking on the ‘Plot’ link for each protein/motif pair (Figure 2). Protein disorder and RLC scores are also visualized in these alignments and the region can be altered to zoom in or out as desired. All alignments can be saved as PNG or high-quality PDF files. Returned motifs are also compared with known literature motifs using CompariMotif (23) (Figure 3). Full-length orthologue alignments for each protein and motif maps, as introduced by the SLiMDisc web server (9) are also available. In addition to the visualizations, all results files normally generated by the commandline implementation of SLiMFinder can be downloaded as plain text for further analysis and manipulations.
Figure 2.
Main results page. Summarized results for each motif are initially displayed. These can be expanded to reveal individual occurrences in each protein for each motif. Alignments can be generated to explore the unmasked and masked sequence context for each motif ‘(M|A)’ or to examine the region around a specific motif occurrence in a single protein ‘(Plot)’. All visualizations can be exported as PNGs or high-quality PDFs.
Figure 3.
CompariMotif results. All motifs returned by SLiMFinder are cross-referenced against known motifs using CompariMotif, enabling easy identification of re-discovered known motifs. All columns are sortable by clicking on their respective headings and more information can be revealed about motifs and their CompariMotif hits by mousing over the appropriate data.
Example analysis
The web server incorporates a full example for a data set of seven proteins containing manually curated, experimentally validated, Dynein light chain binding motifs ([^ P].[KR].TQT) taken from ELM entry LIG_Dynein_DLC8_1 (2) in Uniprot format. A full walkthrough for this data set is provided in the help pages and fully interactive example output is also provided. As previously reported for SLiMFinder (11,17), the SLiMFinder web server returns TQT and K.TQT as significant motifs (P < 0.01).
Getting help
The SLiMFinder web server is supported by an extensive help section, including a quickstart guide and walkthrough with screenshots. Example input files are provided and example input data can be loaded into the input forms. Fully interactive example output (corresponding to running the example input with default parameters) is clearly linked from the help pages (see example analysis). Additional details of the algorithms and options can be found in the SLiMFinder manual, which is also clearly linked from the help pages.
FUTURE WORK
The SLiMFinder server is designed to be flexible and allow easy incorporation of future updates to the main SLiMFinder program (currently version 4.1). The version number is clearly shown on the front page of the web server and is stored in the log file of each run, be it commandline or server based.
Appropriate use of conservation can significantly increase the sensitivity of SLiMFinder (17). Currently, conservation masking is only well supported for metazoan organisms and humans in particular. With time, we hope to expand the range of taxonomic groups for which conservation-based masking is available. These will be added to the Masking options tab and appropriate help pages.
CONCLUSION
Technological advances have greatly increased the coverage of protein interaction networks but it is the ability to add details to these networks—such as predictions of interaction motifs—that will really enable them to drive biological discovery. The P-value returned by SLiMFinder is of fundamental importance to move SLiM discovery out of the domain of a few computational specialists and into the hands of experimental molecular biologists. To facilitate this transition, we have implemented SLiMFinder as an intuitive, interactive web server that provides numerous useful visualizations for data exploration without sacrificing any of the main options offered by the commandline implementation. The SLiMFinder server is available at: http://bioware.ucd.ie/slimfinder.html.
FUNDING
Science Foundation Ireland and the University of Southampton; European Molecular Biology Laboratory, EMBL Interdisciplinary Postdoc (EIPOD) fellowship (to N.E.D.). Funding for open access charge: Science Foundation Ireland (grant 08/IN.1/B1864).
Conflict of interest statement. None declared.
REFERENCES
- 1.Diella F., Haslam N., Chica C., Budd A., Michael S., Brown N.P., Trave G., Gibson T.J. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 2.Gould C.M., Diella F., Via A., Puntervoll P., Gemund C., Chabanis-Davidson S., Michael S., Sayadi A., Bryne J.C., Chica C., et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadaveru K., Vyas J., Schiller M.R. Viral infection and human disease–insights from minimotifs. Front. Biosci. 2008;13:6455–6471. doi: 10.2741/3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neduva V., Russell R.B. Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 2006;17:465–471. doi: 10.1016/j.copbio.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Rajasekaran S., Balla S., Gradie P., Gryk M.R., Kadaveru K., Kundeti V., Maciejewski M.W., Mi T., Rubino N., Vyas J., et al. Minimotif miner 2nd release: a database and web system for motif search. Nucleic Acids Res. 2009;37:D185–D190. doi: 10.1093/nar/gkn865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutman R., Berezin C., Wollman R., Rosenberg Y., Ben-Tal N. QuasiMotiFinder: protein annotation by searching for evolutionarily conserved motif-like patterns. Nucleic Acids Res. 2005;33:W255–W261. doi: 10.1093/nar/gki496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chica C., Labarga A., Gould C.M., Lopez R., Gibson T.J. A tree-based conservation scoring method for short linear motifs in multiple alignments of protein sequences. BMC Bioinformatics. 2008;9:229. doi: 10.1186/1471-2105-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Via A., Gould C.M., Gemund C., Gibson T.J., Helmer-Citterich M. A structure filter for the Eukaryotic Linear Motif Resource. BMC Bioinformatics. 2009;10:351. doi: 10.1186/1471-2105-10-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey N.E., Edwards R.J., Shields D.C. The SLiMDisc server: short, linear motif discovery in proteins. Nucleic Acids Res. 2007;35:W455–W459. doi: 10.1093/nar/gkm400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey N.E., Shields D.C., Edwards R.J. SLiMDisc: short, linear motif discovery, correcting for common evolutionary descent. Nucleic Acids Res. 2006;34:3546–3554. doi: 10.1093/nar/gkl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards R.J., Davey N.E., Shields D.C. SLiMFinder: a probabilistic method for identifying over-represented, convergently evolved, short linear motifs in proteins. PLoS ONE. 2007;2:e967. doi: 10.1371/journal.pone.0000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neduva V., Linding R., Su-Angrand I., Stark A., Masi F.D., Gibson T.J., Lewis J., Serrano L., Russell R.B. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neduva V., Russell R.B. DILIMOT: discovery of linear motifs in proteins. Nucleic Acids Res. 2006;34:W350–W355. doi: 10.1093/nar/gkl159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonassen I., Collins J.F., Higgins D.G. Finding flexible patterns in unaligned protein sequences. Protein Sci. 1995;4:1587–1595. doi: 10.1002/pro.5560040817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frith M.C., Saunders N.F., Kobe B., Bailey T.L. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 2008;4:e1000071. doi: 10.1371/journal.pcbi.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davey N.E., Shields D.C., Edwards R.J. Masking residues using context-specific evolutionary conservation significantly improves short linear motif discovery. Bioinformatics. 2009;25:443–450. doi: 10.1093/bioinformatics/btn664. [DOI] [PubMed] [Google Scholar]
- 18.Davey N.E., Edwards R.J., Shields D.C. Estimation and efficient computation of the true probability of recurrence of short linear protein sequence motifs in unrelated proteins. BMC Bioinformatics. 2010;11:14. doi: 10.1186/1471-2105-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dosztanyi Z., Csizmok V., Tompa P., Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 20.Rigoutsos I., Floratos A. Combinatorial pattern discovery in biological sequences: the TEIRESIAS algorithm. Bioinformatics. 1998;14:55–67. doi: 10.1093/bioinformatics/14.1.55. [DOI] [PubMed] [Google Scholar]
- 21.Neuwald A.F., Green P. Detecting patterns in protein sequences. J. Mol. Biol. 1994;239:698–712. doi: 10.1006/jmbi.1994.1407. [DOI] [PubMed] [Google Scholar]
- 22.Bairoch A., Apweiler R., Wu C.H., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards R.J., Davey N.E., Shields D.C. CompariMotif: quick and easy comparisons of sequence motifs. Bioinformatics. 2008;24:1307–1309. doi: 10.1093/bioinformatics/btn105. [DOI] [PubMed] [Google Scholar]